

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.1 + phase: 0
(773 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
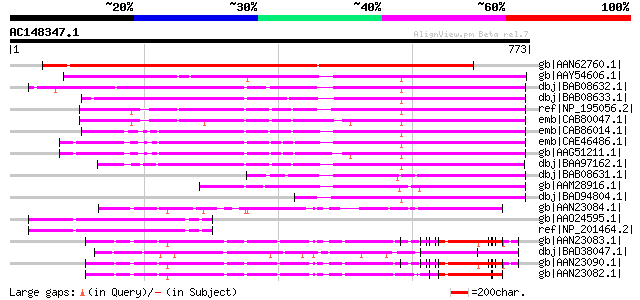
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN62760.1| disease resistance protein-like protein MsR1 [Med... 858 0.0
gb|AAY54606.1| NRG1 [Nicotiana benthamiana] 482 e-134
dbj|BAB08632.1| disease resistance protein-like [Arabidopsis tha... 453 e-126
dbj|BAB08633.1| disease resistance protein-like [Arabidopsis tha... 430 e-118
ref|NP_195056.2| disease resistance protein (CC-NBS-LRR class), ... 366 2e-99
emb|CAB80047.1| putative protein [Arabidopsis thaliana] gi|44902... 348 5e-94
emb|CAB86014.1| disease resistance-like protein [Arabidopsis tha... 337 7e-91
emb|CAE46486.1| CC-NBS-LRR disease resistance protein [Arabidops... 333 1e-89
gb|AAG51211.1| disease resistance protein, putative; 92850-95636... 328 5e-88
dbj|BAA97162.1| disease resistance protein-like [Arabidopsis tha... 320 1e-85
dbj|BAB08631.1| unnamed protein product [Arabidopsis thaliana] g... 276 1e-72
gb|AAM28916.1| NBS/LRR [Pinus taeda] 229 3e-58
dbj|BAD94804.1| putative protein [Arabidopsis thaliana] 228 4e-58
gb|AAN23084.1| putative rp3 protein [Zea mays] 132 3e-29
gb|AAO24595.1| At5g66630 [Arabidopsis thaliana] 131 1e-28
ref|NP_201464.2| LIM domain-containing protein [Arabidopsis thal... 131 1e-28
gb|AAN23083.1| putative rp3 protein [Zea mays] 129 5e-28
dbj|BAD38047.1| putative NBS-LRR resistance protein RGH2 [Oryza ... 129 5e-28
gb|AAN23090.1| putative rp3 protein [Zea mays] gi|23321151|gb|AA... 128 8e-28
gb|AAN23082.1| putative rp3 protein [Zea mays] 128 8e-28
>gb|AAN62760.1| disease resistance protein-like protein MsR1 [Medicago sativa]
Length = 704
Score = 858 bits (2218), Expect = 0.0
Identities = 439/643 (68%), Positives = 520/643 (80%), Gaps = 5/643 (0%)
Query: 49 KDDFDSCVGDDDDKQKMGKDVEGKLRELLKILDKENFGKKISGSILKGPFDVPANPEFTV 108
+++ +S + ++D ++ K +++ L E F +G K PFDVP NP+FTV
Sbjct: 65 REEINSLIEENDAEESACKCSSENDSNVVEKLKNETFE---AGPPFKCPFDVPENPKFTV 121
Query: 109 GLDLQLIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKT 168
GLD+ KLK+E+LR+G STL+LTGLGG+GKTTLATKLC D +V GKF ENIIFVTFSKT
Sbjct: 122 GLDIPFSKLKMELLRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFMENIIFVTFSKT 181
Query: 169 PMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLVEK 228
PMLK IVER+ EHCGYPVPE+Q+DEDAVN LGLLL+K+EGSP+LLVLDDVWP SE LVEK
Sbjct: 182 PMLKTIVERIHEHCGYPVPEFQNDEDAVNRLGLLLKKVEGSPLLLVLDDVWPSSESLVEK 241
Query: 229 FKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEK 288
+FQISD+KILVTSRVAF RF T ILKPLA ED+VTLF HY +EKNSS I DK+L+EK
Sbjct: 242 LQFQISDFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYALMEKNSSDIIDKNLVEK 301
Query: 289 VVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLED 348
VV C+GLPL IKVIATS + RP++LW KIVKELS+G SILDSNTELL RLQKI DVLED
Sbjct: 302 VVRSCQGLPLTIKVIATSLKNRPHDLWRKIVKELSQGHSILDSNTELLTRLQKIFDVLED 361
Query: 349 NAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIA 408
N ECFMDLALFPED RIPVAAL+DMWAELY LDD G +AM+IINKL MNLANV+I
Sbjct: 362 NPTIIECFMDLALFPEDHRIPVAALVDMWAELYRLDDTGIQAMEIINKLGIMNLANVIIP 421
Query: 409 RKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQ 468
RK+ASDT+N YNNHFI+LHD+LRELG Y++T+EP EQRKR +I+ +++K L EKQQ
Sbjct: 422 RKDASDTDNNNYNNHFIMLHDILRELGIYRSTKEPFEQRKRLIIDMHKNK--SGLTEKQQ 479
Query: 469 GTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYT 528
G M ILSK + K PQ++ AR LS+S DETCA DWSQ++PA EVLILNL TKQY+
Sbjct: 480 GLMIRILSKFMRLCVKQNPQQLAARILSVSTDETCALDWSQMEPAQVEVLILNLHTKQYS 539
Query: 529 FPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLKKL 588
PE + KM+KLK LI+ N+ + PSEL N ELLSSL NL++IRLERISVPSF T+KNLKKL
Sbjct: 540 LPEWIGKMSKLKVLIITNYTVHPSELTNFELLSSLQNLEKIRLERISVPSFATVKNLKKL 599
Query: 589 SLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKL 648
SLYMCNTRLAFEKGSILISD FPNLE+L++DYCKD+ LP G+CDIISLKKLS+TNCHK+
Sbjct: 600 SLYMCNTRLAFEKGSILISDAFPNLEELNIDYCKDLLVLPTGICDIISLKKLSVTNCHKI 659
Query: 649 SLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDIS 691
S LP++IGKLENLELLSL SCTDL +P SI +L NL+ LDIS
Sbjct: 660 SSLPEDIGKLENLELLSLSSCTDLEAIPTSIEKLLNLKHLDIS 702
>gb|AAY54606.1| NRG1 [Nicotiana benthamiana]
Length = 850
Score = 482 bits (1241), Expect = e-134
Identities = 289/703 (41%), Positives = 415/703 (58%), Gaps = 39/703 (5%)
Query: 80 LDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRSTLLLTGLGGMGK 139
++ +FG +GS G DVP + VG DL L +LKV++L E ++L+ G GK
Sbjct: 169 MNGNSFGST-NGSGFSGWSDVPQFSDSVVGFDLPLQELKVKLLEEKEKVVVLSAPAGCGK 227
Query: 140 TTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGL 199
TTLA LC +D +K K+++ I FVT SK +K IV +FE GY P++ S+ AV L
Sbjct: 228 TTLAAMLCQEDDIKDKYRD-IFFVTVSKKANIKRIVGEIFEMKGYKGPDFASEHAAVCQL 286
Query: 200 GLLLRKIEGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLA 259
LLR+ P+LLVLDDVW S+ ++E F FQI +KILVTSR F +FD T+ L L+
Sbjct: 287 NNLLRRSTSQPVLLVLDDVWSESDFVIESFIFQIPGFKILVTSRSVFPKFD-TYKLNLLS 345
Query: 260 QEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIV 319
++D+ LF Y+ K+S DL+ K V C G PLA+KV+ S +P +W V
Sbjct: 346 EKDAKALF--YSSAFKDSIPYVQLDLVHKAVRSCCGFPLALKVVGRSLCGQPELIWFNRV 403
Query: 320 KELSRGRSILDSNTELLIRLQKILDVLEDNAIS-------KECFMDLALFPEDQRIPVAA 372
S+ + + + +LL L+ +D L++ + ++C++DL FPED RI AA
Sbjct: 404 MLQSKRQILFPTENDLLRTLRASIDALDEIDLYSSEATTLRDCYLDLGSFPEDHRIHAAA 463
Query: 373 LIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASDTENYYYNNHFIVLHDLLR 432
++DMW E Y LD+DG +AM I +L S NL N+ +ARK+A + N H+I HDLLR
Sbjct: 464 ILDMWVERYNLDEDGMKAMAIFFQLSSQNLVNLALARKDAPAVLGLH-NLHYIQQHDLLR 522
Query: 433 ELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPA 492
EL +Q ++ +E+RKR IN + + W+ + + Q + A
Sbjct: 523 ELVIHQCDEKTVEERKRLYINIKGNDFPK-------------------WWSQQRLQPLQA 563
Query: 493 RTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHGLRPS 552
LSI DE S W V+ EVL+LN +TK Y FP +E+M++LK LIV N+ P+
Sbjct: 564 EVLSIFTDEHFESVWYDVRFPKVEVLVLNFETKTYNFPPFVEQMSQLKTLIVANNYFFPT 623
Query: 553 ELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLAFEKGSILISD 608
+LNN +L S L NLKRI LERISV S T L NL+K+S MC AFE + +S
Sbjct: 624 KLNNFQLCSLL-NLKRISLERISVTSIFTANLQLPNLRKISFIMCEIGEAFENYAANMSY 682
Query: 609 LFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLIS 668
++P L +++++YC D+ +P CD++ LKKLSI CH+L LP+E+GKL NLE+L L S
Sbjct: 683 MWPKLVEMNIEYCSDLVEVPAETCDLVGLKKLSICYCHELVALPEELGKLSNLEVLRLHS 742
Query: 669 CTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTS-CASCELPFSVV 727
CT++ +LP+S+ +L+ L LD+ +C+ L LP + LC+LR + M S ELP SV+
Sbjct: 743 CTNVSKLPESVVKLNRLGFLDVYDCVELDFLPREMDQLCSLRTICMGSRLGFTELPDSVL 802
Query: 728 NLQNLK-VTCDEETAASWESFQSMISNLTIEVPHVEVNLNWLH 769
L L+ V CDEETA+ WE ++ + NL I V ++NLN LH
Sbjct: 803 RLVKLEDVVCDEETASLWEYYKEHLRNLRITVIKEDINLNLLH 845
>dbj|BAB08632.1| disease resistance protein-like [Arabidopsis thaliana]
gi|15240126|ref|NP_201491.1| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395984|sp|Q9FKZ1|DRL42_ARATH Probable disease
resistance protein At5g66900
Length = 809
Score = 453 bits (1166), Expect = e-126
Identities = 276/752 (36%), Positives = 408/752 (53%), Gaps = 46/752 (6%)
Query: 29 IKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDDDKQKMGK----DVEGKLRELLKILDKEN 84
+KW+ K + ++ NKD C D Q + + G L + L K
Sbjct: 89 VKWYEKSK---YTRKIERINKDMLKFCQIDLQLLQHRNQLTLLGLTGNLVNSVDGLSKRM 145
