

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.4 + phase: 1 /pseudo
(776 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
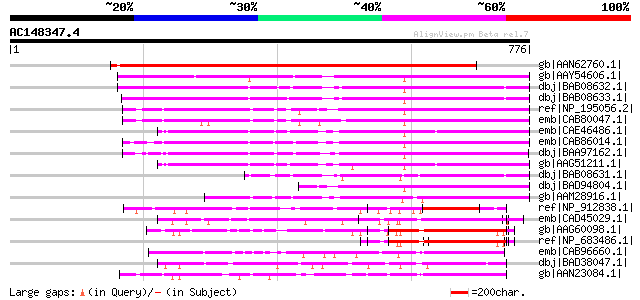
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN62760.1| disease resistance protein-like protein MsR1 [Med... 978 0.0
gb|AAY54606.1| NRG1 [Nicotiana benthamiana] 443 e-122
dbj|BAB08632.1| disease resistance protein-like [Arabidopsis tha... 409 e-112
dbj|BAB08633.1| disease resistance protein-like [Arabidopsis tha... 380 e-104
ref|NP_195056.2| disease resistance protein (CC-NBS-LRR class), ... 337 9e-91
emb|CAB80047.1| putative protein [Arabidopsis thaliana] gi|44902... 319 2e-85
emb|CAE46486.1| CC-NBS-LRR disease resistance protein [Arabidops... 318 6e-85
emb|CAB86014.1| disease resistance-like protein [Arabidopsis tha... 311 5e-83
dbj|BAA97162.1| disease resistance protein-like [Arabidopsis tha... 311 5e-83
gb|AAG51211.1| disease resistance protein, putative; 92850-95636... 311 5e-83
dbj|BAB08631.1| unnamed protein product [Arabidopsis thaliana] g... 259 2e-67
dbj|BAD94804.1| putative protein [Arabidopsis thaliana] 236 2e-60
gb|AAM28916.1| NBS/LRR [Pinus taeda] 229 3e-58
ref|NP_912838.1| unnamed protein product [Oryza sativa (japonica... 157 2e-36
emb|CAD45029.1| NBS-LRR disease resistance protein homologue [Ho... 144 1e-32
gb|AAG60098.1| disease resistance protein, putative [Arabidopsis... 132 3e-29
ref|NP_683486.1| leucine-rich repeat family protein [Arabidopsis... 132 3e-29
emb|CAB96660.1| RPP1 disease resistance protein-like [Arabidopsi... 130 1e-28
dbj|BAD38047.1| putative NBS-LRR resistance protein RGH2 [Oryza ... 128 8e-28
gb|AAN23084.1| putative rp3 protein [Zea mays] 115 4e-24
>gb|AAN62760.1| disease resistance protein-like protein MsR1 [Medicago sativa]
Length = 704
Score = 978 bits (2529), Expect = 0.0
Identities = 495/547 (90%), Positives = 516/547 (93%), Gaps = 2/547 (0%)
Query: 152 KPLWLQSFVGMKKLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLL 211
K W Q G K MENIIFVTFSKTPMLKTIVERIH+HCGY VPEFQ+DEDAVN+LGLL
Sbjct: 158 KLCWDQEVNG--KFMENIIFVTFSKTPMLKTIVERIHEHCGYPVPEFQNDEDAVNRLGLL 215
Query: 212 LKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHED 271
LKKVEGSPLLLVLDDVWPSSESLVEKLQFQ++ FKILVTSRVAFPRFSTTCILKPLAHED
Sbjct: 216 LKKVEGSPLLLVLDDVWPSSESLVEKLQFQISDFKILVTSRVAFPRFSTTCILKPLAHED 275
Query: 272 AVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKEL 331
AVTLFHHYA MEKNSSDIIDKNLVEKVVRSC GLPLTIKVIATSLKNRPHDLW KIVKEL
Sbjct: 276 AVTLFHHYALMEKNSSDIIDKNLVEKVVRSCQGLPLTIKVIATSLKNRPHDLWRKIVKEL 335
Query: 332 SQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYK 391
SQGHSILDSNTELLTRLQKIFDVLEDNP I+ECFMDLALFPEDHRIPVAALVDMWAELY+
Sbjct: 336 SQGHSILDSNTELLTRLQKIFDVLEDNPTIIECFMDLALFPEDHRIPVAALVDMWAELYR 395
Query: 392 LDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKE 451
LDD GIQAMEIINKLGIMNLANVIIPRK+ASDTDNNNYNNHFI+LHDILRELGIY+STKE
Sbjct: 396 LDDTGIQAMEIINKLGIMNLANVIIPRKDASDTDNNNYNNHFIMLHDILRELGIYRSTKE 455
Query: 452 PFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCA 511
PFEQRKRLIIDM+ NKSGL EKQQGLM RI SKFMRLCVKQNPQQLAARILSVSTDETCA
Sbjct: 456 PFEQRKRLIIDMHKNKSGLTEKQQGLMIRILSKFMRLCVKQNPQQLAARILSVSTDETCA 515
Query: 512 LDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQ 571
LDWS M+PAQVEVLILN+HTKQYSLPEWI KMSKL+VLIITNY HPS+L N ELL SLQ
Sbjct: 516 LDWSQMEPAQVEVLILNLHTKQYSLPEWIGKMSKLKVLIITNYTVHPSELTNFELLSSLQ 575
Query: 572 NLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL 631
NLE+IRLERISVPSF T+KNLKKLSLYMCNT LAFEKGSILISDAFPNLEELNIDYCKDL
Sbjct: 576 NLEKIRLERISVPSFATVKNLKKLSLYMCNTRLAFEKGSILISDAFPNLEELNIDYCKDL 635
Query: 632 VVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN 691
+VL TGICDIISLKKL+VTNCHK+SSLP+DIGKLENLELLSLSSCTDLEAIPTSI KLLN
Sbjct: 636 LVLPTGICDIISLKKLSVTNCHKISSLPEDIGKLENLELLSLSSCTDLEAIPTSIEKLLN 695
Query: 692 LKHLDIS 698
LKHLDIS
Sbjct: 696 LKHLDIS 702
>gb|AAY54606.1| NRG1 [Nicotiana benthamiana]
Length = 850
Score = 443 bits (1139), Expect = e-122
Identities = 258/627 (41%), Positives = 370/627 (58%), Gaps = 34/627 (5%)
Query: 162 MKKLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLL 221
+K +I FVT SK +K IV I + GY P+F S+ AV +L LL++ P+L
Sbjct: 240 IKDKYRDIFFVTVSKKANIKRIVGEIFEMKGYKGPDFASEHAAVCQLNNLLRRSTSQPVL 299
Query: 222 LVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQ 281
LVLDDVW S+ ++E FQ+ GFKILVTSR FP+F T L L+ +DA LF Y+
Sbjct: 300 LVLDDVWSESDFVIESFIFQIPGFKILVTSRSVFPKFDTYK-LNLLSEKDAKALF--YSS 356
Query: 282 MEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSN 341
K+S + +LV K VRSC G PL +KV+ SL +P +W V S+ + +
Sbjct: 357 AFKDSIPYVQLDLVHKAVRSCCGFPLALKVVGRSLCGQPELIWFNRVMLQSKRQILFPTE 416
Query: 342 TELLTRLQKIFDVLED-------NPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDD 394
+LL L+ D L++ + +C++DL FPEDHRI AA++DMW E Y LD+
Sbjct: 417 NDLLRTLRASIDALDEIDLYSSEATTLRDCYLDLGSFPEDHRIHAAAILDMWVERYNLDE 476
Query: 395 NGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFE 454
+G++AM I +L NL N+ + RK+A +N H+I HD+LREL I+Q ++ E
Sbjct: 477 DGMKAMAIFFQLSSQNLVNLALARKDAPAV-LGLHNLHYIQQHDLLRELVIHQCDEKTVE 535
Query: 455 QRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDW 514
+RKRL I++ N F + +Q Q L A +LS+ TDE W
Sbjct: 536 ERKRLYINIKGND-----------------FPKWWSQQRLQPLQAEVLSIFTDEHFESVW 578
Query: 515 SHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLE 574
++ +VEVL+LN TK Y+ P ++ +MS+L+ LI+ N F P+KLNN +L SL NL+
Sbjct: 579 YDVRFPKVEVLVLNFETKTYNFPPFVEQMSQLKTLIVANNYFFPTKLNNFQLC-SLLNLK 637
Query: 575 RIRLERISVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKD 630
RI LERISV S T L NL+K+S MC AFE + +S +P L E+NI+YC D
Sbjct: 638 RISLERISVTSIFTANLQLPNLRKISFIMCEIGEAFENYAANMSYMWPKLVEMNIEYCSD 697
Query: 631 LVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLL 690
LV + CD++ LKKL++ CH+L +LP+++GKL NLE+L L SCT++ +P S+ KL
Sbjct: 698 LVEVPAETCDLVGLKKLSICYCHELVALPEELGKLSNLEVLRLHSCTNVSKLPESVVKLN 757
Query: 691 NLKHLDISNCISLSSLPEEFGNLCNLKNLDMAT-CASIELPFSVVNLQNLKTITCDEETA 749
L LD+ +C+ L LP E LC+L+ + M + ELP SV+ L L+ + CDEETA
Sbjct: 758 RLGFLDVYDCVELDFLPREMDQLCSLRTICMGSRLGFTELPDSVLRLVKLEDVVCDEETA 817
Query: 750 ATWEDFQHMLHNMKIEVLHVDVNLNWL 776
+ WE ++ L N++I V+ D+NLN L
Sbjct: 818 SLWEYYKEHLRNLRITVIKEDINLNLL 844
>dbj|BAB08632.1| disease resistance protein-like [Arabidopsis thaliana]
gi|15240126|ref|NP_201491.1| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395984|sp|Q9FKZ1|DRL42_ARATH Probable disease
resistance protein At5g66900
Length = 809
Score = 409 bits (1051), Expect = e-112
Identities = 241/622 (38%), Positives = 360/622 (57%), Gaps = 35/622 (5%)
Query: 162 MKKLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKV-EGSPL 220
+K ++I F S TP + IV+ + QH GY+ F++D A L LL+++ E P+
Sbjct: 213 IKGKFKHIFFNVVSNTPNFRVIVQNLLQHNGYNALTFENDSQAEVGLRKLLEELKENGPI 272
Query: 221 LLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYA 280
LLVLDDVW ++S ++K Q ++ +KILVTSR FP F + LKPL +DA L H+A
Sbjct: 273 LLVLDDVWRGADSFLQKFQIKLPNYKILVTSRFDFPSFDSNYRLKPLEDDDARALLIHWA 332
Query: 281 QMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDS 340
N+S ++L++K+++ CNG P+ I+V+ SLK R + W V+ S+G IL
Sbjct: 333 SRPCNTSPDEYEDLLQKILKRCNGFPIVIEVVGVSLKGRSLNTWKGQVESWSEGEKILGK 392
Query: 341 N-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQA 399
+L LQ FD L+ P + ECF+D+ F ED +I + ++DMW ELY + +
Sbjct: 393 PYPTVLECLQPSFDALD--PNLKECFLDMGSFLEDQKIRASVIIDMWVELYGKGSSILYM 450
Query: 400 MEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRL 459
+ L NL ++ N + ++ YN+ + HDILREL I QS + +RKRL
Sbjct: 451 Y--LEDLASQNLLKLVPLGTN--EHEDGFYNDFLVTQHDILRELAICQSEFKENLERKRL 506
Query: 460 IIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQP 519
+++ N F C+ + A +LS+STD+ + W M
Sbjct: 507 NLEILENT-----------------FPDWCLNT----INASLLSISTDDLFSSKWLEMDC 545
Query: 520 AQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLE 579
VE L+LN+ + Y+LP +I+ M KL+VL ITN+GF+P++L+N L SL NL+RIRLE
Sbjct: 546 PNVEALVLNLSSSDYALPSFISGMKKLKVLTITNHGFYPARLSNFSCLSSLPNLKRIRLE 605
Query: 580 RISVPSFGT----LKNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVVL 634
++S+ L +LKKLSL MC+ F + I++S+A L+E++IDYC DL L
Sbjct: 606 KVSITLLDIPQLQLSSLKKLSLVMCSFGEVFYDTEDIVVSNALSKLQEIDIDYCYDLDEL 665
