

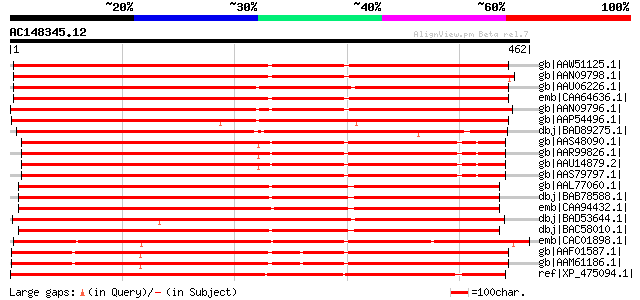
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.12 + phase: 0
(462 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAW51125.1| putative alcohol acyl-transferases [Cucumis melo] 530 e-149
gb|AAN09798.1| benzoyl coenzyme A: benzyl alcohol benzoyl transf... 528 e-148
gb|AAU06226.1| benzoyl-CoA:benzyl alcohol/phenylethanol benzoylt... 528 e-148
emb|CAA64636.1| hsr201 [Nicotiana tabacum] gi|7489142|pir||T0327... 525 e-148
gb|AAN09796.1| benzoyl coenzyme A: benzyl alcohol benzoyl transf... 497 e-139
gb|AAP54496.1| putative hypersensitivity-related (hsr)protein [O... 449 e-124
dbj|BAD89275.1| (-)-13alpha-hydroxymultiflorine/(+)-13alpha- hyd... 442 e-122
gb|AAS48090.1| alcohol acyl transferase [Pyrus communis] 441 e-122
gb|AAR99826.1| alcohol acyl transferase [Malus x domestica] 438 e-121
gb|AAU14879.2| alcohol acyl transferase [Malus x domestica] 437 e-121
gb|AAS79797.1| alcohol acyl transferase [Malus x domestica] 434 e-120
gb|AAL77060.1| putative acyltransferase [Cucumis melo] 433 e-120
dbj|BAB78588.1| alcohol acetyltransferase [Cucumis melo] 431 e-119
emb|CAA94432.1| unknown [Cucumis melo] gi|7484744|pir||T09666 pr... 416 e-114
dbj|BAD53644.1| putative benzoyl coenzyme A, benzyl alcohol benz... 408 e-112
dbj|BAC58010.1| alcohol acyltransferase [Cucumis melo] 404 e-111
emb|CAC01898.1| putative protein [Arabidopsis thaliana] gi|15237... 403 e-111
gb|AAF01587.1| putative hypersensitivity-related gene [Arabidops... 373 e-102
gb|AAM61186.1| putative hypersensitivity-related gene [Arabidops... 371 e-101
ref|XP_475094.1| hypothetical protein [Oryza sativa (japonica cu... 361 2e-98
>gb|AAW51125.1| putative alcohol acyl-transferases [Cucumis melo]
Length = 459
Score = 530 bits (1366), Expect = e-149
Identities = 260/443 (58%), Positives = 338/443 (75%), Gaps = 8/443 (1%)
Query: 4 SSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPV 63
+S L+F V+RSQP+L+ P+ PTPHE K LSDIDDQEGLRF +PVI +RH+P M DP
Sbjct: 2 ASSLVFQVQRSQPQLIPPSDPTPHEFKQLSDIDDQEGLRFQIPVIQFYRHDPRMAGTDPA 61
Query: 64 KVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPF 123
+V+K A+++ LV+YYP AGR+REG GRKL V+CTGEGVMFIEA+AD++L+QFGDALQPPF
Sbjct: 62 RVIKEAIAKALVFYYPFAGRLREGPGRKLFVECTGEGVMFIEADADVSLEQFGDALQPPF 121
Query: 124 PCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEI 183
PC +E L DVP S ++D P+ LIQVTRLKCGGFI AL LNHTM D SGL QF+ A E+
Sbjct: 122 PCLEEPLFDVPNSSGVLDCPLLLIQVTRLKCGGFIFALRLNHTMSDASGLVQFMMAVGEM 181
Query: 184 ARGANQPSIQPVWNREILMARDPPYITCNHREYEQIL-PPNTYIKEEDTTIVVHQSFFFT 242
ARGA PS++PVW R +L ARDPP +TC+HREY++++ T I +D + H+SFFF
Sbjct: 182 ARGATAPSVRPVWQRALLNARDPPKVTCHHREYDEVVDTKGTIIPLDD---MAHRSFFFG 238
Query: 243 SAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANN 302