Query: 85 FGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRSTLLLTGLGGMGKTTLAT 144
+ + + VP + VGLD L +LK +L + TL+++ G GKTTL +
Sbjct: 146 DLLSVPAPVFRDLCSVPKLDKVIVGLDWPLGELKKRLLDDSVVTLVVSAPPGCGKTTLVS 205
Query: 145 KLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLR 204
+LC D +KGKFK +I F S TP ++IV+ L +H GY +++D A GL LL
Sbjct: 206 RLCDDPDIKGKFK-HIFFNVVSNTPNFRVIVQNLLQHNGYNALTFENDSQAEVGLRKLLE 264
Query: 205 KI-EGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDS 263
++ E PILLVLDDVW G++ ++KF+ ++ +YKILVTSR F FD + LKPL +D+
Sbjct: 265 ELKENGPILLVLDDVWRGADSFLQKFQIKLPNYKILVTSRFDFPSFDSNYRLKPLEDDDA 324
Query: 264 VTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELS 323
L H+ N+S +DL++K+++ C G P+ I+V+ S + R W+ V+ S
Sbjct: 325 RALLIHWASRPCNTSPDEYEDLLQKILKRCNGFPIVIEVVGVSLKGRSLNTWKGQVESWS 384
Query: 324 RGRSILDSNTELLIR-LQKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYG 382
G IL ++ LQ D L+ N KECF+D+ F EDQ+I + +IDMW ELYG
Sbjct: 385 EGEKILGKPYPTVLECLQPSFDALDPNL--KECFLDMGSFLEDQKIRASVIIDMWVELYG 442
Query: 383 LDDDGKEAMDIINKLDSMNLANVL-IARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQ 441
K + + L+ + N+L + ++ E+ +YN+ + HD+LREL Q+
Sbjct: 443 -----KGSSILYMYLEDLASQNLLKLVPLGTNEHEDGFYNDFLVTQHDILRELAICQSEF 497
Query: 442 EPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDE 501
+ +RKR + E+ F + A LSIS D+
Sbjct: 498 KENLERKRLNLEILENT-----------------------FPDWCLNTINASLLSISTDD 534
Query: 502 TCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHGLRPSELNNLELLS 561
+S W ++ E L+LNL + Y P + M KLK L + NHG P+ L+N LS
Sbjct: 535 LFSSKWLEMDCPNVEALVLNLSSSDYALPSFISGMKKLKVLTITNHGFYPARLSNFSCLS 594
Query: 562 SLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNT-RLAFEKGSILISDLFPNLEDL 616
SL NLKRIRLE++S+ L +LKKLSL MC+ + ++ I++S+ L+++
Sbjct: 595 SLPNLKRIRLEKVSITLLDIPQLQLSSLKKLSLVMCSFGEVFYDTEDIVVSNALSKLQEI 654
Query: 617 SMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELP 676
+DYC D+ LP + +I+SLK LSITNC+KLS LP+ IG L LE+L L S +L ELP
Sbjct: 655 DIDYCYDLDELPYWISEIVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRLCSSMNLSELP 714
Query: 677 DSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLKVTC 736
++ LSNLR LDIS+C+ L LP++ G L NL+ + M C+ CELP SV NL+NL+V C
Sbjct: 715 EATEGLSNLRFLDISHCLGLRKLPQEIGKLQNLKKISMRKCSGCELPESVTNLENLEVKC 774
Query: 737 DEETAASWESFQSMISNLTIEVPHVEVNLNWL 768
DEET WE + + NL ++ +E NLN L
Sbjct: 775 DEETGLLWERLKPKMRNLRVQEEEIEHNLNLL 806
>dbj|BAB08633.1| disease resistance protein-like [Arabidopsis thaliana]
gi|18087526|gb|AAL58897.1| AT5g66910/MUD21_17
[Arabidopsis thaliana] gi|15240127|ref|NP_201492.1|
disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana] gi|46395983|sp|Q9FKZ0|DRL43_ARATH
Probable disease resistance protein At5g66910
Length = 815
Score = 430 bits (1105), Expect = e-118
Identities = 258/669 (38%), Positives = 378/669 (55%), Gaps = 37/669 (5%)
Query: 108 VGLDLQLIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSK 167
VGLD L++LK ++L S ++++G G GKTTL TKLC D +++G+FK+ I + S
Sbjct: 173 VGLDWPLVELKKKLL--DNSVVVVSGPPGCGKTTLVTKLCDDPEIEGEFKK-IFYSVVSN 229
Query: 168 TPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKI-EGSPILLVLDDVWPGSEDLV 226
TP + IV+ L + G + D A GL LL ++ + ILLVLDDVW GSE L+
Sbjct: 230 TPNFRAIVQNLLQDNGCGAITFDDDSQAETGLRDLLEELTKDGRILLVLDDVWQGSEFLL 289
Query: 227 EKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLI 286
KF+ + DYKILVTS+ F+ T+ L PL E + +L + ++S +DL+
Sbjct: 290 RKFQIDLPDYKILVTSQFDFTSLWPTYHLVPLKYEYARSLLIQWASPPLHTSPDEYEDLL 349
Query: 287 EKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSIL-DSNTELLIRLQKILDV 345
+K+++ C G PL I+V+ S + + LW+ V+ S G +IL ++N + RLQ +V
Sbjct: 350 QKILKRCNGFPLVIEVVGISLKGQALYLWKGQVESWSEGETILGNANPTVRQRLQPSFNV 409
Query: 346 LEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDG-KEAMDIINKLDSMNLAN 404
L+ + KECFMD+ F +DQ+I + +ID+W ELYG + M +N+L S NL
Sbjct: 410 LKPHL--KECFMDMGSFLQDQKIRASLIIDIWMELYGRGSSSTNKFMLYLNELASQNLLK 467
Query: 405 VLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLR 464
++ N E+ +YN + H++LREL +Q+ EPI QRK+ + E
Sbjct: 468 LVHLGTNKR--EDGFYNELLVTQHNILRELAIFQSELEPIMQRKKLNLEIREDNFP---- 521
Query: 465 EKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQT 524
D+ Q + AR LSI D+ +S W ++ E L+LN+ +
Sbjct: 522 ------------------DECLNQPINARLLSIYTDDLFSSKWLEMDCPNVEALVLNISS 563
Query: 525 KQYTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGT--- 581
Y P + +M KLK L + NHG P+ L+N LSSL NLKRIR E++SV
Sbjct: 564 LDYALPSFIAEMKKLKVLTIANHGFYPARLSNFSCLSSLPNLKRIRFEKVSVTLLDIPQL 623
Query: 582 -LKNLKKLSLYMCNT-RLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKK 639
L +LKKLS +MC+ + ++ I +S NL+++ +DYC D+ LP + +++SLK
Sbjct: 624 QLGSLKKLSFFMCSFGEVFYDTEDIDVSKALSNLQEIDIDYCYDLDELPYWIPEVVSLKT 683
Query: 640 LSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSL 699
LSITNC+KLS LP+ IG L LE+L + SC +L ELP++ RLSNLR LDIS+C+ L L
Sbjct: 684 LSITNCNKLSQLPEAIGNLSRLEVLRMCSCMNLSELPEATERLSNLRSLDISHCLGLRKL 743
Query: 700 PEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQSMISNLTIEVP 759
P++ G L L N+ M C+ CELP SV L+NL+V CDE T WE + NL +
Sbjct: 744 PQEIGKLQKLENISMRKCSGCELPDSVRYLENLEVKCDEVTGLLWERLMPEMRNLRVHTE 803
Query: 760 HVEVNLNWL 768
E NL L
Sbjct: 804 ETEHNLKLL 812
>ref|NP_195056.2| disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana]
Length = 816
Score = 366 bits (939), Expect = 2e-99
Identities = 224/676 (33%), Positives = 371/676 (54%), Gaps = 52/676 (7%)
Query: 105 EFTVGLDLQLIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVT 164
+F VGL+L +K+K + ++G+GG+GKTTLA +L D +V+ F+ I+F+T
Sbjct: 179 KFGVGLELGKVKVKKMMFESQGGVFGISGMGGVGKTTLAKELQRDHEVQCHFENRILFLT 238
Query: 165 FSKTPMLKIIVERLFE-----HCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVW 219
S++P+L+ + E ++ G PVP+ D G+ L++LDDVW
Sbjct: 239 VSQSPLLEELRELIWGFLSGCEAGNPVPDCNFPFD-------------GARKLVILDDVW 285
Query: 220 PGSE-DLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSS 278
D + FKF ++ S++ +F T+ ++ L+++++++LF +K+
Sbjct: 286 TTQALDRLTSFKFPGCTTLVVSRSKLTEPKF--TYDVEVLSEDEAISLFCLCAFGQKSIP 343
Query: 279 KIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIR 338
KDL+++V CKGLPLA+KV S +P W+ +++ LS+G DS+ L+R
Sbjct: 344 LGFCKDLVKQVANECKGLPLALKVTGASLNGKPEMYWKGVLQRLSKGEPADDSHESRLLR 403
Query: 339 -LQKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKL 397
++ LD L+ +K+CF+DL FPED++IP+ LI++W EL+ +D+ A I+ L
Sbjct: 404 QMEASLDNLDQT--TKDCFLDLGAFPEDRKIPLDVLINIWIELHDIDEGN--AFAILVDL 459
Query: 398 DSMNLANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNES 457
NL + + S ++Y + F+ HD+LR+L + + + +RKR L+ E
Sbjct: 460 SHKNLLTLGKDPRLGSLYASHY--DIFVTQHDVLRDLALHLSNAGKVNRRKRLLMPKREL 517
Query: 458 KCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEV 517
L G +++ + A+ +SI E W ++ AE+
Sbjct: 518 D-------------------LPGDWERNNDEHYIAQIVSIHTGEMNEMQWFDMEFPKAEI 558
Query: 518 LILNLQTKQYTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVP 577
LILN + +Y P + KM++LK L++IN+G+ P+ L++ + + LS L+ + LER+ VP
Sbjct: 559 LILNFSSDKYVLPPFISKMSRLKVLVIINNGMSPAVLHDFSIFAHLSKLRSLWLERVHVP 618
Query: 578 SFGT----LKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCD 633
LKNL K+SL +C +F++ + ++D+FP L DL++D+C D+ ALP+ +C
Sbjct: 619 QLSNSTTPLKNLHKMSLILCKINKSFDQTGLDVADIFPKLGDLTIDHCDDLVALPSSICG 678
Query: 634 IISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+ SL LSITNC +L LP+ + KL+ LE+L L +C +L LP I L L+ LDIS C