Query: 635 QTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKH 694
I +I+SLK L++TNC+KLS LP+ IG L LE+L L S +L +P + L NL+
Sbjct: 666 PYWISEIVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRLCSSMNLSELPEATEGLSNLRF 725
Query: 695 LDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWED 754
LDIS+C+ L LP+E G L NLK + M C+ ELP SV NL+NL+ + CDEET WE
Sbjct: 726 LDISHCLGLRKLPQEIGKLQNLKKISMRKCSGCELPESVTNLENLE-VKCDEETGLLWER 784
Query: 755 FQHMLHNMKIEVLHVDVNLNWL 776
+ + N++++ ++ NLN L
Sbjct: 785 LKPKMRNLRVQEEEIEHNLNLL 806
>dbj|BAB08633.1| disease resistance protein-like [Arabidopsis thaliana]
gi|18087526|gb|AAL58897.1| AT5g66910/MUD21_17
[Arabidopsis thaliana] gi|15240127|ref|NP_201492.1|
disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana] gi|46395983|sp|Q9FKZ0|DRL43_ARATH
Probable disease resistance protein At5g66910
Length = 815
Score = 380 bits (977), Expect = e-104
Identities = 231/618 (37%), Positives = 348/618 (55%), Gaps = 33/618 (5%)
Query: 167 ENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKV-EGSPLLLVLD 225
+ I + S TP + IV+ + Q G F D A L LL+++ + +LLVLD
Sbjct: 220 KKIFYSVVSNTPNFRAIVQNLLQDNGCGAITFDDDSQAETGLRDLLEELTKDGRILLVLD 279
Query: 226 DVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKN 285
DVW SE L+ K Q + +KILVTS+ F T L PL +E A +L +A +
Sbjct: 280 DVWQGSEFLLRKFQIDLPDYKILVTSQFDFTSLWPTYHLVPLKYEYARSLLIQWASPPLH 339
Query: 286 SSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSIL-DSNTEL 344
+S ++L++K+++ CNG PL I+V+ SLK + LW V+ S+G +IL ++N +
Sbjct: 340 TSPDEYEDLLQKILKRCNGFPLVIEVVGISLKGQALYLWKGQVESWSEGETILGNANPTV 399
Query: 345 LTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQA-MEII 403
RLQ F+VL+ P + ECFMD+ F +D +I + ++D+W ELY + M +
Sbjct: 400 RQRLQPSFNVLK--PHLKECFMDMGSFLQDQKIRASLIIDIWMELYGRGSSSTNKFMLYL 457
Query: 404 NKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDM 463
N+L NL ++ + ++ YN + H+ILREL I+QS EP QRK+L +++
Sbjct: 458 NELASQNLLKLV--HLGTNKREDGFYNELLVTQHNILRELAIFQSELEPIMQRKKLNLEI 515
Query: 464 NNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVE 523
+ F C+ Q + AR+LS+ TD+ + W M VE
Sbjct: 516 REDN-----------------FPDECLNQ---PINARLLSIYTDDLFSSKWLEMDCPNVE 555
Query: 524 VLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISV 583
L+LNI + Y+LP +IA+M KL+VL I N+GF+P++L+N L SL NL+RIR E++SV
Sbjct: 556 ALVLNISSLDYALPSFIAEMKKLKVLTIANHGFYPARLSNFSCLSSLPNLKRIRFEKVSV 615
Query: 584 PSFGT----LKNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVVLQTGI 638
L +LKKLS +MC+ F + I +S A NL+E++IDYC DL L I
Sbjct: 616 TLLDIPQLQLGSLKKLSFFMCSFGEVFYDTEDIDVSKALSNLQEIDIDYCYDLDELPYWI 675
Query: 639 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 698
+++SLK L++TNC+KLS LP+ IG L LE+L + SC +L +P + +L NL+ LDIS
Sbjct: 676 PEVVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRMCSCMNLSELPEATERLSNLRSLDIS 735
Query: 699 NCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHM 758
+C+ L LP+E G L L+N+ M C+ ELP SV L+NL+ + CDE T WE
Sbjct: 736 HCLGLRKLPQEIGKLQKLENISMRKCSGCELPDSVRYLENLE-VKCDEVTGLLWERLMPE 794
Query: 759 LHNMKIEVLHVDVNLNWL 776
+ N+++ + NL L
Sbjct: 795 MRNLRVHTEETEHNLKLL 812
>ref|NP_195056.2| disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana]
Length = 816
Score = 337 bits (864), Expect = 9e-91
Identities = 207/616 (33%), Positives = 336/616 (53%), Gaps = 43/616 (6%)
Query: 169 IIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVW 228
I+F+T S++P+L+ + E I F S +A N + +G+ L++LDDVW
Sbjct: 234 ILFLTVSQSPLLEELRELIWG--------FLSGCEAGNPVPDCNFPFDGARKLVILDDVW 285
Query: 229 PSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSD 288
++++L F+ G LV SR T ++ L+ ++A++LF A +K+
Sbjct: 286 -TTQALDRLTSFKFPGCTTLVVSRSKLTEPKFTYDVEVLSEDEAISLFCLCAFGQKSIPL 344
Query: 289 IIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSN-TELLTR 347
K+LV++V C GLPL +KV SL +P W +++ LS+G DS+ + LL +
Sbjct: 345 GFCKDLVKQVANECKGLPLALKVTGASLNGKPEMYWKGVLQRLSKGEPADDSHESRLLRQ 404
Query: 348 LQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLG 407
++ D L+ K +CF+DL FPED +IP+ L+++W EL+ +D+ A I+ L
Sbjct: 405 MEASLDNLDQTTK--DCFLDLGAFPEDRKIPLDVLINIWIELHDIDEGN--AFAILVDLS 460
Query: 408 IMNLANVIIPRKNASDTDNNNYNNH---FIILHDILRELGIYQSTKEPFEQRKRLIIDMN 464
NL + + Y +H F+ HD+LR+L ++ S +RKRL+ M
Sbjct: 461 HKNLLTL-----GKDPRLGSLYASHYDIFVTQHDVLRDLALHLSNAGKVNRRKRLL--MP 513
Query: 465 NNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEV 524
+ L + + N + A+I+S+ T E + W M+ + E+
Sbjct: 514 KRELDLPGDWE---------------RNNDEHYIAQIVSIHTGEMNEMQWFDMEFPKAEI 558
Query: 525 LILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVP 584
LILN + +Y LP +I+KMS+L+VL+I N G P+ L++ + L L + LER+ VP
Sbjct: 559 LILNFSSDKYVLPPFISKMSRLKVLVIINNGMSPAVLHDFSIFAHLSKLRSLWLERVHVP 618
Query: 585 SFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICD 640
LKNL K+SL +C +F++ + ++D FP L +L ID+C DLV L + IC
Sbjct: 619 QLSNSTTPLKNLHKMSLILCKINKSFDQTGLDVADIFPKLGDLTIDHCDDLVALPSSICG 678
Query: 641 IISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNC 700
+ SL L++TNC +L LP+++ KL+ LE+L L +C +L+ +P I +L LK+LDIS C
Sbjct: 679 LTSLSCLSITNCPRLGELPKNLSKLQALEILRLYACPELKTLPGEICELPGLKYLDISQC 738
Query: 701 ISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLH 760
+SLS LPEE G L L+ +DM C + P S V+L++L+ + CD + A WE+ + +
Sbjct: 739 VSLSCLPEEIGKLKKLEKIDMRECCFSDRPSSAVSLKSLRHVICDTDVAFMWEEVEKAVP 798
Query: 761 NMKIEVLHVDVNLNWL 776
+KIE +L+WL
Sbjct: 799 GLKIEAAEKCFSLDWL 814
>emb|CAB80047.1| putative protein [Arabidopsis thaliana] gi|4490297|emb|CAB38788.1|
putative protein [Arabidopsis thaliana]
gi|46396028|sp|Q9SZA7|DRL29_ARATH Probable disease
resistance protein At4g33300
Length = 855
Score = 319 bits (818), Expect = 2e-85
Identities = 206/644 (31%), Positives = 342/644 (52%), Gaps = 60/644 (9%)
Query: 169 IIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVW 228
I+F+T S++P+L+ + E I F S +A N + +G+ L++LDDVW
Sbjct: 234 ILFLTVSQSPLLEELRELIWG--------FLSGCEAGNPVPDCNFPFDGARKLVILDDVW 285
Query: 229 PSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKN--- 285
++++L F+ G LV SR T ++ L+ ++A++LF A +K+
Sbjct: 286 -TTQALDRLTSFKFPGCTTLVVSRSKLTEPKFTYDVEVLSEDEAISLFCLCAFGQKSIPL 344
Query: 286 --SSDIIDKNLVE-----------KVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELS 332
D++ ++ ++ +V C GLPL +KV SL +P W +++ LS
Sbjct: 345 GFCKDLVKQHNIQSFSILRVLCLAQVANECKGLPLALKVTGASLNGKPEMYWKGVLQRLS 404
Query: 333 QGHSILDSN-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYK 391
+G DS+ + LL +++ D L+ K +CF+DL FPED +IP+ L+++W EL+
Sbjct: 405 KGEPADDSHESRLLRQMEASLDNLDQTTK--DCFLDLGAFPEDRKIPLDVLINIWIELHD 462
Query: 392 LDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNH---FIILHDILRELGIYQS 448
+D+ A I+ L NL + + Y +H F+ HD+LR+L ++ S
Sbjct: 463 IDEGN--AFAILVDLSHKNLLTL-----GKDPRLGSLYASHYDIFVTQHDVLRDLALHLS 515
Query: 449 TKEPFEQRKRLII-----------DMNNNKSGLAEKQQGLMTRIFSKF-MRLCVKQNPQQ 496
+RKRL++ + NN++ +A+ + FS + L + N Q
Sbjct: 516 NAGKVNRRKRLLMPKRELDLPGDWERNNDEHYIAQIVSIHTGKCFSTLQIFLVLSCNNHQ 575
Query: 497 LAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGF 556
+ E + W M+ + E+LILN + +Y LP +I+KMS+L+VL+I N G
Sbjct: 576 ------DHNVGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLVIINNGM 629
Query: 557 HPSKLNNIELLGSLQNLERIRLERISVPSFGT----LKNLKKLSLYMCNTILAFEKGSIL 612
P+ L++ + L L + LER+ VP LKNL K+SL +C +F++ +
Sbjct: 630 SPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQTGLD 689
Query: 613 ISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLS 672
++D FP L +L ID+C DLV L + IC + SL L++TNC +L LP+++ KL+ LE+L
Sbjct: 690 VADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQALEILR 749
Query: 673 LSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFS 732
L +C +L+ +P I +L LK+LDIS C+SLS LPEE G L L+ +DM C + P S
Sbjct: 750 LYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCFSDRPSS 809
Query: 733 VVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
V+L++L+ + CD + A WE+ + + +KIE +L+WL
Sbjct: 810 AVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 853
>emb|CAE46486.1| CC-NBS-LRR disease resistance protein [Arabidopsis thaliana]
gi|30692890|ref|NP_174620.2| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395988|sp|Q9FW44|ADR1_ARATH Disease resistance
protein ADR1 (Activated disease resistance protein 1)
Length = 787
Score = 318 bits (814), Expect = 6e-85
Identities = 192/562 (34%), Positives = 321/562 (56%), Gaps = 33/562 (5%)
Query: 221 LLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYA 280
L++LDDVW + ESL ++L ++ G LV SR TT ++ L ++A++L A
Sbjct: 251 LVILDDVW-TRESL-DRLMSKIRGSTTLVVSRSKLADPRTTYNVELLKKDEAMSLLCLCA 308
Query: 281 QMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDS 340
+K+ +K LV++VV C GLPL++KV+ SLKN+P W +VK L +G + ++
Sbjct: 309 FEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPERYWEGVVKRLLRGEAADET 368
Query: 341 N-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQA 399
+ + + +++ + L+ PKI +CF+D+ FPED +IP+ L +W E + +D+ A
Sbjct: 369 HESRVFAHMEESLENLD--PKIRDCFLDMGAFPEDKKIPLDLLTSVWVERHDIDEE--TA 424
Query: 400 MEIINKLGIMNLANVIIPRKNASDTDNN-NYNNHFIILHDILRELGIYQSTKEPFEQRKR 458
+ +L NL ++ N D + Y + F+ HD+LR+L ++ S + +R+R
Sbjct: 425 FSFVLRLADKNLLTIV---NNPRFGDVHIGYYDVFVTQHDVLRDLALHMSNRVDVNRRER 481
Query: 459 LIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQ 518
L++ K + ++ R + K + A+I+S+ T E ++W M
Sbjct: 482 LLMP----------KTEPVLPREWEK-------NKDEPFDAKIVSLHTGEMDEMNWFDMD 524
Query: 519 PAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRL 578
+ EVLILN + Y LP +I KMS+LRVL+I N G P++L+ + +L L + L
Sbjct: 525 LPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINNGMSPARLHGFSIFANLAKLRSLWL 584
Query: 579 ERISVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVL 634
+R+ VP + LKNL K+ L C +F + S IS FP+L +L ID+C DL+ L
Sbjct: 585 KRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDISKIFPSLSDLTIDHCDDLLEL 644
Query: 635 QTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKH 694
++ I I SL L++TNC ++ LP+++ +++LE L L +C +L ++P + +L LK+
Sbjct: 645 KS-IFGITSLNSLSITNCPRILELPKNLSNVQSLERLRLYACPELISLPVEVCELPCLKY 703
Query: 695 LDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWED 754
+DIS C+SL SLPE+FG L +L+ +DM C+ + LP SV L +L+ + CDEET++ WE
Sbjct: 704 VDISQCVSLVSLPEKFGKLGSLEKIDMRECSLLGLPSSVAALVSLRHVICDEETSSMWEM 763
Query: 755 FQHMLHNMKIEVLHVDVNLNWL 776
+ ++ + IEV ++WL
Sbjct: 764 VKKVVPELCIEVAKKCFTVDWL 785
>emb|CAB86014.1| disease resistance-like protein [Arabidopsis thaliana]
gi|21281177|gb|AAM45000.1| putative disease resistance
[Arabidopsis thaliana] gi|15292721|gb|AAK92729.1|
putative disease resistance [Arabidopsis thaliana]
gi|9758447|dbj|BAB08976.1| unnamed protein product
[Arabidopsis thaliana] gi|15238281|ref|NP_196092.1|
disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana] gi|46396009|sp|Q9LZ25|DRL30_ARATH
Probable disease resistance protein At5g04720
Length = 811
Score = 311 bits (797), Expect = 5e-83
Identities = 195/614 (31%), Positives = 328/614 (52%), Gaps = 44/614 (7%)
Query: 169 IIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVW 228
++F+T S++P L E + H + +++ A + S L++LDDVW
Sbjct: 234 VLFLTVSQSPNL----EELRAHIWGFLTSYEAGVGAT---------LPESRKLVILDDVW 280
Query: 229 PSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSD 288
+ ESL + + + G LV SR T ++ L +A LF +K
Sbjct: 281 -TRESLDQLMFENIPGTTTLVVSRSKLADSRVTYDVELLNEHEATALFCLSVFNQKLVPS 339
Query: 289 IIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSN-TELLTR 347
++LV++VV C GLPL++KVI SLK RP W V+ LS+G +++ + + +
Sbjct: 340 GFSQSLVKQVVGECKGLPLSLKVIGASLKERPEKYWEGAVERLSRGEPADETHESRVFAQ 399
Query: 348 LQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLG 407
++ + L+ PK +CF+ L FPED +IP+ L+++ EL+ L+D A +I L
Sbjct: 400 IEATLENLD--PKTRDCFLVLGAFPEDKKIPLDVLINVLVELHDLED--ATAFAVIVDLA 455
Query: 408 IMNLANVII-PRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNN 466
NL ++ PR T +Y + F+ HD+LR++ + S R+RL++
Sbjct: 456 NRNLLTLVKDPRFGHMYT---SYYDIFVTQHDVLRDVALRLSNHGKVNNRERLLMP---- 508
Query: 467 KSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLI 526
K++ ++ R + + N + AR++S+ T E +DW M+ + EVLI
Sbjct: 509 ------KRESMLPREWER-------NNDEPYKARVVSIHTGEMTQMDWFDMELPKAEVLI 555
Query: 527 LNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSF 586
L+ + +Y LP +IAKM KL L+I N G P++L++ + +L L+ + L+R+ VP
Sbjct: 556 LHFSSDKYVLPPFIAKMGKLTALVIINNGMSPARLHDFSIFTNLAKLKSLWLQRVHVPEL 615
Query: 587 GT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDII 642
+ L+NL KLSL C + ++ + I+ FP L +L ID+C DL+ L + IC I
Sbjct: 616 SSSTVPLQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTIDHCDDLLELPSTICGIT 675
Query: 643 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 702
SL +++TNC ++ LP+++ KL+ L+LL L +C +L ++P I +L LK++DIS C+S
Sbjct: 676 SLNSISITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVEICELPRLKYVDISQCVS 735
Query: 703 LSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNM 762
LSSLPE+ G + L+ +D C+ +P SVV L +L+ + CD E WE Q + +
Sbjct: 736 LSSLPEKIGKVKTLEKIDTRECSLSSIPNSVVLLTSLRHVICDREALWMWEKVQKAVAGL 795
Query: 763 KIEVLHVDVNLNWL 776
++E + +WL
Sbjct: 796 RVEAAEKSFSRDWL 809
>dbj|BAA97162.1| disease resistance protein-like [Arabidopsis thaliana]
gi|15238054|ref|NP_199539.1| disease resistance protein
(NBS-LRR class), putative [Arabidopsis thaliana]
gi|46396005|sp|Q9LVT1|DRL39_ARATH Putative disease
resistance protein At5g47280
Length = 623
Score = 311 bits (797), Expect = 5e-83
Identities = 202/614 (32%), Positives = 337/614 (53%), Gaps = 43/614 (7%)
Query: 169 IIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEG-SPLLLVLDDV 227
++F+T S++P L+ + I +F + +A G L + G + L++LDDV
Sbjct: 43 VLFLTVSQSPNLEELRSLIR--------DFLTGHEA--GFGTALPESVGHTRKLVILDDV 92
Query: 228 WPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSS 287
+ ESL ++L F + G LV S+ TT ++ L DA +LF A +K+
Sbjct: 93 -RTRESL-DQLMFNIPGTTTLVVSQSKLVDPRTTYDVELLNEHDATSLFCLSAFNQKSVP 150
Query: 288 DIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSN-TELLT 346
K+LV++VV GLPL++KV+ SL +RP W V+ LS+G + +++ +++
Sbjct: 151 SGFSKSLVKQVVGESKGLPLSLKVLGASLNDRPETYWAIAVERLSRGEPVDETHESKVFA 210
Query: 347 RLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKL 406
+++ + L+ PK ECF+D+ FPE +IPV L++M +++ L+D A +++ L
Sbjct: 211 QIEATLENLD--PKTKECFLDMGAFPEGKKIPVDVLINMLVKIHDLEDAA--AFDVLVDL 266
Query: 407 GIMNLANVII-PRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNN 465
NL ++ P A T +Y + F+ HD+LR++ ++ + + +R RL++
Sbjct: 267 ANRNLLTLVKDPTFVAMGT---SYYDIFVTQHDVLRDVALHLTNRGKVSRRDRLLMPKRE 323
Query: 466 NKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVL 525
T + S++ R N + AR++S+ T E +DW M + EVL
Sbjct: 324 -------------TMLPSEWER----SNDEPYNARVVSIHTGEMTEMDWFDMDFPKAEVL 366
Query: 526 ILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPS 585
I+N + Y LP +IAKM LRV +I N G P+ L++ + SL NL + LER+ VP
Sbjct: 367 IVNFSSDNYVLPPFIAKMGMLRVFVIINNGTSPAHLHDFPIPTSLTNLRSLWLERVHVPE 426
Query: 586 FGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDI 641
+ LKNL KL L +C +F++ +I I+ FP L ++ IDYC DL L + IC I
Sbjct: 427 LSSSMIPLKNLHKLYLIICKINNSFDQTAIDIAQIFPKLTDITIDYCDDLAELPSTICGI 486
Query: 642 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCI 701
SL +++TNC + LP++I KL+ L+LL L +C +L+++P I +L L ++DIS+C+
Sbjct: 487 TSLNSISITNCPNIKELPKNISKLQALQLLRLYACPELKSLPVEICELPRLVYVDISHCL 546
Query: 702 SLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHN 761
SLSSLPE+ GN+ L+ +DM C+ +P S V+L +L +TC E W++ + +
Sbjct: 547 SLSSLPEKIGNVRTLEKIDMRECSLSSIPSSAVSLTSLCYVTCYREALWMWKEVEKAVPG 606
Query: 762 MKIEVLHVDVNLNW 775
++IE N+ W
Sbjct: 607 LRIEATEKWFNMTW 620
>gb|AAG51211.1| disease resistance protein, putative; 92850-95636 [Arabidopsis
thaliana] gi|10998939|gb|AAG26078.1| disease resistance
protein, putative [Arabidopsis thaliana]
Length = 797
Score = 311 bits (797), Expect = 5e-83
Identities = 192/572 (33%), Positives = 322/572 (55%), Gaps = 43/572 (7%)
Query: 221 LLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYA 280
L++LDDVW + ESL ++L ++ G LV SR TT ++ L ++A++L A
Sbjct: 251 LVILDDVW-TRESL-DRLMSKIRGSTTLVVSRSKLADPRTTYNVELLKKDEAMSLLCLCA 308
Query: 281 QMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDS 340
+K+ +K LV++VV C GLPL++KV+ SLKN+P W +VK L +G + ++
Sbjct: 309 FEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPERYWEGVVKRLLRGEAADET 368
Query: 341 N-TELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQA 399
+ + + +++ + L+ PKI +CF+D+ FPED +IP+ L +W E + +D+ A