Sbjct: 679 LTSLSCLSITNCPRLGELPKNLSKLQALEILRLYACPELKTLPGEICELPGLKYLDISQC 738
Query: 694 ISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLK-VTCDEETAASWESFQSMIS 752
+SLS LPE+ G L L +DM C + P S V+L++L+ V CD + A WE + +
Sbjct: 739 VSLSCLPEEIGKLKKLEKIDMRECCFSDRPSSAVSLKSLRHVICDTDVAFMWEEVEKAVP 798
Query: 753 NLTIEVPHVEVNLNWL 768
L IE +L+WL
Sbjct: 799 GLKIEAAEKCFSLDWL 814
>emb|CAB80047.1| putative protein [Arabidopsis thaliana] gi|4490297|emb|CAB38788.1|
putative protein [Arabidopsis thaliana]
gi|46396028|sp|Q9SZA7|DRL29_ARATH Probable disease
resistance protein At4g33300
Length = 855
Score = 348 bits (892), Expect = 5e-94
Identities = 227/705 (32%), Positives = 374/705 (52%), Gaps = 71/705 (10%)
Query: 105 EFTVGLDLQLIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVT 164
+F VGL+L +K+K + ++G+GG+GKTTLA +L D +V+ F+ I+F+T
Sbjct: 179 KFGVGLELGKVKVKKMMFESQGGVFGISGMGGVGKTTLAKELQRDHEVQCHFENRILFLT 238
Query: 165 FSKTPMLKIIVERLFE-----HCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVW 219
S++P+L+ + E ++ G PVP+ D G+ L++LDDVW
Sbjct: 239 VSQSPLLEELRELIWGFLSGCEAGNPVPDCNFPFD-------------GARKLVILDDVW 285
Query: 220 PGSE-DLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSS 278
D + FKF ++ S++ +F T+ ++ L+++++++LF +K+
Sbjct: 286 TTQALDRLTSFKFPGCTTLVVSRSKLTEPKF--TYDVEVLSEDEAISLFCLCAFGQKSIP 343
Query: 279 KIPDKDLIEK----------------VVEHCKGLPLAIKVIATSFRYRPYELWEKIVKEL 322
KDL+++ V CKGLPLA+KV S +P W+ +++ L
Sbjct: 344 LGFCKDLVKQHNIQSFSILRVLCLAQVANECKGLPLALKVTGASLNGKPEMYWKGVLQRL 403
Query: 323 SRGRSILDSNTELLIR-LQKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELY 381
S+G DS+ L+R ++ LD L+ +K+CF+DL FPED++IP+ LI++W EL+
Sbjct: 404 SKGEPADDSHESRLLRQMEASLDNLDQT--TKDCFLDLGAFPEDRKIPLDVLINIWIELH 461
Query: 382 GLDDDGKEAMDIINKLDSMNLANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQ 441
+D+ A I+ L NL + + S ++Y + F+ HD+LR+L + +
Sbjct: 462 DIDEGN--AFAILVDLSHKNLLTLGKDPRLGSLYASHY--DIFVTQHDVLRDLALHLSNA 517
Query: 442 EPIEQRKRQLINTNESKCDQRL-REKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISID 500
+ +RKR L+ E R + +A I+S G TL I +
Sbjct: 518 GKVNRRKRLLMPKRELDLPGDWERNNDEHYIAQIVSIHTGKCFS---------TLQIFLV 568
Query: 501 ETCAS------------DWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHG 548
+C + W ++ AE+LILN + +Y P + KM++LK L++IN+G
Sbjct: 569 LSCNNHQDHNVGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLVIINNG 628
Query: 549 LRPSELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLAFEKGSI 604
+ P+ L++ + + LS L+ + LER+ VP LKNL K+SL +C +F++ +
Sbjct: 629 MSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQTGL 688
Query: 605 LISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELL 664
++D+FP L DL++D+C D+ ALP+ +C + SL LSITNC +L LP+ + KL+ LE+L
Sbjct: 689 DVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQALEIL 748
Query: 665 SLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPF 724
L +C +L LP I L L+ LDIS C+SLS LPE+ G L L +DM C + P
Sbjct: 749 RLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCFSDRPS 808
Query: 725 SVVNLQNLK-VTCDEETAASWESFQSMISNLTIEVPHVEVNLNWL 768
S V+L++L+ V CD + A WE + + L IE +L+WL
Sbjct: 809 SAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 853
>emb|CAB86014.1| disease resistance-like protein [Arabidopsis thaliana]
gi|21281177|gb|AAM45000.1| putative disease resistance
[Arabidopsis thaliana] gi|15292721|gb|AAK92729.1|
putative disease resistance [Arabidopsis thaliana]
gi|9758447|dbj|BAB08976.1| unnamed protein product
[Arabidopsis thaliana] gi|15238281|ref|NP_196092.1|
disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana] gi|46396009|sp|Q9LZ25|DRL30_ARATH
Probable disease resistance protein At5g04720
Length = 811
Score = 337 bits (865), Expect = 7e-91
Identities = 215/670 (32%), Positives = 361/670 (53%), Gaps = 49/670 (7%)
Query: 108 VGLDLQLIKLKVEILR--EGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTF 165
VGLDL K+K + + +G + ++G+ G GKTTLA +L D++V+G F ++F+T
Sbjct: 180 VGLDLGKRKVKEMLFKSIDGERLIGISGMSGSGKTTLAKELARDEEVRGHFGNKVLFLTV 239
Query: 166 SKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDL 225
S++P L E L H + Y++ G+G L + S L++LDDVW + +
Sbjct: 240 SQSPNL----EELRAHIWGFLTSYEA------GVGATLPE---SRKLVILDDVW--TRES 284
Query: 226 VEKFKFQ-ISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKD 284
+++ F+ I LV SR + T+ ++ L + ++ LF +K +
Sbjct: 285 LDQLMFENIPGTTTLVVSRSKLADSRVTYDVELLNEHEATALFCLSVFNQKLVPSGFSQS 344
Query: 285 LIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSN-TELLIRLQKIL 343
L+++VV CKGLPL++KVI S + RP + WE V+ LSRG +++ + + +++ L
Sbjct: 345 LVKQVVGECKGLPLSLKVIGASLKERPEKYWEGAVERLSRGEPADETHESRVFAQIEATL 404
Query: 344 DVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLA 403
+ L+ +++CF+ L FPED++IP+ LI++ EL+ L+D A +I L + NL
Sbjct: 405 ENLDPK--TRDCFLVLGAFPEDKKIPLDVLINVLVELHDLED--ATAFAVIVDLANRNLL 460
Query: 404 NVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRL 463
++ + +YY + F+ HD+LR++ + + R+R L+ ES +
Sbjct: 461 TLVKDPRFGHMYTSYY--DIFVTQHDVLRDVALRLSNHGKVNNRERLLMPKRESMLPRE- 517
Query: 464 REKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQ 523
+++ + AR +SI E DW ++ AEVLIL+
Sbjct: 518 ------------------WERNNDEPYKARVVSIHTGEMTQMDWFDMELPKAEVLILHFS 559
Query: 524 TKQYTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGT-- 581
+ +Y P + KM KL AL++IN+G+ P+ L++ + ++L+ LK + L+R+ VP +
Sbjct: 560 SDKYVLPPFIAKMGKLTALVIINNGMSPARLHDFSIFTNLAKLKSLWLQRVHVPELSSST 619
Query: 582 --LKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKK 639
L+NL KLSL C + ++ + I+ +FP L DL++D+C D+ LP+ +C I SL
Sbjct: 620 VPLQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTIDHCDDLLELPSTICGITSLNS 679
Query: 640 LSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSL 699
+SITNC ++ LP+ + KL+ L+LL L +C +L LP I L L+ +DIS C+SLSSL
Sbjct: 680 ISITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVEICELPRLKYVDISQCVSLSSL 739
Query: 700 PEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLK-VTCDEETAASWESFQSMISNLTIEV 758
PE G + L +D C+ +P SVV L +L+ V CD E WE Q ++ L +E
Sbjct: 740 PEKIGKVKTLEKIDTRECSLSSIPNSVVLLTSLRHVICDREALWMWEKVQKAVAGLRVEA 799
Query: 759 PHVEVNLNWL 768
+ +WL
Sbjct: 800 AEKSFSRDWL 809
>emb|CAE46486.1| CC-NBS-LRR disease resistance protein [Arabidopsis thaliana]
gi|30692890|ref|NP_174620.2| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395988|sp|Q9FW44|ADR1_ARATH Disease resistance
protein ADR1 (Activated disease resistance protein 1)
Length = 787
Score = 333 bits (854), Expect = 1e-89
Identities = 212/702 (30%), Positives = 378/702 (53%), Gaps = 58/702 (8%)
Query: 74 RELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRSTLL-LT 132
R + ++L + N ++++ + ++PE L+L K+K + + + L ++
Sbjct: 135 RNMDRLLTERNDSLSFPETMME--IETVSDPEIQTVLELGKKKVKEMMFKFTDTHLFGIS 192
Query: 133 GLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSD 192
G+ G GKTTLA +L DD V+G FK ++F+T S++P FE+ + E+ D
Sbjct: 193 GMSGSGKTTLAIELSKDDDVRGLFKNKVLFLTVSRSPN--------FENLESCIREFLYD 244
Query: 193 EDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKT 252
G+ RK L++LDDVW + + +++ +I LV SR + T
Sbjct: 245 -------GVHQRK------LVILDDVW--TRESLDRLMSKIRGSTTLVVSRSKLADPRTT 289
Query: 253 FILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPY 312
+ ++ L ++++++L +K+ +K L+++VV+ CKGLPL++KV+ S + +P
Sbjct: 290 YNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPE 349
Query: 313 ELWEKIVKELSRGRSILDSN-TELLIRLQKILDVLEDNAISKECFMDLALFPEDQRIPVA 371
WE +VK L RG + +++ + + +++ L+ L+ ++CF+D+ FPED++IP+
Sbjct: 350 RYWEGVVKRLLRGEAADETHESRVFAHMEESLENLDPKI--RDCFLDMGAFPEDKKIPLD 407
Query: 372 ALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASDTENYYYNNHFIVLHDLL 431
L +W E + +D++ A + +L NL + + D YY+ F+ HD+L
Sbjct: 408 LLTSVWVERHDIDEE--TAFSFVLRLADKNLLTI-VNNPRFGDVHIGYYDV-FVTQHDVL 463
Query: 432 RELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVP 491
R+L + + + + +R+R L+ E + ++K K +
Sbjct: 464 RDLALHMSNRVDVNRRERLLMPKTEPVLPRE-------------------WEKNKDEPFD 504
Query: 492 ARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHGLRP 551
A+ +S+ E +W + AEVLILN + Y P + KM++L+ L++IN+G+ P
Sbjct: 505 AKIVSLHTGEMDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINNGMSP 564
Query: 552 SELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLAFEKGSILIS 607
+ L+ + ++L+ L+ + L+R+ VP + LKNL K+ L C + +F + S IS
Sbjct: 565 ARLHGFSIFANLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDIS 624
Query: 608 DLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLI 667
+FP+L DL++D+C D+ L + + I SL LSITNC ++ LP+ + +++LE L L
Sbjct: 625 KIFPSLSDLTIDHCDDLLELKS-IFGITSLNSLSITNCPRILELPKNLSNVQSLERLRLY 683
Query: 668 SCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVV 727
+C +L+ LP + L L+ +DIS C+SL SLPE FG L +L +DM C+ LP SV
Sbjct: 684 ACPELISLPVEVCELPCLKYVDISQCVSLVSLPEKFGKLGSLEKIDMRECSLLGLPSSVA 743
Query: 728 NLQNLK-VTCDEETAASWESFQSMISNLTIEVPHVEVNLNWL 768
L +L+ V CDEET++ WE + ++ L IEV ++WL
Sbjct: 744 ALVSLRHVICDEETSSMWEMVKKVVPELCIEVAKKCFTVDWL 785
>gb|AAG51211.1| disease resistance protein, putative; 92850-95636 [Arabidopsis
thaliana] gi|10998939|gb|AAG26078.1| disease resistance
protein, putative [Arabidopsis thaliana]
Length = 797
Score = 328 bits (840), Expect = 5e-88
Identities = 215/706 (30%), Positives = 381/706 (53%), Gaps = 56/706 (7%)
Query: 74 RELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRSTLL-LT 132
R + ++L + N ++++ + ++PE L+L K+K + + + L ++
Sbjct: 135 RNMDRLLTERNDSLSFPETMME--IETVSDPEIQTVLELGKKKVKEMMFKFTDTHLFGIS 192
Query: 133 GLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSD 192
G+ G GKTTLA +L DD V+G FK ++F+T S++P FE+ + E+ D
Sbjct: 193 GMSGSGKTTLAIELSKDDDVRGLFKNKVLFLTVSRSPN--------FENLESCIREFLYD 244
Query: 193 EDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKT 252
G+ RK L++LDDVW + + +++ +I LV SR + T
Sbjct: 245 -------GVHQRK------LVILDDVW--TRESLDRLMSKIRGSTTLVVSRSKLADPRTT 289
Query: 253 FILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPY 312
+ ++ L ++++++L +K+ +K L+++VV+ CKGLPL++KV+ S + +P
Sbjct: 290 YNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPE 349
Query: 313 ELWEKIVKELSRGRSILDSN-TELLIRLQKILDVLEDNAISKECFMDLALFPEDQRIPVA 371
WE +VK L RG + +++ + + +++ L+ L+ ++CF+D+ FPED++IP+
Sbjct: 350 RYWEGVVKRLLRGEAADETHESRVFAHMEESLENLDPKI--RDCFLDMGAFPEDKKIPLD 407
Query: 372 ALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASDTENYYYNNHFIVLHDLL 431
L +W E + +D++ A + +L NL + + D YY+ F+ HD+L
Sbjct: 408 LLTSVWVERHDIDEE--TAFSFVLRLADKNLLTI-VNNPRFGDVHIGYYDV-FVTQHDVL 463
Query: 432 RELGNYQNTQEPIEQRKRQLINTNESKCDQRL-REKQQGTMAHILSKLIGWFDKPKPQKV 490
R+L + + + + +R+R L+ E + + K + A I+S G
Sbjct: 464 RDLALHMSNRVDVNRRERLLMPKTEPVLPREWEKNKDEPFDAKIVSLHTG---------- 513
Query: 491 PARTLSISIDETCASD---WSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINH 547
S++++E D W + AEVLILN + Y P + KM++L+ L++IN+
Sbjct: 514 ---KTSLTLNEFGEMDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINN 570
Query: 548 GLRPSELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLAFEKGS 603
G+ P+ L+ + ++L+ L+ + L+R+ VP + LKNL K+ L C + +F + S
Sbjct: 571 GMSPARLHGFSIFANLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTS 630
Query: 604 ILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLEL 663
IS +FP+L DL++D+C D+ L + + I SL LSITNC ++ LP+ + +++LE
Sbjct: 631 FDISKIFPSLSDLTIDHCDDLLELKS-IFGITSLNSLSITNCPRILELPKNLSNVQSLER 689
Query: 664 LSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELP 723
L L +C +L+ LP + L L+ +DIS C+SL SLPE FG L +L +DM C+ LP
Sbjct: 690 LRLYACPELISLPVEVCELPCLKYVDISQCVSLVSLPEKFGKLGSLEKIDMRECSLLGLP 749
Query: 724 FSVVNLQNLK-VTCDEETAASWESFQSMISNLTIEVPHVEVNLNWL 768
SV L +L+ V CDEET++ WE + ++ L IEV ++WL
Sbjct: 750 SSVAALVSLRHVICDEETSSMWEMVKKVVPELCIEVAKKCFTVDWL 795
>dbj|BAA97162.1| disease resistance protein-like [Arabidopsis thaliana]
gi|15238054|ref|NP_199539.1| disease resistance protein
(NBS-LRR class), putative [Arabidopsis thaliana]
gi|46396005|sp|Q9LVT1|DRL39_ARATH Putative disease
resistance protein At5g47280
Length = 623
Score = 320 bits (820), Expect = 1e-85
Identities = 203/644 (31%), Positives = 341/644 (52%), Gaps = 44/644 (6%)
Query: 131 LTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQ 190
++G+ G GKT LA +L D++V+G F ++F+T S++P L E + ++
Sbjct: 14 ISGMIGSGKTILAKELARDEEVRGHFANRVLFLTVSQSPNL--------EELRSLIRDFL 65
Query: 191 SDEDAVNGLGLLLRKIEG-SPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRF 249
+ +A G G L + G + L++LDDV + + +++ F I LV S+
Sbjct: 66 TGHEA--GFGTALPESVGHTRKLVILDDV--RTRESLDQLMFNIPGTTTLVVSQSKLVDP 121
Query: 250 DKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRY 309
T+ ++ L + D+ +LF +K+ K L+++VV KGLPL++KV+ S
Sbjct: 122 RTTYDVELLNEHDATSLFCLSAFNQKSVPSGFSKSLVKQVVGESKGLPLSLKVLGASLND 181
Query: 310 RPYELWEKIVKELSRGRSILDSN-TELLIRLQKILDVLEDNAISKECFMDLALFPEDQRI 368
RP W V+ LSRG + +++ +++ +++ L+ L+ +KECF+D+ FPE ++I
Sbjct: 182 RPETYWAIAVERLSRGEPVDETHESKVFAQIEATLENLDPK--TKECFLDMGAFPEGKKI 239
Query: 369 PVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASDTENYYYNNHFIVLH 428
PV LI+M +++ L+D A D++ L + NL ++ + +YY + F+ H
Sbjct: 240 PVDVLINMLVKIHDLEDAA--AFDVLVDLANRNLLTLVKDPTFVAMGTSYY--DIFVTQH 295
Query: 429 DLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQ 488
D+LR++ + + + +R R L+ E+ +++ +
Sbjct: 296 DVLRDVALHLTNRGKVSRRDRLLMPKRETMLPSE-------------------WERSNDE 336
Query: 489 KVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHG 548
AR +SI E DW + AEVLI+N + Y P + KM L+ ++IN+G
Sbjct: 337 PYNARVVSIHTGEMTEMDWFDMDFPKAEVLIVNFSSDNYVLPPFIAKMGMLRVFVIINNG 396
Query: 549 LRPSELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLAFEKGSI 604
P+ L++ + +SL+NL+ + LER+ VP + LKNL KL L +C +F++ +I
Sbjct: 397 TSPAHLHDFPIPTSLTNLRSLWLERVHVPELSSSMIPLKNLHKLYLIICKINNSFDQTAI 456
Query: 605 LISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELL 664
I+ +FP L D+++DYC D+ LP+ +C I SL +SITNC + LP+ I KL+ L+LL
Sbjct: 457 DIAQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISKLQALQLL 516
Query: 665 SLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPF 724
L +C +L LP I L L +DIS+C+SLSSLPE GN+ L +DM C+ +P
Sbjct: 517 RLYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMRECSLSSIPS 576
Query: 725 SVVNLQNL-KVTCDEETAASWESFQSMISNLTIEVPHVEVNLNW 767
S V+L +L VTC E W+ + + L IE N+ W
Sbjct: 577 SAVSLTSLCYVTCYREALWMWKEVEKAVPGLRIEATEKWFNMTW 620
>dbj|BAB08631.1| unnamed protein product [Arabidopsis thaliana]
gi|15240125|ref|NP_201490.1| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395985|sp|Q9FKZ2|DRL41_ARATH Probable disease
resistance protein At5g66890
Length = 415
Score = 276 bits (707), Expect = 1e-72
Identities = 166/424 (39%), Positives = 240/424 (56%), Gaps = 36/424 (8%)
Query: 353 KECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNA 412