Sbjct: 369 HESRVFAHMEESLENLD--PKIRDCFLDMGAFPEDKKIPLDLLTSVWVERHDIDEE--TA 424
Query: 400 MEIINKLGIMNLANVIIPRKNASDTDNN-NYNNHFIILHDILRELGIYQSTKEPFEQRKR 458
+ +L NL ++ N D + Y + F+ HD+LR+L ++ S + +R+R
Sbjct: 425 FSFVLRLADKNLLTIV---NNPRFGDVHIGYYDVFVTQHDVLRDLALHMSNRVDVNRRER 481
Query: 459 LIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCA------- 511
L++ K + ++ R + K + A+I+S+ T +T
Sbjct: 482 LLMP----------KTEPVLPREWEK-------NKDEPFDAKIVSLHTGKTSLTLNEFGE 524
Query: 512 ---LDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLG 568
++W M + EVLILN + Y LP +I KMS+LRVL+I N G P++L+ +
Sbjct: 525 MDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINNGMSPARLHGFSIFA 584
Query: 569 SLQNLERIRLERISVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELN 624
+L L + L+R+ VP + LKNL K+ L C +F + S IS FP+L +L
Sbjct: 585 NLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDISKIFPSLSDLT 644
Query: 625 IDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPT 684
ID+C DL+ L++ I I SL L++TNC ++ LP+++ +++LE L L +C +L ++P
Sbjct: 645 IDHCDDLLELKS-IFGITSLNSLSITNCPRILELPKNLSNVQSLERLRLYACPELISLPV 703
Query: 685 SIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITC 744
+ +L LK++DIS C+SL SLPE+FG L +L+ +DM C+ + LP SV L +L+ + C
Sbjct: 704 EVCELPCLKYVDISQCVSLVSLPEKFGKLGSLEKIDMRECSLLGLPSSVAALVSLRHVIC 763
Query: 745 DEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
DEET++ WE + ++ + IEV ++WL
Sbjct: 764 DEETSSMWEMVKKVVPELCIEVAKKCFTVDWL 795
>dbj|BAB08631.1| unnamed protein product [Arabidopsis thaliana]
gi|15240125|ref|NP_201490.1| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395985|sp|Q9FKZ2|DRL41_ARATH Probable disease
resistance protein At5g66890
Length = 415
Score = 259 bits (663), Expect = 2e-67
Identities = 164/435 (37%), Positives = 245/435 (55%), Gaps = 41/435 (9%)
Query: 352 FDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNL 411
FD L N + ECF+D+A F ED RI + ++D+W+ Y G + M + L NL
Sbjct: 9 FDALPHN--LRECFLDMASFLEDQRIIASTIIDLWSASY-----GKEGMNNLQDLASRNL 61
Query: 412 ANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFE--QRKRLIIDMNNNKSG 469
++ +N + ++ YN + ++LRE I Q KE +RKRL +++ +NK
Sbjct: 62 LKLLPIGRN--EYEDGFYNELLVKQDNVLREFAINQCLKESSSIFERKRLNLEIQDNK-- 117
Query: 470 LAEKQQGLMTRIFSKFMRLCVKQNPQQ---LAARILSVSTDETCALDWSHMQPAQVEVLI 526
F C+ NP+Q + A + S+STD++ A W M VE L+
Sbjct: 118 ---------------FPNWCL--NPKQPIVINASLFSISTDDSFASSWFEMDCPNVEALV 160
Query: 527 LNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISV--- 583
LNI + Y+LP +IA M +L+V+II N+G P+KL N+ L SL NL+RIR E++S+
Sbjct: 161 LNISSSNYALPNFIATMKELKVVIIINHGLEPAKLTNLSCLSSLPNLKRIRFEKVSISLL 220
Query: 584 --PSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDI 641
P G LK+L+KLSL+ C+ + A + +S+ +L+E+ IDYC +L L I +
Sbjct: 221 DIPKLG-LKSLEKLSLWFCHVVDALNELED-VSETLQSLQEIEIDYCYNLDELPYWISQV 278
Query: 642 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCI 701
+SLKKL+VTNC+KL + + IG L +LE L LSSC L +P +I +L NL+ LD+S
Sbjct: 279 VSLKKLSVTNCNKLCRVIEAIGDLRDLETLRLSSCASLLELPETIDRLDNLRFLDVSGGF 338
Query: 702 SLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHN 761
L +LP E G L L+ + M C ELP SV NL+NL+ + CDE+TA W+ + + N
Sbjct: 339 QLKNLPLEIGKLKKLEKISMKDCYRCELPDSVKNLENLE-VKCDEDTAFLWKILKPEMKN 397
Query: 762 MKIEVLHVDVNLNWL 776
+ I + NLN L
Sbjct: 398 LTITEEKTEHNLNLL 412
>dbj|BAD94804.1| putative protein [Arabidopsis thaliana]
Length = 366
Score = 236 bits (603), Expect = 2e-60
Identities = 127/348 (36%), Positives = 200/348 (56%), Gaps = 21/348 (6%)
Query: 433 FIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQ 492
F+ HD+LR+L ++ S +RKRL+ M + L + +
Sbjct: 34 FVTQHDVLRDLALHLSNAGKVNRRKRLL--MPKRELDLPGDWE---------------RN 76
Query: 493 NPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIIT 552
N + A+I+S+ T E + W M+ + E+LILN + +Y LP +I+KMS+L+VL+I
Sbjct: 77 NDEHYIAQIVSIHTGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLVII 136
Query: 553 NYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGT----LKNLKKLSLYMCNTILAFEK 608
N G P+ L++ + L L + LER+ VP LKNL K+SL +C +F++
Sbjct: 137 NNGMSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQ 196
Query: 609 GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENL 668
+ ++D FP L +L ID+C DLV L + IC + SL L++TNC +L LP+++ KL+ L
Sbjct: 197 TGLDVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQAL 256
Query: 669 ELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE 728
E+L L +C +L+ +P I +L LK+LDIS C+SLS LPEE G L L+ +DM C +
Sbjct: 257 EILRLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCFSD 316
Query: 729 LPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
P S V+L++L+ + CD + A WE+ + + +KIE +L+WL
Sbjct: 317 RPSSAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 364
>gb|AAM28916.1| NBS/LRR [Pinus taeda]
Length = 479
Score = 229 bits (584), Expect = 3e-58
Identities = 156/500 (31%), Positives = 257/500 (51%), Gaps = 42/500 (8%)
Query: 292 KNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTE-LLTRLQK 350
++LV++V C GLPL +KVI +SL+ + +W ++LS+ SI + E LL RL+
Sbjct: 5 EDLVKQVAAECKGLPLALKVIGSSLRGKRRPIWINAERKLSKSESISEYYKESLLKRLET 64
Query: 351 IFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMN 410
DVL+D K +CF+DL FP+ + V L+D+W + +++ A E++ +L N
Sbjct: 65 SIDVLDDKHK--QCFLDLGAFPKGRKFSVETLLDIWVYVRQME--WTDAFEVLLELASRN 120
Query: 411 LANVI-IPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSG 469
L N+ P A D + + HD++R+L +Y ++++ KRL +K
Sbjct: 121 LLNLTGYPGSGA--IDYSCASELTFSQHDVMRDLALYLASQDNIISPKRLFTPSKEDK-- 176
Query: 470 LAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNI 529
+ T S Q A+ +S+ T DW + +VE L L
Sbjct: 177 -------IPTEWLSTL-------KDQASRAQFVSIYTGAMQEQDWCQIDFPEVEALALFF 222
Query: 530 HTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSF--- 586
QY LP ++ + KL+VLI+ NY + + + S + + L ++ VP+
Sbjct: 223 SANQYCLPTFLHRTPKLKVLIVYNYSSMRANIIGLPRFSSPIQIRSVFLHKLIVPASLYE 282
Query: 587 --GTLKNLKKLSLYMCNTILAFEKGSILISDA------FPNLEELNIDYCKDLVVLQTGI 638
+ + L+KLS+ +C + G+ + D FPN+ E+NID+C DL L +
Sbjct: 283 NCRSWERLEKLSVCLCEGL-----GNSSLVDMELEPLNFPNITEINIDHCSDLGELPLKL 337
Query: 639 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 698
C++ SL++L+VTNCH + +LP D+G+L++L +L LS+C L +P SI KL L++LDIS
Sbjct: 338 CNLTSLQRLSVTNCHLIQNLPDDMGRLKSLRVLRLSACPSLSRLPPSICKLGQLEYLDIS 397
Query: 699 NCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVV--NLQNLKTITCDEETAATWEDFQ 756
C L LP EF L NL+ LDM C+ ++ +V+ +L+ + D+E A
Sbjct: 398 LCRCLQDLPSEFDQLSNLETLDMRECSGLKKVPTVIQSSLKRVVISDSDKEYEAWXSIKA 457
Query: 757 HMLHNMKIEVLHVDVNLNWL 776
LH + I+V+ +L WL
Sbjct: 458 STLHTLTIDVVPEIFSLAWL 477
>ref|NP_912838.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|9558523|dbj|BAB03441.1| NBS-LRR disease resistance
protein -like [Oryza sativa (japonica cultivar-group)]
Length = 1292
Score = 157 bits (396), Expect = 2e-36
Identities = 168/636 (26%), Positives = 285/636 (44%), Gaps = 103/636 (16%)
Query: 170 IFVTFSKTPMLKTIVERIH-------QHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLL 222
I+V+ S L T+V+ +H + C + + E L + G+ LL
Sbjct: 219 IWVSMSAGFSLATLVQAVHPIVAAPSERCDLATTTTTNLEAIARFLSMAFT---GNKYLL 275
Query: 223 VLDDVWPSSESLVEKLQFQMTG----FKILVTSRVAFPRFSTTCI----LKPLAHEDAVT 274
VLDDVW S E+L+ + G KI+VT+R + LK L+ ED
Sbjct: 276 VLDDVWSESHDEWERLRLLLRGGKRGSKIIVTTRSRRIGMMVGTVPPLMLKSLSDEDCWE 335
Query: 275 LFHHYAQMEKNSSDIIDK--NLVEKVVRSCNGLPLTIKVIATSLK-NRPHDLWHKIV-KE 330
LF A E+ ++ K + +++V C G+PL K + + L+ R + W + E
Sbjct: 336 LFKRKA-FEEADEELYPKLVRIGKEIVPKCGGVPLAAKALGSMLRFKRNEESWIAVRDSE 394
Query: 331 LSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELY 390
+ Q LD +L L+ +D + P + +CF ++FP +H I L+ W L
Sbjct: 395 IWQ----LDKEETILPSLKLSYDQMP--PVLKQCFAYCSVFPRNHEIDKGKLIQQWVALG 448
Query: 391 KLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTK 450
++ + + +K A D + ++ L+E+ + +K
Sbjct: 449 FVEPSKYGCQPVSDK---------------ADDCFEH------LLWMSFLQEVDQHDLSK 487
Query: 451 EPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDE-T 509
+ E R+ +++ LA+ G +I S + N + A R S+ D +
Sbjct: 488 KGLEVDGRVKYKIHDLVHDLAQSVAGDEVQIIS-----AKRVNGRTEACRYASLHDDMGS 542
Query: 510 CALDWSHMQPAQVE-----------------VLILNIHTKQY-SLPEWIAKMSKLRVL-- 549
+ WS ++ + + +L++ Q LP+ + K+ LR L