+ECF+D+A F EDQRI + +ID+W+ YG KE M+ + L S NL +L +N
Sbjct: 17 RECFLDMASFLEDQRIIASTIIDLWSASYG-----KEGMNNLQDLASRNLLKLLPIGRN- 70
Query: 413 SDTENYYYNNHFIVLHDLLRELGNYQNTQEP--IEQRKRQLINTNESKCDQRLREKQQGT 470
+ E+ +YN + ++LRE Q +E I +RKR + ++K
Sbjct: 71 -EYEDGFYNELLVKQDNVLREFAINQCLKESSSIFERKRLNLEIQDNKFPN--------- 120
Query: 471 MAHILSKLIGWFDKPK-PQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTF 529
W PK P + A SIS D++ AS W ++ E L+LN+ + Y
Sbjct: 121 ----------WCLNPKQPIVINASLFSISTDDSFASSWFEMDCPNVEALVLNISSSNYAL 170
Query: 530 PELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISV-----PSFGTLKN 584
P + M +LK +I+INHGL P++L NL LSSL NLKRIR E++S+ P G LK+
Sbjct: 171 PNFIATMKELKVVIIINHGLEPAKLTNLSCLSSLPNLKRIRFEKVSISLLDIPKLG-LKS 229
Query: 585 LKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITN 644
L+KLSL+ C+ A + +S+ +L+++ +DYC ++ LP + ++SLKKLS+TN
Sbjct: 230 LEKLSLWFCHVVDALNELED-VSETLQSLQEIEIDYCYNLDELPYWISQVVSLKKLSVTN 288
Query: 645 CHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFG 704
C+KL + + IG L +LE L L SC L+ELP++I RL NLR LD+S L +LP + G
Sbjct: 289 CNKLCRVIEAIGDLRDLETLRLSSCASLLELPETIDRLDNLRFLDVSGGFQLKNLPLEIG 348
Query: 705 NLCNLRNLDMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQSMISNLTIEVPHVEVN 764
L L + M C CELP SV NL+NL+V CDE+TA W+ + + NLTI E N
Sbjct: 349 KLKKLEKISMKDCYRCELPDSVKNLENLEVKCDEDTAFLWKILKPEMKNLTITEEKTEHN 408
Query: 765 LNWL 768
LN L
Sbjct: 409 LNLL 412
>gb|AAM28916.1| NBS/LRR [Pinus taeda]
Length = 479
Score = 229 bits (584), Expect = 3e-58
Identities = 154/499 (30%), Positives = 253/499 (49%), Gaps = 39/499 (7%)
Query: 283 KDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTE-LLIRLQK 341
+DL+++V CKGLPLA+KVI +S R + +W ++LS+ SI + E LL RL+
Sbjct: 5 EDLVKQVAAECKGLPLALKVIGSSLRGKRRPIWINAERKLSKSESISEYYKESLLKRLET 64
Query: 342 ILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMN 401
+DVL+D K+CF+DL FP+ ++ V L+D+W +Y + +A +++ +L S N
Sbjct: 65 SIDVLDDK--HKQCFLDLGAFPKGRKFSVETLLDIW--VYVRQMEWTDAFEVLLELASRN 120
Query: 402 LANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQ 461
L N L + + + HD++R+L Y +Q+ I KR + E K
Sbjct: 121 LLN-LTGYPGSGAIDYSCASELTFSQHDVMRDLALYLASQDNIISPKRLFTPSKEDKIPT 179
Query: 462 RLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILN 521
W K Q A+ +SI DW Q+ E L L
Sbjct: 180 E------------------WLSTLKDQASRAQFVSIYTGAMQEQDWCQIDFPEVEALALF 221
Query: 522 LQTKQYTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSF-- 579
QY P + + KLK LIV N+ + + L SS ++ + L ++ VP+
Sbjct: 222 FSANQYCLPTFLHRTPKLKVLIVYNYSSMRANIIGLPRFSSPIQIRSVFLHKLIVPASLY 281
Query: 580 ---GTLKNLKKLSLYMCNTRLAFEKGSILISDL----FPNLEDLSMDYCKDMTALPNGVC 632
+ + L+KLS+ +C S++ +L FPN+ ++++D+C D+ LP +C
Sbjct: 282 ENCRSWERLEKLSVCLCE---GLGNSSLVDMELEPLNFPNITEINIDHCSDLGELPLKLC 338
Query: 633 DIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISN 692
++ SL++LS+TNCH + LP ++G+L++L +L L +C L LP SI +L L LDIS
Sbjct: 339 NLTSLQRLSVTNCHLIQNLPDDMGRLKSLRVLRLSACPSLSRLPPSICKLGQLEYLDISL 398
Query: 693 CISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVV--NLQNLKVTCDEETAASWESFQ-S 749
C L LP +F L NL LDM C+ + +V+ +L+ + ++ ++ +W S + S
Sbjct: 399 CRCLQDLPSEFDQLSNLETLDMRECSGLKKVPTVIQSSLKRVVISDSDKEYEAWXSIKAS 458
Query: 750 MISNLTIEVPHVEVNLNWL 768
+ LTI+V +L WL
Sbjct: 459 TLHTLTIDVVPEIFSLAWL 477
>dbj|BAD94804.1| putative protein [Arabidopsis thaliana]
Length = 366
Score = 228 bits (582), Expect = 4e-58
Identities = 124/350 (35%), Positives = 195/350 (55%), Gaps = 24/350 (6%)
Query: 424 FIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFD 483
F+ HD+LR+L + + + +RKR L+ E L G ++
Sbjct: 34 FVTQHDVLRDLALHLSNAGKVNRRKRLLMPKRELD-------------------LPGDWE 74
Query: 484 KPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALI 543
+ + A+ +SI E W ++ AE+LILN + +Y P + KM++LK L+
Sbjct: 75 RNNDEHYIAQIVSIHTGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLV 134
Query: 544 VINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLAF 599
+IN+G+ P+ L++ + + LS L+ + LER+ VP LKNL K+SL +C +F
Sbjct: 135 IINNGMSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSF 194
Query: 600 EKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLE 659
++ + ++D+FP L DL++D+C D+ ALP+ +C + SL LSITNC +L LP+ + KL+
Sbjct: 195 DQTGLDVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQ 254
Query: 660 NLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS 719
LE+L L +C +L LP I L L+ LDIS C+SLS LPE+ G L L +DM C
Sbjct: 255 ALEILRLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCF 314
Query: 720 CELPFSVVNLQNLK-VTCDEETAASWESFQSMISNLTIEVPHVEVNLNWL 768
+ P S V+L++L+ V CD + A WE + + L IE +L+WL
Sbjct: 315 SDRPSSAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 364
>gb|AAN23084.1| putative rp3 protein [Zea mays]
Length = 944
Score = 132 bits (333), Expect = 3e-29
Identities = 171/638 (26%), Positives = 273/638 (41%), Gaps = 115/638 (18%)
Query: 133 GLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSD 192
GLGG GKTTLA KL +D K E +++V S+ ++ +VE+LF+ + SD
Sbjct: 197 GLGGSGKTTLA-KLVFNDGNIIKHFEVVLWVHVSREFAVEKLVEKLFK----AIAGDMSD 251
Query: 193 EDAVNGLGLLLR-KIEGSPILLVLDDVWPGSEDLVEKFKFQI--------SDYKILVTSR 243
+ + + K+ G L VLDDVW +ED VE +F + S + SR
Sbjct: 252 HPPLQHVSRTISDKLVGKRFLAVLDDVW--TEDRVEWEQFMVHLKSGAPGSSILLTTRSR 309
Query: 244 VAFSRFDKTFI--LKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIE---KVVEHCKGLPL 298
D ++ L L++EDS +F+ + + K D + ++ ++VE C G+PL
Sbjct: 310 KVAEAVDSSYAYNLPFLSKEDSWKVFQQCFGI---ALKALDPEFLQTGKEIVEKCGGVPL 366
Query: 299 AIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDN-------AI 351
AIKVIA +K + RSI DSN +LDV +D ++
Sbjct: 367 AIKVIAGVLHG---------IKGIEEWRSICDSN---------LLDVQDDEHRVFACLSL 408
Query: 352 S--------KECFMDLALFPEDQRIPVAALIDMW-AELYGLDDDGKEAMDI-INKLDSMN 401
S K CF+ ++FP I LI W A + + ++A D+ I DS+
Sbjct: 409 SFVHLPDHLKPCFLHCSIFPRGYVINRRHLISQWIAHGFVPTNQARQAEDVGIGYFDSLL 468
Query: 402 LANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQ 461
L T ++HDL R++ + E I TN K +
Sbjct: 469 KVGFLQDHVQIWSTRGEVTCKMHDLVHDLARQILRDEFVSE---------IETN--KQIK 517
Query: 462 RLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILN 521
R R L+ G D KV A L
Sbjct: 518 RCRYLS-------LTSCTGKLDNKLCGKVRA---------------------------LY 543
Query: 522 LQTKQYTFPELMEKMNKLKALIV--INHGLRPSELNNLELLSSLSNLKRIRLERISVPSF 579
+ ++ F + M K ++ +I+ I P ++ E L L + + E + +
Sbjct: 544 VCGRELEFDKTMNKQCCVRTIILKYITDDSLPLFVSKFEYLGYLE-ISDVNCEALP-EAL 601
Query: 580 GTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKK 639
NL+ L + C+ RLA SI L L ++ + +LP + D +L++
Sbjct: 602 SRCWNLQALHVLNCS-RLAVVPESI---GKLKKLRTLELNGVSSIKSLPQSIGDCDNLRR 657
Query: 640 LSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELP--DSIGRLSNLRLLDISNCISLS 697
L + C + +P +GKLENL +LS++ C L +LP DS G+L NL+ + + C +L
Sbjct: 658 LYLEECRGIEDIPNSLGKLENLRILSIVDCVSLQKLPPSDSFGKLLNLQTITFNLCYNLR 717
Query: 698 SLPEDFGNLCNLRNLDMTSCASC-ELPFSVVNLQNLKV 734
+LP+ +L +L ++D+ C ELP + NL+NLKV
Sbjct: 718 NLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKV 755
>gb|AAO24595.1| At5g66630 [Arabidopsis thaliana]
Length = 702
Score = 131 bits (329), Expect = 1e-28
Identities = 93/276 (33%), Positives = 144/276 (51%), Gaps = 14/276 (5%)
Query: 29 IKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDDDKQKMGKDVEGKLRELLKILDKENFGKK 88
I+W+S+ K+ ++ K N+D C + Q + + + + K +
Sbjct: 91 IQWYSIAKKALYTREIKAINQDFLKFCQIELQLIQHRNQLQYMRSMGMASVSTKADLLSD 150
Query: 89 ISGSILKGPFDVPANPEFTVGLDLQ--LIKLKVEILREGRSTLLLTGLGGMGKTTLATKL 146
I K + A PE L+ L++LK + +G T++++ +GKTTL TKL
Sbjct: 151 IGNEFSK--LCLVAQPEVVTKFWLKRPLMELKKMLFEDGVVTVVVSAPYALGKTTLVTKL 208
Query: 147 CLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKI 206
C D VK KFK+ I F++ SK P +++I +L EH G EY++D DA+ + LL+++
Sbjct: 209 CHDADVKEKFKQ-IFFISVSKFPNVRLIGHKLLEHIGCKANEYENDLDAMLYIQQLLKQL 267
Query: 207 -EGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVT 265
ILLVLDDVW E L++KF Q+ DYKILVTSR F+ F TF LKPL ++
Sbjct: 268 GRNGSILLVLDDVWAEEESLLQKFLIQLPDYKILVTSRFEFTSFGPTFHLKPLIDDE--- 324
Query: 266 LFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIK 301
E+E+N K+P+ + + C G A+K
Sbjct: 325 -VECRDEIEEN-EKLPE---VNPPLSMCGGCNSAVK 355
>ref|NP_201464.2| LIM domain-containing protein [Arabidopsis thaliana]
Length = 702
Score = 131 bits (329), Expect = 1e-28
Identities = 93/276 (33%), Positives = 144/276 (51%), Gaps = 14/276 (5%)
Query: 29 IKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDDDKQKMGKDVEGKLRELLKILDKENFGKK 88
I+W+S+ K+ ++ K N+D C + Q + + + + K +
Sbjct: 91 IQWYSIAKKALYTREIKAINQDFLKFCQIELQLIQHRNQLQYMRSMGMASVSTKADLLSD 150
Query: 89 ISGSILKGPFDVPANPEFTVGLDLQ--LIKLKVEILREGRSTLLLTGLGGMGKTTLATKL 146
I K + A PE L+ L++LK + +G T++++ +GKTTL TKL
Sbjct: 151 IGNEFSK--LCLVAQPEVVTKFWLKRPLMELKKMLFEDGVVTVVVSAPYALGKTTLVTKL 208
Query: 147 CLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKI 206
C D VK KFK+ I F++ SK P +++I +L EH G EY++D DA+ + LL+++
Sbjct: 209 CHDADVKEKFKQ-IFFISVSKFPNVRLIGHKLLEHIGCKANEYENDLDAMLYIQQLLKQL 267
Query: 207 -EGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVT 265
ILLVLDDVW E L++KF Q+ DYKILVTSR F+ F TF LKPL ++
Sbjct: 268 GRNGSILLVLDDVWAEEESLLQKFLIQLPDYKILVTSRFEFTSFGPTFHLKPLIDDE--- 324
Query: 266 LFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIK 301
E+E+N K+P+ + + C G A+K
Sbjct: 325 -VECRDEIEEN-EKLPE---VNPPLSMCGGCNSAVK 355
>gb|AAN23083.1| putative rp3 protein [Zea mays]
Length = 1251
Score = 129 bits (323), Expect = 5e-28
Identities = 168/624 (26%), Positives = 267/624 (41%), Gaps = 55/624 (8%)
Query: 113 QLIKLKVEILREGRSTLL-LTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPML 171
Q+I +E + R ++ + GLGG GKTTLA ++ D + F E +++V S+ +
Sbjct: 178 QIISKLIETDSQQRIKIVAVIGLGGSGKTTLAKQVFNDGNIIKHF-EVLLWVHVSREFAV 236
Query: 172 KIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLR-KIEGSPILLVLDDVWPGSEDLVEKFK 230
+ +VE+LFE + + SD + + + K+ G L VLDDVW +ED VE +
Sbjct: 237 EKLVEKLFE----AIAGHMSDHLPLQHVSRTISDKLVGKRFLAVLDDVW--TEDRVEWER 290
Query: 231 FQI--------SDYKILVTSRVAFSRFDKTFI--LKPLAQEDSVTLFRHYTEVEKNSSKI 280
F + S + SR D ++ L L++EDS +F+ + +
Sbjct: 291 FMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYDLPFLSKEDSWKVFQQCFRIAIQALDT 350
Query: 281 PDKDLIEKVVEHCKGLPLAIKVIATSFR-YRPYELWEKIVKELSRGRSILDSNTELLIRL 339
++V+ C G+PLAIKVIA + E W+ I S + D + L
Sbjct: 351 EFLQAGIEIVDKCGGVPLAIKVIAGVLHGMKGIEEWQSICN--SNLLDVHDDEHRVFACL 408
Query: 340 QKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMW-AELYGLDDDGKEAMDI-INKL 397
L D+ K CF+ ++FP + LI W A + + ++A D+ I
Sbjct: 409 WLSFVHLPDHL--KPCFLHCSIFPRGYVLNRCHLISQWIAHGFIPTNQARQAEDVGIGYF 466
Query: 398 DSMNLANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNES 457
DS L V + D Y +HDL+ +L E + + I TN
Sbjct: 467 DS--LLKVGFLQDQDRDQNLYTRGEVTCKMHDLVHDLARKILRDEFVSE-----IETN-- 517
Query: 458 KCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEV 517
K +R R LS G D KV A + D + +
Sbjct: 518 KQIKRCRYLS-------LSSCTGKLDNKLCGKVHALYV---CGRELEFDRTMNKQCYVRT 567
Query: 518 LILNLQTKQYTFPELMEKMNKLKALIV--INHGLRPSELNNLELLSSLSNLKRIRLERIS 575
+IL T + + P + K L L + +N P L+ L +L L +L +
Sbjct: 568 IILKYITAE-SLPLFVSKFEYLGYLEISDVNCEALPEALSRCWNLQALHVLACSKL-AVV 625
Query: 576 VPSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDII 635
S G LK L+ L L ++ + + I D NL L ++ C+ + +PN + +
Sbjct: 626 PESIGKLKKLRTLELNGVSSIKSLPES---IGDC-DNLRRLYLEGCRGIEDIPNSLGKLE 681
Query: 636 SLKKLSITNCHKLSLL--PQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+L+ LSI C L L GKL NL+ ++ SC +L LP + LS+L ++D+ C
Sbjct: 682 NLRILSIVACFSLKKLSPSASFGKLLNLQTITFKSCFNLRNLPQCMTSLSHLEMVDLGYC 741
Query: 694 ISLSSLPEDFGNLCNLRNLDMTSC 717
L LPE GNL NL+ L++ C
Sbjct: 742 FELVELPEGIGNLRNLKVLNLKKC 765
Score = 79.3 bits (194), Expect = 4e-13
Identities = 52/149 (34%), Positives = 78/149 (51%), Gaps = 13/149 (8%)
Query: 621 CKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIG 680
C ++ LPN + ++ SL+ L + CH L LP++IG+L +L+ L +I T L LP+S+
Sbjct: 1103 CDNLRVLPNWLVELKSLQSLEVLFCHALQQLPEQIGELCSLQHLHIIYLTSLTCLPESMQ 1162
Query: 681 RLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS-CELPFSVVNLQNLKV----- 734
RL++LR LD+ C +L+ LPE G L L+ L++ C LP S+ L L+
Sbjct: 1163 RLTSLRTLDMFGCGALTQLPEWLGELSALQKLNLGGCRGLTSLPRSIQCLTALEELFIGG 1222
Query: 735 ------TCDEETAASWESFQSMISNLTIE 757
C E W S I NL +E
Sbjct: 1223 NPDLLRRCREGVGEDW-PLVSHIQNLRLE 1250
Score = 68.6 bits (166), Expect = 8e-10
Identities = 35/94 (37%), Positives = 52/94 (55%)
Query: 626 ALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNL 685
ALP + +L+ L + C KL+++P+ IGKL+ L L L + + LP+SIG NL
Sbjct: 600 ALPEALSRCWNLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKSLPESIGDCDNL 659
Query: 686 RLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS 719
R L + C + +P G L NLR L + +C S
Sbjct: 660 RRLYLEGCRGIEDIPNSLGKLENLRILSIVACFS 693
Score = 67.8 bits (164), Expect = 1e-09
Identities = 35/97 (36%), Positives = 59/97 (60%), Gaps = 4/97 (4%)
Query: 639 KLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSS 698
++S NC L P+ + + NL+ L +++C+ L +P+SIG+L LR L+++ S+ S
Sbjct: 592 EISDVNCEAL---PEALSRCWNLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKS 648
Query: 699 LPEDFGNLCNLRNLDMTSCASCE-LPFSVVNLQNLKV 734
LPE G+ NLR L + C E +P S+ L+NL++
Sbjct: 649 LPESIGDCDNLRRLYLEGCRGIEDIPNSLGKLENLRI 685
Score = 67.0 bits (162), Expect = 2e-09
Identities = 41/112 (36%), Positives = 62/112 (54%), Gaps = 1/112 (0%)
Query: 613 LEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDL 672
L L + C +T LP + +L +L I +C L +LP + +L++L+ L ++ C L
Sbjct: 1071 LHTLEIFKCTGLTHLPESIHCPTTLCRLVIRSCDNLRVLPNWLVELKSLQSLEVLFCHAL 1130
Query: 673 VELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSC-ASCELP 723
+LP+ IG L +L+ L I SL+ LPE L +LR LDM C A +LP
Sbjct: 1131 QQLPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLRTLDMFGCGALTQLP 1182
Score = 60.1 bits (144), Expect = 3e-07
Identities = 32/101 (31%), Positives = 54/101 (52%)
Query: 612 NLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTD 671
+L+ L + +C + LP + ++ SL+ L I L+ LP+ + +L +L L + C
Sbjct: 1118 SLQSLEVLFCHALQQLPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLRTLDMFGCGA 1177
Query: 672 LVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNL 712
L +LP+ +G LS L+ L++ C L+SLP L L L
Sbjct: 1178 LTQLPEWLGELSALQKLNLGGCRGLTSLPRSIQCLTALEEL 1218
Score = 50.8 bits (120), Expect = 2e-04
Identities = 41/136 (30%), Positives = 69/136 (50%), Gaps = 9/136 (6%)
Query: 582 LKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLS 641
LK+L+ L + C+ A ++ I +L +L+ L + Y +T LP + + SL+ L
Sbjct: 1116 LKSLQSLEVLFCH---ALQQLPEQIGELC-SLQHLHIIYLTSLTCLPESMQRLTSLRTLD 1171
Query: 642 ITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISL----- 696
+ C L+ LP+ +G+L L+ L+L C L LP SI L+ L L I L
Sbjct: 1172 MFGCGALTQLPEWLGELSALQKLNLGGCRGLTSLPRSIQCLTALEELFIGGNPDLLRRCR 1231