Sbjct: 543 TDVLWSMLRKVRAFHSWGRSLDINLFLHSRFLRVLDLRGSQIMELPQSVGKLKHLRYLDL 602
Query: 550 ---IITNYGFHPSKLNNIELLG---------------SLQNLERIRLERI---SVP-SFG 587
+I+ S L+N++ L +L+NLE + L S+P S G
Sbjct: 603 SSSLISTLPNCISSLHNLQTLHLYNCINLNVLPMSVCALENLEILNLSACNFHSLPDSIG 662
Query: 588 TLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKL 647
L+NL+ L+L +C+ ++ +L LN+ C +L +L IC + +L L
Sbjct: 663 HLQNLQDLNLSLCSFLVTLPSSI----GTLQSLHLLNLKGCGNLEILPDTICSLQNLHFL 718
Query: 648 NVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLP 707
N++ C L +LP++IG L NL L+LS CTDLE+IPTSIG++ +L LD+S+C SLS LP
Sbjct: 719 NLSRCGVLQALPKNIGNLSNLLHLNLSQCTDLESIPTSIGRIKSLHILDLSHCSSLSELP 778
Query: 708 EEFGNLCNLKNLDMA-TCASIELPFSVVNLQNLKTI 742
G L L+ L ++ +S+ LP S +L NL+T+
Sbjct: 779 GSIGGLHELQILILSHHASSLALPVSTSHLPNLQTL 814
Score = 78.2 bits (191), Expect = 1e-12
Identities = 66/233 (28%), Positives = 104/233 (44%), Gaps = 30/233 (12%)
Query: 536 LPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI----SVP-SFGTLK 590
LP+ I + L L ++ G + NI G+L NL + L + S+P S G +K
Sbjct: 705 LPDTICSLQNLHFLNLSRCGVLQALPKNI---GNLSNLLHLNLSQCTDLESIPTSIGRIK 761
Query: 591 NLKKLSLYMCNT-----------------ILAFEKGSILI---SDAFPNLEELNIDYCKD 630
+L L L C++ IL+ S+ + + PNL+ L++ +
Sbjct: 762 SLHILDLSHCSSLSELPGSIGGLHELQILILSHHASSLALPVSTSHLPNLQTLDLSWNLS 821
Query: 631 LVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLL 690
L L I ++ SLK L + C L LP+ I L LE L+ C +L +P + ++
Sbjct: 822 LEELPESIGNLHSLKTLILFQCWSLRKLPESITNLMMLESLNFVGCENLAKLPDGMTRIT 881
Query: 691 NLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTIT 743
NLKHL C SL LP FG L+ L + + S+ L++L +T
Sbjct: 882 NLKHLRNDQCRSLKQLPNGFGRWTKLETLSLLMIG--DKHSSITELKDLNNLT 932
Score = 67.4 bits (163), Expect = 2e-09
Identities = 36/85 (42%), Positives = 52/85 (60%)
Query: 618 PNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCT 677
P LEEL I+YC+ L VL I + +L+KL ++NC +L +LP+ IG L LE L +S C
Sbjct: 1182 PKLEELTIEYCEMLRVLAEPIRYLTTLRKLKISNCTELDALPEWIGDLVALESLQISCCP 1241
Query: 678 DLEAIPTSIGKLLNLKHLDISNCIS 702
L +IP + L L+ L ++ C S
Sbjct: 1242 KLVSIPKGLQHLTALEELTVTACSS 1266
Score = 47.0 bits (110), Expect = 0.002
Identities = 30/103 (29%), Positives = 49/103 (47%), Gaps = 12/103 (11%)
Query: 668 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS- 726
LE L++ C L + I L L+ L ISNC L +LPE G+L L++L ++ C
Sbjct: 1184 LEELTIEYCEMLRVLAEPIRYLTTLRKLKISNCTELDALPEWIGDLVALESLQISCCPKL 1243
Query: 727 IELPFSVVNLQNLKTIT-----------CDEETAATWEDFQHM 758
+ +P + +L L+ +T C ++T W H+
Sbjct: 1244 VSIPKGLQHLTALEELTVTACSSELNENCRKDTGKDWFKICHI 1286
>emb|CAD45029.1| NBS-LRR disease resistance protein homologue [Hordeum vulgare]
Length = 1262
Score = 144 bits (362), Expect = 1e-32
Identities = 154/608 (25%), Positives = 265/608 (43%), Gaps = 97/608 (15%)
Query: 221 LLVLDDVWPSSESLVEKLQFQM---TGFKILVTSR-VAFPRFSTTC-----ILKPLAHED 271
L+VLDD+W + +++L+ + T K+LVT+R + R +L PL ++
Sbjct: 268 LIVLDDLWETGYFQLDQLKLMLNVSTKMKVLVTTRSIDIARKMGNVGVEPYMLDPLDNDM 327
Query: 272 AVTLFHHYAQMEKNSSDIIDKNLVE----KVVRSCNGLPLTIKVIATSLKNRPHDLWHKI 327
+ ++ + DK +E K+ R C GLPL + + L W I
Sbjct: 328 CWRIIKQSSRFQSRP----DKEQLEPNGQKIARKCGGLPLAAQALGFLLSGMDLSEWEAI 383
Query: 328 VKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWA 387
+S S++ +L L+ ++ L P + CF +FP+ H I L+ W
Sbjct: 384 C--ISDIWDEPFSDSTVLPSLKLSYNTL--TPYMRLCFAYCGIFPKGHNISKDYLIHQWI 439
Query: 388 ELYKLD-DNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILR----- 441
L ++ N A+++ K L + +T N ++HD+ R
Sbjct: 440 ALGFIEPSNKFSAIQLGGKYVRQFLGMSFLHHSKLPETFGNAMFTMHDLVHDLARSVITE 499
Query: 442 ELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQ-GLMTRIFSKFMRLCVKQNPQ----- 495
EL ++ + + K I + +++ + MT IF +R+ + +
Sbjct: 500 ELVVFDAEIVSDNRIKEYCIYASLTNCNISDHNKVRKMTTIFPPKLRVMHFSDCKLHGSA 559
Query: 496 ---QLAARILSVSTDETCALDWSHMQPAQVEVLILN-IHTKQYSLPEWIAKMSKLRVLII 551
Q R+L +S + Q Q+EVLI + +Q+ PE I ++SKL L +
Sbjct: 560 FSFQKCLRVLDLSGCSIKDFASALGQLKQLEVLIAQKLQDRQF--PESITRLSKLHYLNL 617
Query: 552 ---------------------------TNYGFHPSKLNNI------------------EL 566
TN P L + E
Sbjct: 618 SGSRGISEIPSSVGKLVSLVHLDLSYCTNVKVIPKALGILRNLQTLDLSWCEKLESLPES 677
Query: 567 LGSLQNLERIRL----ERISVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLE 621
LGS+QNL+R+ L E ++P S G+LK+++ L L C + + + + N++
Sbjct: 678 LGSVQNLQRLNLSNCFELEALPESLGSLKDVQTLDLSSCYKLESLPESL----GSLKNVQ 733
Query: 622 ELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEA 681
L++ C LV L + + +L+ ++++ C KL + P+ G LENL++L+LS+C +LE+
Sbjct: 734 TLDLSRCYKLVSLPKNLGRLKNLRTIDLSGCKKLETFPESFGSLENLQILNLSNCFELES 793
Query: 682 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLK 740
+P S G L NL+ L++ C L SLPE G L NL+ LD + C +E +P S+ L NL+
Sbjct: 794 LPESFGSLKNLQTLNLVECKKLESLPESLGGLKNLQTLDFSVCHKLESVPESLGGLNNLQ 853
Query: 741 TI---TCD 745
T+ CD
Sbjct: 854 TLKLSVCD 861
Score = 129 bits (325), Expect = 3e-28
Identities = 89/257 (34%), Positives = 148/257 (56%), Gaps = 17/257 (6%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI 581
++ L ++ K S+PE + ++ L+ L ++ + ++ ++ LGSL+NL+ + L
Sbjct: 828 LQTLDFSVCHKLESVPESLGGLNNLQTLKLS---VCDNLVSLLKSLGSLKNLQTLDLSGC 884
Query: 582 ----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQT 636
S+P S G+L+NL+ L+L C + + + NL+ LNI +C +LV L
Sbjct: 885 KKLESLPESLGSLENLQILNLSNCFKLESLPESL----GRLKNLQTLNISWCTELVFLPK 940
Query: 637 GICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLD 696
+ ++ +L +L+++ C KL SLP +G LENLE L+LS C LE++P S+G L NL+ LD
Sbjct: 941 NLGNLKNLPRLDLSGCMKLESLPDSLGSLENLETLNLSKCFKLESLPESLGGLQNLQTLD 1000
Query: 697 ISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTIT---CDE-ETAAT 751
+ C L SLPE G L NL+ L ++ C +E LP S+ L+NL+T+T CD+ E+
Sbjct: 1001 LLVCHKLESLPESLGGLKNLQTLQLSFCHKLESLPESLGGLKNLQTLTLSVCDKLESLPE 1060
Query: 752 WEDFQHMLHNMKIEVLH 768
LH +K++V +
Sbjct: 1061 SLGSLKNLHTLKLQVCY 1077
Score = 127 bits (319), Expect = 1e-27
Identities = 86/228 (37%), Positives = 133/228 (57%), Gaps = 15/228 (6%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 580
+E L L+ K SLPE + + L+ L + KL ++ E LG L+NL+ ++L
Sbjct: 972 LETLNLSKCFKLESLPESLGGLQNLQTLDLLVC----HKLESLPESLGGLKNLQTLQLSF 1027
Query: 581 I----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 635
S+P S G LKNL+ L+L +C+ + + + + NL L + C L L
Sbjct: 1028 CHKLESLPESLGGLKNLQTLTLSVCDKLESLPESL----GSLKNLHTLKLQVCYKLKSLP 1083
Query: 636 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 695
+ I +L LN++ CH L S+P+ +G LENL++L+LS+C LE+IP S+G L NL+ L
Sbjct: 1084 ESLGSIKNLHTLNLSVCHNLESIPESVGSLENLQILNLSNCFKLESIPKSLGSLKNLQTL 1143
Query: 696 DISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 742
+S C L SLP+ GNL NL+ LD++ C +E LP S+ +L+NL+T+
Sbjct: 1144 ILSWCTRLVSLPKNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTL 1191
Score = 120 bits (300), Expect = 2e-25
Identities = 82/230 (35%), Positives = 133/230 (57%), Gaps = 19/230 (8%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 580
++ L L + K SLPE + + L+ L ++ KL ++ E LG L+NL+ + L
Sbjct: 996 LQTLDLLVCHKLESLPESLGGLKNLQTLQLS----FCHKLESLPESLGGLKNLQTLTLSV 1051
Query: 581 I----SVP-SFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVV 633
S+P S G+LKNL L L +C + + + GSI NL LN+ C +L
Sbjct: 1052 CDKLESLPESLGSLKNLHTLKLQVCYKLKSLPESLGSI------KNLHTLNLSVCHNLES 1105
Query: 634 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 693
+ + + +L+ LN++NC KL S+P+ +G L+NL+ L LS CT L ++P ++G L NL+
Sbjct: 1106 IPESVGSLENLQILNLSNCFKLESIPKSLGSLKNLQTLILSWCTRLVSLPKNLGNLKNLQ 1165
Query: 694 HLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 742
LD+S C L SLP+ G+L NL+ L+++ C +E LP + +L+ L+T+
Sbjct: 1166 TLDLSGCKKLESLPDSLGSLENLQTLNLSNCFKLESLPEILGSLKKLQTL 1215
Score = 119 bits (297), Expect = 5e-25
Identities = 79/228 (34%), Positives = 135/228 (58%), Gaps = 15/228 (6%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 580
++ L L+ K SLPE + + L+ L ++ KL ++ E LGSL+NL ++L+
Sbjct: 1020 LQTLQLSFCHKLESLPESLGGLKNLQTLTLSVC----DKLESLPESLGSLKNLHTLKLQV 1075
Query: 581 I----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 635
S+P S G++KNL L+L +C+ + + + + NL+ LN+ C L +
Sbjct: 1076 CYKLKSLPESLGSIKNLHTLNLSVCHNLESIPESV----GSLENLQILNLSNCFKLESIP 1131
Query: 636 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 695
+ + +L+ L ++ C +L SLP+++G L+NL+ L LS C LE++P S+G L NL+ L
Sbjct: 1132 KSLGSLKNLQTLILSWCTRLVSLPKNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTL 1191
Query: 696 DISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 742
++SNC L SLPE G+L L+ L++ C +E LP S+ +L++L+T+
Sbjct: 1192 NLSNCFKLESLPEILGSLKKLQTLNLFRCGKLESLPESLGSLKHLQTL 1239
Score = 118 bits (296), Expect = 6e-25
Identities = 84/230 (36%), Positives = 130/230 (56%), Gaps = 19/230 (8%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRL-- 578
++ L L+ K SLPE + + L++L ++N KL ++ E LG L+NL+ + +
Sbjct: 876 LQTLDLSGCKKLESLPESLGSLENLQILNLSNC----FKLESLPESLGRLKNLQTLNISW 931
Query: 579 --ERISVP-SFGTLKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVV 633
E + +P + G LKNL +L L C + L GS+ NLE LN+ C L
Sbjct: 932 CTELVFLPKNLGNLKNLPRLDLSGCMKLESLPDSLGSL------ENLETLNLSKCFKLES 985
Query: 634 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 693
L + + +L+ L++ CHKL SLP+ +G L+NL+ L LS C LE++P S+G L NL+
Sbjct: 986 LPESLGGLQNLQTLDLLVCHKLESLPESLGGLKNLQTLQLSFCHKLESLPESLGGLKNLQ 1045
Query: 694 HLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 742
L +S C L SLPE G+L NL L + C ++ LP S+ +++NL T+
Sbjct: 1046 TLTLSVCDKLESLPESLGSLKNLHTLKLQVCYKLKSLPESLGSIKNLHTL 1095
Score = 116 bits (290), Expect = 3e-24
Identities = 81/227 (35%), Positives = 124/227 (53%), Gaps = 16/227 (7%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRL-- 578
V+ L L+ K SLP+ + ++ LR + ++ KL E GSL+NL+ + L
Sbjct: 732 VQTLDLSRCYKLVSLPKNLGRLKNLRTIDLSGC----KKLETFPESFGSLENLQILNLSN 787
Query: 579 --ERISVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 635
E S+P SFG+LKNL+ L+L C + + + NL+ L+ C L +
Sbjct: 788 CFELESLPESFGSLKNLQTLNLVECKKLESLPESL----GGLKNLQTLDFSVCHKLESVP 843
Query: 636 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 695
+ + +L+ L ++ C L SL + +G L+NL+ L LS C LE++P S+G L NL+ L
Sbjct: 844 ESLGGLNNLQTLKLSVCDNLVSLLKSLGSLKNLQTLDLSGCKKLESLPESLGSLENLQIL 903
Query: 696 DISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTI 742
++SNC L SLPE G L NL+ L+++ C EL F NL NLK +
Sbjct: 904 NLSNCFKLESLPESLGRLKNLQTLNISWCT--ELVFLPKNLGNLKNL 948
Score = 116 bits (290), Expect = 3e-24
Identities = 79/225 (35%), Positives = 129/225 (57%), Gaps = 9/225 (4%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI 581
V+ L L+ K SLPE + + ++ L ++ S N+ L +L+ ++ +++
Sbjct: 708 VQTLDLSSCYKLESLPESLGSLKNVQTLDLSRCYKLVSLPKNLGRLKNLRTIDLSGCKKL 767
Query: 582 SV--PSFGTLKNLKKLSLYMCNTILAFEKGSILIS-DAFPNLEELNIDYCKDLVVLQTGI 638
SFG+L+NL+ L+L C FE S+ S + NL+ LN+ CK L L +
Sbjct: 768 ETFPESFGSLENLQILNLSNC-----FELESLPESFGSLKNLQTLNLVECKKLESLPESL 822
Query: 639 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 698
+ +L+ L+ + CHKL S+P+ +G L NL+ L LS C +L ++ S+G L NL+ LD+S
Sbjct: 823 GGLKNLQTLDFSVCHKLESVPESLGGLNNLQTLKLSVCDNLVSLLKSLGSLKNLQTLDLS 882
Query: 699 NCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 742
C L SLPE G+L NL+ L+++ C +E LP S+ L+NL+T+
Sbjct: 883 GCKKLESLPESLGSLENLQILNLSNCFKLESLPESLGRLKNLQTL 927
Score = 110 bits (276), Expect = 1e-22
Identities = 77/222 (34%), Positives = 124/222 (55%), Gaps = 15/222 (6%)
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNI-ELLGSLQNLERIRLER 580
++ L L++ K SLPE + + L L + KL ++ E LGS++NL + L
Sbjct: 1044 LQTLTLSVCDKLESLPESLGSLKNLHTLKLQVC----YKLKSLPESLGSIKNLHTLNLSV 1099
Query: 581 I----SVP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 635
S+P S G+L+NL+ L+L C + + K + NL+ L + +C LV L
Sbjct: 1100 CHNLESIPESVGSLENLQILNLSNCFKLESIPKSL----GSLKNLQTLILSWCTRLVSLP 1155
Query: 636 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHL 695
+ ++ +L+ L+++ C KL SLP +G LENL+ L+LS+C LE++P +G L L+ L
Sbjct: 1156 KNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTLNLSNCFKLESLPEILGSLKKLQTL 1215
Query: 696 DISNCISLSSLPEEFGNLCNLKNLDMATCASIE-LPFSVVNL 736
++ C L SLPE G+L +L+ L + C +E LP S+ NL
Sbjct: 1216 NLFRCGKLESLPESLGSLKHLQTLVLIDCPKLEYLPKSLENL 1257
>gb|AAG60098.1| disease resistance protein, putative [Arabidopsis thaliana]
Length = 1398
Score = 132 bits (333), Expect = 3e-29
Identities = 86/216 (39%), Positives = 126/216 (57%), Gaps = 19/216 (8%)
Query: 536 LPEWIAKMSKLRVLIITNYGFHPSKLNNIEL-LGSLQNLERIRLERISV-----PSFGTL 589
LP I + L+ L ++ S L + L +G+L NL+ + L S S G L
Sbjct: 874 LPSSIGNLINLKKLDLSGC----SSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNL 929
Query: 590 KNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKL 647
NLK L+L C++++ G+++ NL+EL + C LV L + I ++I+LKKL
Sbjct: 930 INLKTLNLSECSSLVELPSSIGNLI------NLQELYLSECSSLVELPSSIGNLINLKKL 983
Query: 648 NVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLP 707
+++ C L LP IG L NL+ L+LS C+ L +P+SIG L+NL+ L +S C SL LP
Sbjct: 984 DLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELP 1043
Query: 708 EEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTI 742
GNL NLK LD++ C+S +ELP S+ NL NLKT+
Sbjct: 1044 SSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTL 1079
Score = 132 bits (331), Expect = 6e-29
Identities = 85/222 (38%), Positives = 124/222 (55%), Gaps = 23/222 (10%)
Query: 536 LPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERIS--------VPSFG 587
LP I + L++L + + +E+ S+ NL ++L +S S G
Sbjct: 826 LPSSIGNLISLKILYLKRIS------SLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIG 879
Query: 588 TLKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLK 645
L NLKKL L C+++ L G+++ NL+EL + C LV L + I ++I+LK
Sbjct: 880 NLINLKKLDLSGCSSLVELPLSIGNLI------NLQELYLSECSSLVELPSSIGNLINLK 933
Query: 646 KLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSS 705
LN++ C L LP IG L NL+ L LS C+ L +P+SIG L+NLK LD+S C SL
Sbjct: 934 TLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVE 993
Query: 706 LPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTITCDE 746
LP GNL NLK L+++ C+S +ELP S+ NL NL+ + E
Sbjct: 994 LPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSE 1035
Score = 124 bits (311), Expect = 1e-26
Identities = 161/632 (25%), Positives = 260/632 (40%), Gaps = 127/632 (20%)
Query: 205 VNKLGLLLKKVEGSPLLLVLDDVWPSSESLVEKLQFQMT-----GFKILVTSR-----VA 254
+++LG V G +L+VLD V + LV+ L G +I++T++ A
Sbjct: 345 IHQLGTAQNFVMGKKVLIVLDGV----DQLVQLLAMPKAVCLGPGSRIIITTQDQQLLKA 400
Query: 255 FPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIAT 314
F + P HE A+ +F +A + D +K L KV R LPL ++V+ +
Sbjct: 401 FQIKHIYNVDFPPDHE-ALQIFCIHAFGHDSPDDGFEK-LATKVTRLAGNLPLGLRVMGS 458
Query: 315 SLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPED 374
+ + W EL + LD E+ + L+ +DVL+D K + F+ +A F D
Sbjct: 459 HFRGMSKEDWK---GELPRLRIRLDG--EIGSILKFSYDVLDDEDK--DLFLHIACFFND 511
Query: 375 HRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFI 434
I D + G+Q V++ R S+ +N +
Sbjct: 512 EGID-HTFEDTLRHKFSNVQRGLQ---------------VLVQRSLISEDLTQPMHNLLV 555
Query: 435 IL-HDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQN 493
L +I+R +Y+ K F + I ++ + +G +E G+ ++ L +
Sbjct: 556 QLGREIVRNQSVYEPGKRQFLVDGKEICEVLTSHTG-SESVIGINFEVYWSMDELNISDR 614
Query: 494 PQQLAARILSVSTDETC------------------ALDWSHMQPAQVE--------VLIL 527
+ + + DE L W + + V I+
Sbjct: 615 VFEGMSNLQFFRFDENSYGRLHLPQGLNYLPPKLRILHWDYYPMTSLPSKFNLKFLVKII 674
Query: 528 NIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNN------------------IELLGS 569
H++ L E I + L+V+ + Y H +L N IEL S
Sbjct: 675 LKHSELEKLWEGIQPLVNLKVMDL-RYSSHLKELPNLSTAINLLEMVLSDCSSLIELPSS 733
Query: 570 LQNLERIR----------------------LERISV----------PSFGTLKNLKKLSL 597
+ N I+ L R+ + S G L NL +L L
Sbjct: 734 IGNATNIKSLDIQGCSSLLKLPSSIGNLITLPRLDLMGCSSLVELPSSIGNLINLPRLDL 793
Query: 598 YMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKL 655
C++++ G+++ NLE C L+ L + I ++ISLK L + L
Sbjct: 794 MGCSSLVELPSSIGNLI------NLEAFYFHGCSSLLELPSSIGNLISLKILYLKRISSL 847
Query: 656 SSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN 715
+P IG L NL+LL+LS C+ L +P+SIG L+NLK LD+S C SL LP GNL N
Sbjct: 848 VEIPSSIGNLINLKLLNLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLIN 907
Query: 716 LKNLDMATCAS-IELPFSVVNLQNLKTITCDE 746
L+ L ++ C+S +ELP S+ NL NLKT+ E
Sbjct: 908 LQELYLSECSSLVELPSSIGNLINLKTLNLSE 939
Score = 122 bits (306), Expect = 4e-26
Identities = 76/184 (41%), Positives = 111/184 (60%), Gaps = 15/184 (8%)