Query: 697 SSLPEDFGNLCNLRNL 712
+ ED+ + +++NL
Sbjct: 1232 EGVGEDWPLVSHIQNL 1247
Score = 50.4 bits (119), Expect = 2e-04
Identities = 34/101 (33%), Positives = 53/101 (51%), Gaps = 1/101 (0%)
Query: 634 IISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+ L L I C L+ LP+ I L L + SC +L LP+ + L +L+ L++ C
Sbjct: 1068 LTGLHTLEIFKCTGLTHLPESIHCPTTLCRLVIRSCDNLRVLPNWLVELKSLQSLEVLFC 1127
Query: 694 ISLSSLPEDFGNLCNLRNLDMTSCASCE-LPFSVVNLQNLK 733
+L LPE G LC+L++L + S LP S+ L +L+
Sbjct: 1128 HALQQLPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLR 1168
>dbj|BAD38047.1| putative NBS-LRR resistance protein RGH2 [Oryza sativa (japonica
cultivar-group)]
Length = 1216
Score = 129 bits (323), Expect = 5e-28
Identities = 169/681 (24%), Positives = 286/681 (41%), Gaps = 78/681 (11%)
Query: 127 STLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPV 186
S + + GLGGMGKTTLA + D + + +V S L IV + H
Sbjct: 195 SIIPVVGLGGMGKTTLAKAVYTDKET--HMFDVKAWVHVSMEFQLNKIVSGIISHVEGST 252
Query: 187 PEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWPGS----EDLVEKFKFQISDYKILVTS 242
P +D + L R + L++LDD+W E L+E + KI+VT+
Sbjct: 253 PANIADLQYLK--SQLDRILCNKLYLIILDDLWEEGWSKLEKLMEMLQSGKKGSKIIVTT 310
Query: 243 R----------VAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEH 292
R + S F +K + F +N D+ +++ +
Sbjct: 311 RSEKVVNTLSTIRLSYFHTVDPIKLVGMSIDECWFIMKPRNMENCEFSDLVDIGKEIAQR 370
Query: 293 CKGLPLAIKVIA-TSFRYRPYELWEKIVKELSRGRSILDS-NTELLIRLQKILDVLEDNA 350
C G+PL K + ++R E W +I + +ILD+ + E I +L
Sbjct: 371 CSGVPLVAKALGYVMQKHRTREEWMEI-----KNSNILDTKDDEEGILKGLLLSYYHMPP 425
Query: 351 ISKECFMDLALFPEDQRIPVAALIDMWAEL-YGLDDDG----KEAMDIINKLDSMNLANV 405
K CFM ++FP I LI W L + D DG K AM+ +N+L M+ +
Sbjct: 426 QLKLCFMYCSMFPMSHVIDHDCLIQQWIALGFIQDTDGQPLQKVAMEYVNELLGMSFLTI 485
Query: 406 LIARKNASDTENYYYNNHFIVLHDLLREL-----GNYQNTQEPIEQRKRQ---------- 450
+ + + + +HD++ EL GN + E R +
Sbjct: 486 FTS-PTVLASRMLFKPTLKLHMHDMVHELARHVAGNEFSHTNGAENRNTKRDNLNFHYHL 544
Query: 451 LINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQV 510
L+N NE+ K T L G P++ + TL + + + S++
Sbjct: 545 LLNQNETSS----AYKSLATKVRAL-HFRGCDKMHLPKQAFSHTLCLRVLDLGGRQVSEL 599
Query: 511 QPALAEVLILNL----QTKQYTFPELMEKMNKLKALIVINHGLR--PSELNNLEL----- 559
++ ++ +L + +F + + L+ALI+ N L+ P+ + L+
Sbjct: 600 PSSVYKLKLLRYLDASSLRISSFSKSFNHLLNLQALILSNTYLKTLPTNIGCLQKLQYFD 659
Query: 560 LSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMCN--TRLAFEKGSILISDLFPNLEDLS 617
LS +NL + SFG L +L L+L C+ L G++ L+ LS
Sbjct: 660 LSGCANLNEL------PTSFGDLSSLLFLNLASCHELEALPMSFGNL------NRLQFLS 707
Query: 618 MDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPD 677
+ C + +LP C + L L +++C+ L LP I +L LE L++ SC+ + LP+
Sbjct: 708 LSDCYKLNSLPESCCQLHDLAHLDLSDCYNLGKLPDCIDQLSKLEYLNMTSCSKVQALPE 767
Query: 678 SIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMT-SCASCELPFSVVNLQNLKVTC 736
S+ +L+ LR L++S C+ L +LP G+L L++LD+ S +LP S+ N+ LK
Sbjct: 768 SLCKLTMLRHLNLSYCLRLENLPSCIGDL-QLQSLDIQGSFLLRDLPNSIFNMSTLKTVD 826
Query: 737 DEETAASWESFQSMISNLTIE 757
T + + NL +E
Sbjct: 827 GTFTYLVSSKVEKLRENLKLE 847
Score = 53.1 bits (126), Expect = 3e-05
Identities = 33/87 (37%), Positives = 50/87 (56%), Gaps = 1/87 (1%)
Query: 612 NLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLEN-LELLSLISCT 670
++E L++ + ALP + SL +LSI C +L LP+ +G LE +S+ +C
Sbjct: 1110 SIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCP 1169
Query: 671 DLVELPDSIGRLSNLRLLDISNCISLS 697
L LP+SI RL+ L+ L I+NC LS
Sbjct: 1170 MLSSLPESIRRLTKLKKLRITNCPVLS 1196
Score = 47.0 bits (110), Expect = 0.002
Identities = 28/83 (33%), Positives = 45/83 (53%), Gaps = 1/83 (1%)
Query: 636 SLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGR-LSNLRLLDISNCI 694
S++ L++ + L LP+ I +L LS++ C +L LP+ +G + L + I C
Sbjct: 1110 SIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCP 1169
Query: 695 SLSSLPEDFGNLCNLRNLDMTSC 717
LSSLPE L L+ L +T+C
Sbjct: 1170 MLSSLPESIRRLTKLKKLRITNC 1192
Score = 41.2 bits (95), Expect = 0.13
Identities = 19/44 (43%), Positives = 28/44 (63%)
Query: 606 ISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLS 649
+ D F LE++S+D C +++LP + + LKKL ITNC LS
Sbjct: 1153 LGDYFTCLEEISIDTCPMLSSLPESIRRLTKLKKLRITNCPVLS 1196
Score = 35.8 bits (81), Expect = 5.5
Identities = 26/78 (33%), Positives = 40/78 (50%), Gaps = 2/78 (2%)
Query: 658 LENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCN-LRNLDMTS 716
L ++E L+L+S L LP++I ++L L I C L +LPE G+ L + + +
Sbjct: 1108 LSSIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDT 1167
Query: 717 CAS-CELPFSVVNLQNLK 733
C LP S+ L LK
Sbjct: 1168 CPMLSSLPESIRRLTKLK 1185
>gb|AAN23090.1| putative rp3 protein [Zea mays] gi|23321151|gb|AAN23085.1| putative
rp3 protein [Zea mays] gi|23321143|gb|AAN23081.1|
putative rp3 protein [Zea mays]
Length = 1251
Score = 128 bits (321), Expect = 8e-28
Identities = 168/624 (26%), Positives = 267/624 (41%), Gaps = 55/624 (8%)
Query: 113 QLIKLKVEILREGRSTLL-LTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPML 171
Q+I +E + R ++ + GLGG GKTTLA ++ D + F E +++V S+ +
Sbjct: 178 QIISKLIETDSQQRIKIVAVIGLGGSGKTTLAKQVFNDGNIIKHF-EVLLWVHVSREFAV 236
Query: 172 KIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLR-KIEGSPILLVLDDVWPGSEDLVEKFK 230
+ +VE+LFE + + SD + + + K+ G L VLDDVW +ED VE +
Sbjct: 237 EKLVEKLFE----AIAGHMSDHLPLQHVSRTISDKLVGKRFLAVLDDVW--TEDRVEWER 290
Query: 231 FQI--------SDYKILVTSRVAFSRFDKTFI--LKPLAQEDSVTLFRHYTEVEKNSSKI 280
F + S + SR D ++ L L++EDS +F+ + +
Sbjct: 291 FMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYDLPFLSKEDSWKVFQQCFGIAIQALDT 350
Query: 281 PDKDLIEKVVEHCKGLPLAIKVIATSFR-YRPYELWEKIVKELSRGRSILDSNTELLIRL 339
++V+ C G+PLAIKVIA + E W+ I S + D + L
Sbjct: 351 EFLQAGIEIVDKCGGVPLAIKVIAGVLHGMKGIEEWQSICN--SNLLDVHDDEHRVFACL 408
Query: 340 QKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMW-AELYGLDDDGKEAMDI-INKL 397
L D+ K CF+ ++FP + LI W A + + ++A D+ I
Sbjct: 409 WLSFVHLPDHL--KPCFLHCSIFPRGYVLNRCHLISQWIAHGFIPTNQARQAEDVGIGYF 466
Query: 398 DSMNLANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNES 457
DS L V + D Y +HDL+ +L E + + I TN
Sbjct: 467 DS--LLKVGFLQDQDRDQNLYTRGEVTCKMHDLVHDLARKILRDEFVSE-----IETN-- 517
Query: 458 KCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEV 517
K +R R LS G D KV A + D + +
Sbjct: 518 KQIKRCRYLS-------LSSCTGKLDNKLCGKVHALYV---CGRELEFDRTMNKQCYVRT 567
Query: 518 LILNLQTKQYTFPELMEKMNKLKALIV--INHGLRPSELNNLELLSSLSNLKRIRLERIS 575
+IL T + + P + K L L + +N P L+ L +L L +L +
Sbjct: 568 IILKYITAE-SLPLFVSKFEYLGYLEISDVNCEALPEALSRCWNLQALHVLACSKL-AVV 625
Query: 576 VPSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDII 635
S G LK L+ L L ++ + + I D NL L ++ C+ + +PN + +
Sbjct: 626 PESIGKLKKLRTLELNGVSSIKSLPES---IGDC-DNLRRLYLEGCRGIEDIPNSLGKLE 681
Query: 636 SLKKLSITNCHKLSLL--PQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+L+ LSI C L L GKL NL+ ++ SC +L LP + LS+L ++D+ C
Sbjct: 682 NLRILSIVACFSLKKLSPSASFGKLLNLQTITFKSCFNLRNLPQCMTSLSHLEMVDLGYC 741
Query: 694 ISLSSLPEDFGNLCNLRNLDMTSC 717
L LPE GNL NL+ L++ C
Sbjct: 742 FELVELPEGIGNLRNLKVLNLKKC 765
Score = 79.3 bits (194), Expect = 4e-13
Identities = 52/149 (34%), Positives = 78/149 (51%), Gaps = 13/149 (8%)
Query: 621 CKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIG 680
C ++ LPN + ++ SL+ L + CH L LP++IG+L +L+ L +I T L LP+S+
Sbjct: 1103 CDNLRVLPNWLVELKSLQSLEVLFCHALQQLPEQIGELCSLQHLHIIYLTSLTCLPESMQ 1162