Query: 567 LGSLQNLERIRLERISV-----PSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 619
+G+L NL+ + L S S G L NL++L L C++++ G+++ N
Sbjct: 926 IGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLI------N 979
Query: 620 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 679
L++L++ C LV L I ++I+LK LN++ C L LP IG L NL+ L LS C+ L
Sbjct: 980 LKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSL 1039
Query: 680 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQN 738
+P+SIG L+NLK LD+S C SL LP GNL NLK L+++ C+S +ELP S+ NL N
Sbjct: 1040 VELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNL-N 1098
Query: 739 LKTI 742
LK +
Sbjct: 1099 LKKL 1102
Score = 116 bits (290), Expect = 3e-24
Identities = 76/203 (37%), Positives = 113/203 (55%), Gaps = 19/203 (9%)
Query: 567 LGSLQNLERIRLERIS----VP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLE 621
+G+L NL+++ L S +P S G L NLK L+L C++++ S NL+
Sbjct: 1046 IGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS-----SIGNLNLK 1100
Query: 622 ELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEA 681
+L++ C LV L + I ++I+LKKL+++ C L LP IG L NL+ L LS C+ L
Sbjct: 1101 KLDLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVE 1160
Query: 682 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI----ELPFSVVNL- 736
+P+SIG L+NL+ L +S C SL LP GNL NLK LD+ C + +LP S+ L
Sbjct: 1161 LPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLV 1220
Query: 737 ----QNLKTITCDEETAATWEDF 755
++L+T+ C W F
Sbjct: 1221 AESCESLETLACSFPNPQVWLKF 1243
>ref|NP_683486.1| leucine-rich repeat family protein [Arabidopsis thaliana]
Length = 703
Score = 132 bits (333), Expect = 3e-29
Identities = 86/216 (39%), Positives = 126/216 (57%), Gaps = 19/216 (8%)
Query: 536 LPEWIAKMSKLRVLIITNYGFHPSKLNNIEL-LGSLQNLERIRLERISV-----PSFGTL 589
LP I + L+ L ++ S L + L +G+L NL+ + L S S G L
Sbjct: 179 LPSSIGNLINLKKLDLSGC----SSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNL 234
Query: 590 KNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKL 647
NLK L+L C++++ G+++ NL+EL + C LV L + I ++I+LKKL
Sbjct: 235 INLKTLNLSECSSLVELPSSIGNLI------NLQELYLSECSSLVELPSSIGNLINLKKL 288
Query: 648 NVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLP 707
+++ C L LP IG L NL+ L+LS C+ L +P+SIG L+NL+ L +S C SL LP
Sbjct: 289 DLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELP 348
Query: 708 EEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTI 742
GNL NLK LD++ C+S +ELP S+ NL NLKT+
Sbjct: 349 SSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTL 384
Score = 132 bits (331), Expect = 6e-29
Identities = 85/222 (38%), Positives = 124/222 (55%), Gaps = 23/222 (10%)
Query: 536 LPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERIS--------VPSFG 587
LP I + L++L + + +E+ S+ NL ++L +S S G
Sbjct: 131 LPSSIGNLISLKILYLKRIS------SLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIG 184
Query: 588 TLKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLK 645
L NLKKL L C+++ L G+++ NL+EL + C LV L + I ++I+LK
Sbjct: 185 NLINLKKLDLSGCSSLVELPLSIGNLI------NLQELYLSECSSLVELPSSIGNLINLK 238
Query: 646 KLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSS 705
LN++ C L LP IG L NL+ L LS C+ L +P+SIG L+NLK LD+S C SL
Sbjct: 239 TLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVE 298
Query: 706 LPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTITCDE 746
LP GNL NLK L+++ C+S +ELP S+ NL NL+ + E
Sbjct: 299 LPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSE 340
Score = 122 bits (306), Expect = 4e-26
Identities = 76/184 (41%), Positives = 111/184 (60%), Gaps = 15/184 (8%)
Query: 567 LGSLQNLERIRLERISV-----PSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 619
+G+L NL+ + L S S G L NL++L L C++++ G+++ N
Sbjct: 231 IGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLI------N 284
Query: 620 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 679
L++L++ C LV L I ++I+LK LN++ C L LP IG L NL+ L LS C+ L
Sbjct: 285 LKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSL 344
Query: 680 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQN 738
+P+SIG L+NLK LD+S C SL LP GNL NLK L+++ C+S +ELP S+ NL N
Sbjct: 345 VELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNL-N 403
Query: 739 LKTI 742
LK +
Sbjct: 404 LKKL 407
Score = 117 bits (294), Expect = 1e-24
Identities = 83/230 (36%), Positives = 123/230 (53%), Gaps = 17/230 (7%)
Query: 525 LILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRL----ER 580
++L+ + LP I + ++ L I S L +G+L L R+ L
Sbjct: 24 MVLSDCSSLIELPSSIGNATNIKSLDIQGCS---SLLKLPSSIGNLITLPRLDLMGCSSL 80
Query: 581 ISVPS-FGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVLQTG 637
+ +PS G L NL +L L C++++ G+++ NLE C L+ L +
Sbjct: 81 VELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLI------NLEAFYFHGCSSLLELPSS 134
Query: 638 ICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDI 697
I ++ISLK L + L +P IG L NL+LL+LS C+ L +P+SIG L+NLK LD+
Sbjct: 135 IGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIGNLINLKKLDL 194
Query: 698 SNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLKTITCDE 746
S C SL LP GNL NL+ L ++ C+S +ELP S+ NL NLKT+ E
Sbjct: 195 SGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLKTLNLSE 244
Score = 116 bits (290), Expect = 3e-24
Identities = 76/203 (37%), Positives = 113/203 (55%), Gaps = 19/203 (9%)
Query: 567 LGSLQNLERIRLERIS----VP-SFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLE 621
+G+L NL+++ L S +P S G L NLK L+L C++++ S NL+
Sbjct: 351 IGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS-----SIGNLNLK 405
Query: 622 ELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEA 681
+L++ C LV L + I ++I+LKKL+++ C L LP IG L NL+ L LS C+ L
Sbjct: 406 KLDLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVE 465
Query: 682 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI----ELPFSVVNL- 736
+P+SIG L+NL+ L +S C SL LP GNL NLK LD+ C + +LP S+ L
Sbjct: 466 LPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLV 525
Query: 737 ----QNLKTITCDEETAATWEDF 755
++L+T+ C W F
Sbjct: 526 AESCESLETLACSFPNPQVWLKF 548
Score = 91.3 bits (225), Expect = 1e-16
Identities = 50/125 (40%), Positives = 70/125 (56%), Gaps = 1/125 (0%)
Query: 619 NLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTD 678
NL E+ + C L+ L + I + ++K L++ C L LP IG L L L L C+
Sbjct: 20 NLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSIGNLITLPRLDLMGCSS 79
Query: 679 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQ 737
L +P+SIG L+NL LD+ C SL LP GNL NL+ C+S +ELP S+ NL
Sbjct: 80 LVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLINLEAFYFHGCSSLLELPSSIGNLI 139
Query: 738 NLKTI 742
+LK +
Sbjct: 140 SLKIL 144
Score = 83.6 bits (205), Expect = 2e-14
Identities = 46/115 (40%), Positives = 67/115 (58%), Gaps = 5/115 (4%)
Query: 627 YCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSI 686
+ K+L L T I +L ++ +++C L LP IG N++ L + C+ L +P+SI
Sbjct: 8 HLKELPNLSTAI----NLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSI 63
Query: 687 GKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCAS-IELPFSVVNLQNLK 740
G L+ L LD+ C SL LP GNL NL LD+ C+S +ELP S+ NL NL+
Sbjct: 64 GNLITLPRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLINLE 118
>emb|CAB96660.1| RPP1 disease resistance protein-like [Arabidopsis thaliana]
gi|15238999|ref|NP_196686.1| disease resistance protein
(TIR-NBS-LRR class), putative [Arabidopsis thaliana]
Length = 1189
Score = 130 bits (328), Expect = 1e-28
Identities = 148/572 (25%), Positives = 265/572 (45%), Gaps = 75/572 (13%)
Query: 208 LGLLLKKVEGSPLLLVLDDVWPSSE--SLVEKLQFQMTGFKILVTSR--VAFPRFSTTCI 263
LG+ +++ +L+VLD V S + ++ ++ + G +I++T++ F I
Sbjct: 332 LGVAQDRLKDKKVLVVLDGVNQSVQLDAMAKEAWWFGPGSRIIITTQDQKLFRAHGINHI 391
Query: 264 LKP--LAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPH 321
K E+A+ +F YA + + D +NL KV+ LPL ++++ + +
Sbjct: 392 YKVDFPPTEEALQIFCMYAFGQNSPKDGF-QNLAWKVINLAGNLPLGLRIMGSYFRGMSR 450
Query: 322 DLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAA 381
+ W K + L S LD++ + + + +D L+D K + F+ +A F I +
Sbjct: 451 EEWKKSLPRLE---SSLDADIQSILKFS--YDALDDEDKNL--FLHIACFFNGKEIKI-- 501
Query: 382 LVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILH---D 438
L + A+ + +E+ +L NV+ + S ++ H ++ +
Sbjct: 502 LEEHLAKKF---------VEVRQRL------NVLAEKSLISFSNWGTIEMHKLLAKLGGE 546
Query: 439 ILRELGIYQSTKEPFEQRKRLIIDMNNNKSG------------LAEKQQGLMTRIFS--- 483
I+R I++ + F I D+ N + + E++ + R+F
Sbjct: 547 IVRNQSIHEPGQRQFLFDGEEICDVLNGDAAGSKSVIGIDFHYIIEEEFDMNERVFEGMS 606
Query: 484 --KFMRLCVKQNPQQLAARILSVSTDETCALDWSHM------QPAQVEVLI-LNI-HTKQ 533
+F+R + QL+ R LS + + LDW + VE LI LN+ H+K
Sbjct: 607 NLQFLRFDCDHDTLQLS-RGLSYLSRKLQLLDWIYFPMTCLPSTVNVEFLIELNLTHSKL 665
Query: 534 YSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLER----ISVPS-FGT 588
L E + + LR + ++ + L + L + NL ++ L I +PS G
Sbjct: 666 DMLWEGVKPLHNLRQMDLS----YSVNLKELPDLSTAINLRKLILSNCSSLIKLPSCIGN 721
Query: 589 LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 648
NL+ L L C++++ DA NL++L + YC +LV L + I + I+L++L+
Sbjct: 722 AINLEDLDLNGCSSLVELPS----FGDAI-NLQKLLLRYCSNLVELPSSIGNAINLRELD 776
Query: 649 VTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 708
+ C L LP IG NL +L L+ C++L +P+SIG +NL+ LD+ C L LP
Sbjct: 777 LYYCSSLIRLPSSIGNAINLLILDLNGCSNLLELPSSIGNAINLQKLDLRRCAKLLELPS 836
Query: 709 EFGNLCNLKNLDMATCAS-IELPFSVVNLQNL 739
GN NL+NL + C+S +ELP S+ N NL
Sbjct: 837 SIGNAINLQNLLLDDCSSLLELPSSIGNATNL 868
>dbj|BAD38047.1| putative NBS-LRR resistance protein RGH2 [Oryza sativa (japonica
cultivar-group)]
Length = 1216
Score = 128 bits (321), Expect = 8e-28
Identities = 149/577 (25%), Positives = 255/577 (43%), Gaps = 82/577 (14%)
Query: 221 LLVLDDVWPSS----ESLVEKLQFQMTGFKILVTSR----------VAFPRFSTTCILKP 266
L++LDD+W E L+E LQ G KI+VT+R + F T +K
Sbjct: 276 LIILDDLWEEGWSKLEKLMEMLQSGKKGSKIIVTTRSEKVVNTLSTIRLSYFHTVDPIKL 335
Query: 267 LAHE-DAVTLFHHYAQMEK-NSSDIIDKNLVEKVVRSCNGLPLTIKVIATSL-KNRPHDL 323
+ D ME SD++D + +++ + C+G+PL K + + K+R +
Sbjct: 336 VGMSIDECWFIMKPRNMENCEFSDLVD--IGKEIAQRCSGVPLVAKALGYVMQKHRTREE 393
Query: 324 WHKIVKELSQGHSILDSNTELLTRLQKIF-DVLEDNPKIMECFMDLALFPEDHRIPVAAL 382
W +I + +ILD+ + L+ + P++ CFM ++FP H I L
Sbjct: 394 WMEI-----KNSNILDTKDDEEGILKGLLLSYYHMPPQLKLCFMYCSMFPMSHVIDHDCL 448
Query: 383 VDMWAELYKLDDNGIQ-----AMEIINKL-GIMNLANVIIPRKNASDT-DNNNYNNHFII 435
+ W L + D Q AME +N+L G+ L P AS H
Sbjct: 449 IQQWIALGFIQDTDGQPLQKVAMEYVNELLGMSFLTIFTSPTVLASRMLFKPTLKLH--- 505
Query: 436 LHDILRELGIYQSTKE-----PFEQRKRLIIDMNN------NKSGLAEKQQGLMTRIFSK 484
+HD++ EL + + E E R ++N N++ + + L T++ +
Sbjct: 506 MHDMVHELARHVAGNEFSHTNGAENRNTKRDNLNFHYHLLLNQNETSSAYKSLATKVRAL 565
Query: 485 FMRLCVKQN-PQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQY--------- 534
R C K + P+Q + L + LD Q +++ + + +Y
Sbjct: 566 HFRGCDKMHLPKQAFSHTLCLRV-----LDLGGRQVSELPSSVYKLKLLRYLDASSLRIS 620
Query: 535 SLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERIS-----VPSFGTL 589
S + + L+ LI++N N +G LQ L+ L + SFG L
Sbjct: 621 SFSKSFNHLLNLQALILSNTYLKTLPTN----IGCLQKLQYFDLSGCANLNELPTSFGDL 676
Query: 590 KNLKKLSLYMCNTILAFEKGSILISDAFPNLEEL---NIDYCKDLVVLQTGICDIISLKK 646
+L L+L C+ + A +F NL L ++ C L L C + L
Sbjct: 677 SSLLFLNLASCHELEALPM-------SFGNLNRLQFLSLSDCYKLNSLPESCCQLHDLAH 729
Query: 647 LNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSL 706
L++++C+ L LP I +L LE L+++SC+ ++A+P S+ KL L+HL++S C+ L +L
Sbjct: 730 LDLSDCYNLGKLPDCIDQLSKLEYLNMTSCSKVQALPESLCKLTMLRHLNLSYCLRLENL 789
Query: 707 PEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLKTI 742
P G+L L++LD+ + +LP S+ N+ LKT+
Sbjct: 790 PSCIGDL-QLQSLDIQGSFLLRDLPNSIFNMSTLKTV 825
Score = 47.4 bits (111), Expect = 0.002
Identities = 28/83 (33%), Positives = 42/83 (49%), Gaps = 1/83 (1%)
Query: 643 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN-LKHLDISNCI 701
S++ L + + L +LP+ I +L LS+ C +LE +P +G L+ + I C
Sbjct: 1110 SIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCP 1169
Query: 702 SLSSLPEEFGNLCNLKNLDMATC 724
LSSLPE L LK L + C
Sbjct: 1170 MLSSLPESIRRLTKLKKLRITNC 1192
Score = 39.7 bits (91), Expect = 0.38
Identities = 27/80 (33%), Positives = 41/80 (50%), Gaps = 2/80 (2%)
Query: 665 LENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN-LKNLDMAT 723
L ++E L+L S L A+P +I +L L I C L +LPE G+ L+ + + T
Sbjct: 1108 LSSIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDT 1167
Query: 724 CASI-ELPFSVVNLQNLKTI 742
C + LP S+ L LK +
Sbjct: 1168 CPMLSSLPESIRRLTKLKKL 1187
Score = 38.1 bits (87), Expect = 1.1
Identities = 31/96 (32%), Positives = 49/96 (50%), Gaps = 7/96 (7%)
Query: 561 LNNIELLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNL 620
L++IE L +L ++ +R ++ F +L +LS+ C + + + D F L
Sbjct: 1108 LSSIESL-TLMSIAGLRALPEAIQCF---TSLWRLSILGCGELETLPEW---LGDYFTCL 1160
Query: 621 EELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLS 656
EE++ID C L L I + LKKL +TNC LS
Sbjct: 1161 EEISIDTCPMLSSLPESIRRLTKLKKLRITNCPVLS 1196
Score = 35.8 bits (81), Expect = 5.5
Identities = 20/61 (32%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 617 FPNLEELNIDYCKDLVVLQTGICDIIS-LKKLNVTNCHKLSSLPQDIGKLENLELLSLSS 675
F +L L+I C +L L + D + L+++++ C LSSLP+ I +L L+ L +++
Sbjct: 1132 FTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCPMLSSLPESIRRLTKLKKLRITN 1191
Query: 676 C 676
C
Sbjct: 1192 C 1192
>gb|AAN23084.1| putative rp3 protein [Zea mays]
Length = 944
Score = 115 bits (289), Expect = 4e-24
Identities = 150/614 (24%), Positives = 260/614 (41%), Gaps = 84/614 (13%)
Query: 164 KLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLK-KVEGSPLLL 222
K E +++V S+ ++ +VE++ + ++ SD + + + K+ G L
Sbjct: 218 KHFEVVLWVHVSREFAVEKLVEKLFK----AIAGDMSDHPPLQHVSRTISDKLVGKRFLA 273
Query: 223 VLDDVWPSSESLVEKLQFQM------TGFKILVTSR---VAFPRFSTTCILKP-LAHEDA 272
VLDDVW +E VE QF + G IL+T+R VA S+ P L+ ED+
Sbjct: 274 VLDDVW--TEDRVEWEQFMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYNLPFLSKEDS 331
Query: 273 VTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELS 332
+F + + D +++V C G+PL IKVIA L +K +
Sbjct: 332 WKVFQQCFGIALKALDPEFLQTGKEIVEKCGGVPLAIKVIAGVLHG---------IKGIE 382
Query: 333 QGHSILDSNT--------ELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVD 384
+ SI DSN + L F L D+ K CF+ ++FP + I L+
Sbjct: 383 EWRSICDSNLLDVQDDEHRVFACLSLSFVHLPDHLK--PCFLHCSIFPRGYVINRRHLIS 440
Query: 385 MWA--------ELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIIL 436
W + + +D GI + + K+G + I + ++ ++
Sbjct: 441 QWIAHGFVPTNQARQAEDVGIGYFDSLLKVGFLQDHVQIWSTRGEVTCKMHD------LV 494
Query: 437 HDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQ 496
HD+ R++ R + ++ NK + L + +LC K
Sbjct: 495 HDLARQI-----------LRDEFVSEIETNKQIKRCRYLSLTSCTGKLDNKLCGKVRALY 543
Query: 497 LAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGF 556
+ R L D + + V +IL T SLP +++K L L I++
Sbjct: 544 VCGRELE--------FDKTMNKQCCVRTIILKYITDD-SLPLFVSKFEYLGYLEISDVNC 594
Query: 557 H--PSKLNNIELLGSLQNLERIRLERISV--PSFGTLKNLKKLSLYMCNTILAFEKGSIL 612
P L+ +LQ L + R++V S G LK L+ L L ++I + +
Sbjct: 595 EALPEALSRC---WNLQALHVLNCSRLAVVPESIGKLKKLRTLELNGVSSIKSLPQS--- 648
Query: 613 ISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLP--QDIGKLENLEL 670
I D NL L ++ C+ + + + + +L+ L++ +C L LP GKL NL+
Sbjct: 649 IGDC-DNLRRLYLEECRGIEDIPNSLGKLENLRILSIVDCVSLQKLPPSDSFGKLLNLQT 707
Query: 671 LSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIE-L 729
++ + C +L +P + L++L+ +D+ C L LPE GNL NLK L++ C + L
Sbjct: 708 ITFNLCYNLRNLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKVLNLKKCKKLRGL 767
Query: 730 PFSVVNLQNLKTIT 743
P L L+ ++
Sbjct: 768 PAGCGKLTRLQQLS 781
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.333 0.145 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,194,942,558
Number of Sequences: 2540612
Number of extensions: 47731700
Number of successful extensions: 246482
Number of sequences better than 10.0: 3948
Number of HSP's better than 10.0 without gapping: 1498
Number of HSP's successfully gapped in prelim test: 2490
Number of HSP's that attempted gapping in prelim test: 217822
Number of HSP's gapped (non-prelim): 16413
length of query: 776
length of database: 863,360,394
effective HSP length: 136
effective length of query: 640
effective length of database: 517,837,162
effective search space: 331415783680
effective search space used: 331415783680
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 79 (35.0 bits)
Medicago: description of AC148347.4