Query: 681 RLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS-CELPFSVVNLQNLKV----- 734
RL++LR LD+ C +L+ LPE G L L+ L++ C LP S+ L L+
Sbjct: 1163 RLTSLRTLDMFGCGALTQLPEWLGELSALQKLNLGGCRGLTSLPRSIQCLTALEELFIGG 1222
Query: 735 ------TCDEETAASWESFQSMISNLTIE 757
C E W S I NL +E
Sbjct: 1223 NPDLLRRCREGVGEDW-PLVSHIQNLRLE 1250
Score = 68.6 bits (166), Expect = 8e-10
Identities = 35/94 (37%), Positives = 52/94 (55%)
Query: 626 ALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNL 685
ALP + +L+ L + C KL+++P+ IGKL+ L L L + + LP+SIG NL
Sbjct: 600 ALPEALSRCWNLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKSLPESIGDCDNL 659
Query: 686 RLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS 719
R L + C + +P G L NLR L + +C S
Sbjct: 660 RRLYLEGCRGIEDIPNSLGKLENLRILSIVACFS 693
Score = 67.8 bits (164), Expect = 1e-09
Identities = 35/97 (36%), Positives = 59/97 (60%), Gaps = 4/97 (4%)
Query: 639 KLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSS 698
++S NC L P+ + + NL+ L +++C+ L +P+SIG+L LR L+++ S+ S
Sbjct: 592 EISDVNCEAL---PEALSRCWNLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKS 648
Query: 699 LPEDFGNLCNLRNLDMTSCASCE-LPFSVVNLQNLKV 734
LPE G+ NLR L + C E +P S+ L+NL++
Sbjct: 649 LPESIGDCDNLRRLYLEGCRGIEDIPNSLGKLENLRI 685
Score = 67.0 bits (162), Expect = 2e-09
Identities = 41/112 (36%), Positives = 62/112 (54%), Gaps = 1/112 (0%)
Query: 613 LEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDL 672
L L + C +T LP + +L +L I +C L +LP + +L++L+ L ++ C L
Sbjct: 1071 LHTLEIFKCTGLTHLPESIHCPTTLCRLVIRSCDNLRVLPNWLVELKSLQSLEVLFCHAL 1130
Query: 673 VELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSC-ASCELP 723
+LP+ IG L +L+ L I SL+ LPE L +LR LDM C A +LP
Sbjct: 1131 QQLPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLRTLDMFGCGALTQLP 1182
Score = 60.1 bits (144), Expect = 3e-07
Identities = 32/101 (31%), Positives = 54/101 (52%)
Query: 612 NLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTD 671
+L+ L + +C + LP + ++ SL+ L I L+ LP+ + +L +L L + C
Sbjct: 1118 SLQSLEVLFCHALQQLPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLRTLDMFGCGA 1177
Query: 672 LVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNL 712
L +LP+ +G LS L+ L++ C L+SLP L L L
Sbjct: 1178 LTQLPEWLGELSALQKLNLGGCRGLTSLPRSIQCLTALEEL 1218
Score = 50.8 bits (120), Expect = 2e-04
Identities = 41/136 (30%), Positives = 69/136 (50%), Gaps = 9/136 (6%)
Query: 582 LKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLS 641
LK+L+ L + C+ A ++ I +L +L+ L + Y +T LP + + SL+ L
Sbjct: 1116 LKSLQSLEVLFCH---ALQQLPEQIGELC-SLQHLHIIYLTSLTCLPESMQRLTSLRTLD 1171
Query: 642 ITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISL----- 696
+ C L+ LP+ +G+L L+ L+L C L LP SI L+ L L I L
Sbjct: 1172 MFGCGALTQLPEWLGELSALQKLNLGGCRGLTSLPRSIQCLTALEELFIGGNPDLLRRCR 1231
Query: 697 SSLPEDFGNLCNLRNL 712
+ ED+ + +++NL
Sbjct: 1232 EGVGEDWPLVSHIQNL 1247
Score = 50.4 bits (119), Expect = 2e-04
Identities = 34/101 (33%), Positives = 53/101 (51%), Gaps = 1/101 (0%)
Query: 634 IISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+ L L I C L+ LP+ I L L + SC +L LP+ + L +L+ L++ C
Sbjct: 1068 LTGLHTLEIFKCTGLTHLPESIHCPTTLCRLVIRSCDNLRVLPNWLVELKSLQSLEVLFC 1127
Query: 694 ISLSSLPEDFGNLCNLRNLDMTSCASCE-LPFSVVNLQNLK 733
+L LPE G LC+L++L + S LP S+ L +L+
Sbjct: 1128 HALQQLPEQIGELCSLQHLHIIYLTSLTCLPESMQRLTSLR 1168
>gb|AAN23082.1| putative rp3 protein [Zea mays]
Length = 948
Score = 128 bits (321), Expect = 8e-28
Identities = 168/624 (26%), Positives = 267/624 (41%), Gaps = 55/624 (8%)
Query: 113 QLIKLKVEILREGRSTLL-LTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPML 171
Q+I +E + R ++ + GLGG GKTTLA ++ D + F E +++V S+ +
Sbjct: 178 QIISKLIETDSQQRIKIVAVIGLGGSGKTTLAKQVFNDGNIIKHF-EVLLWVHVSREFAV 236
Query: 172 KIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLR-KIEGSPILLVLDDVWPGSEDLVEKFK 230
+ +VE+LFE + + SD + + + K+ G L VLDDVW +ED VE +
Sbjct: 237 EKLVEKLFE----AIAGHMSDHLPLQHVSRTISDKLVGKRFLAVLDDVW--TEDRVEWER 290
Query: 231 FQI--------SDYKILVTSRVAFSRFDKTFI--LKPLAQEDSVTLFRHYTEVEKNSSKI 280
F + S + SR D ++ L L++EDS +F+ + +
Sbjct: 291 FMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYDLPFLSKEDSWKVFQQCFGIAIQALDT 350
Query: 281 PDKDLIEKVVEHCKGLPLAIKVIATSFR-YRPYELWEKIVKELSRGRSILDSNTELLIRL 339
++V+ C G+PLAIKVIA + E W+ I S + D + L
Sbjct: 351 EFLQAGIEIVDKCGGVPLAIKVIAGVLHGMKGIEEWQSICN--SNLLDVHDDEHRVFACL 408
Query: 340 QKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMW-AELYGLDDDGKEAMDI-INKL 397
L D+ K CF+ ++FP + LI W A + + ++A D+ I
Sbjct: 409 WLSFVHLPDHL--KPCFLHCSIFPRGYVLNRCHLISQWIAHGFIPTNQARQAEDVGIGYF 466
Query: 398 DSMNLANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNES 457
DS L V + D Y +HDL+ +L E + + I TN
Sbjct: 467 DS--LLKVGFLQDQDRDQNLYTRGEVTCKMHDLVHDLARKILRDEFVSE-----IETN-- 517
Query: 458 KCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEV 517
K +R R LS G D KV A + D + +
Sbjct: 518 KQIKRCRYLS-------LSSCTGKLDNKLCGKVHALYV---CGRELEFDRTMNKQCYVRT 567
Query: 518 LILNLQTKQYTFPELMEKMNKLKALIV--INHGLRPSELNNLELLSSLSNLKRIRLERIS 575
+IL T + + P + K L L + +N P L+ L +L L +L +
Sbjct: 568 IILKYITAE-SLPLFVSKFEYLGYLEISDVNCEALPEALSRCWNLQALHVLACSKL-AVV 625
Query: 576 VPSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDII 635
S G LK L+ L L ++ + + I D NL L ++ C+ + +PN + +
Sbjct: 626 PESIGKLKKLRTLELNGVSSIKSLPES---IGDC-DNLRRLYLEGCRGIEDIPNSLGKLE 681
Query: 636 SLKKLSITNCHKLSLL--PQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+L+ LSI C L L GKL NL+ ++ SC +L LP + LS+L ++D+ C
Sbjct: 682 NLRILSIVACFSLKKLSPSASFGKLLNLQTITFKSCFNLRNLPQCMTSLSHLEMVDLGYC 741
Query: 694 ISLSSLPEDFGNLCNLRNLDMTSC 717
L LPE GNL NL+ L++ C
Sbjct: 742 FELVELPEGIGNLRNLKVLNLKKC 765
Score = 68.6 bits (166), Expect = 8e-10
Identities = 35/94 (37%), Positives = 52/94 (55%)
Query: 626 ALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNL 685
ALP + +L+ L + C KL+++P+ IGKL+ L L L + + LP+SIG NL
Sbjct: 600 ALPEALSRCWNLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKSLPESIGDCDNL 659
Query: 686 RLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS 719
R L + C + +P G L NLR L + +C S
Sbjct: 660 RRLYLEGCRGIEDIPNSLGKLENLRILSIVACFS 693
Score = 67.8 bits (164), Expect = 1e-09
Identities = 35/97 (36%), Positives = 59/97 (60%), Gaps = 4/97 (4%)
Query: 639 KLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSS 698
++S NC L P+ + + NL+ L +++C+ L +P+SIG+L LR L+++ S+ S
Sbjct: 592 EISDVNCEAL---PEALSRCWNLQALHVLACSKLAVVPESIGKLKKLRTLELNGVSSIKS 648
Query: 699 LPEDFGNLCNLRNLDMTSCASCE-LPFSVVNLQNLKV 734
LPE G+ NLR L + C E +P S+ L+NL++
Sbjct: 649 LPESIGDCDNLRRLYLEGCRGIEDIPNSLGKLENLRI 685
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,277,182,539
Number of Sequences: 2540612
Number of extensions: 55107492
Number of successful extensions: 189450
Number of sequences better than 10.0: 3955
Number of HSP's better than 10.0 without gapping: 1283
Number of HSP's successfully gapped in prelim test: 2710
Number of HSP's that attempted gapping in prelim test: 164306
Number of HSP's gapped (non-prelim): 14653
length of query: 773
length of database: 863,360,394
effective HSP length: 136
effective length of query: 637
effective length of database: 517,837,162
effective search space: 329862272194
effective search space used: 329862272194
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC148347.1