+ I+AIR+ +P HL +C++++++TAC W RT +LQ +P+E+VR++CIVN+RS+F N
Sbjct: 239 PSEISAIRKALPSHLRQCSSFEVLTACLWRFRTISLQPDPEEEVRVLCIVNSRSKF---N 295
Query: 303 RSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKER 362
GYYGN AF A+TTA +LC N LGYA+EL+RKAKA VTE YM S+AD MVIK R
Sbjct: 296 PPLPTGYYGNAFAFPVALTTAGKLCQNPLGYALELVRKAKADVTEDYMKSVADLMVIKGR 355
Query: 363 CLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG-GTFLVTHKNAKGEEG 421
FT RT +VSD TRA F +V+FGWG+A+YGG AKGG+G+ G +F + KN KGE G
Sbjct: 356 PHFTVVRTYLVSDVTRAGFEDVDFGWGKAMYGGPAKGGVGAIPGVASFYIPFKNKKGERG 415
Query: 422 LILPICLPPEDMKRFVKELDDML 444
+++P+CLP M+RFVKELD +L
Sbjct: 416 ILVPLCLPAPAMERFVKELDALL 438
>gb|AAN09798.1| benzoyl coenzyme A: benzyl alcohol benzoyl transferase [Nicotiana
tabacum]
Length = 460
Score = 528 bits (1361), Expect = e-148
Identities = 265/454 (58%), Positives = 336/454 (73%), Gaps = 14/454 (3%)
Query: 4 SSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPV 63
SS L+FTVRR +PEL+ PA PTP E+K LSDIDDQEGLRF +PVI + + SM KDPV
Sbjct: 6 SSELVFTVRRQKPELIAPAKPTPREIKFLSDIDDQEGLRFQIPVIQFYHKDSSMGRKDPV 65
Query: 64 KVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPF 123
KV+K A++ TLV+YYP AGR+REG GRKLMVDCTGEG+MF+EA+AD+TL+QFGD LQPPF
Sbjct: 66 KVIKKAIAETLVFYYPFAGRLREGNGRKLMVDCTGEGIMFVEADADVTLEQFGDELQPPF 125
Query: 124 PCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEI 183
PC +E+L DVP S +++ P+ LIQVTRL+CGGFI AL LNHTM D GL QF++A E+
Sbjct: 126 PCLEELLYDVPDSAGVLNCPLLLIQVTRLRCGGFIFALRLNHTMSDAPGLVQFMTAVGEM 185
Query: 184 ARGANQPSIQPVWNREILMARDPPYITCNHREYEQIL-PPNTYIKEEDTTIVVHQSFFFT 242
ARGA+ PSI PVW RE+L AR+PP +TC H EY+++ T I +D +VH+SFFF
Sbjct: 186 ARGASAPSILPVWCRELLNARNPPQVTCTHHEYDEVRDTKGTIIPLDD---MVHKSFFFG 242
Query: 243 SAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANN 302
+ ++A+RR VP HL +C+ ++L+TA W CRT +L+ +P+E+VR +CIVNARSRF N
Sbjct: 243 PSEVSALRRFVPHHLRKCSTFELLTAVLWRCRTMSLKPDPEEEVRALCIVNARSRF---N 299
Query: 303 RSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKER 362
GYYGN AF AVTTA +L N LGYA+EL++K K+ VTE+YM S+AD MV+K R
Sbjct: 300 PPLPTGYYGNAFAFPVAVTTAAKLSKNPLGYALELVKKTKSDVTEEYMKSVADLMVLKGR 359
Query: 363 CLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG-GTFLVTHKNAKGEEG 421
FT RT +VSD TR F EV+FGWG+AVYGG AKGG+G+ G +F + KN KGE G
Sbjct: 360 PHFTVVRTFLVSDVTRGGFGEVDFGWGKAVYGGPAKGGVGAIPGVASFYIPFKNKKGENG 419
Query: 422 LILPICLPPEDMKRFVKELDDM------LGNQNY 449
+++PICLP M+ FVKELD M L N NY
Sbjct: 420 IVVPICLPGFAMETFVKELDGMLKVDAPLDNSNY 453
>gb|AAU06226.1| benzoyl-CoA:benzyl alcohol/phenylethanol benzoyltransferase; BPBT
[Petunia x hybrida] gi|49798480|gb|AAT68601.1| benzoyl
coenzyme A: benzyl alcohol benzoyl transferase [Petunia
x hybrida]
Length = 460
Score = 528 bits (1361), Expect = e-148
Identities = 261/442 (59%), Positives = 332/442 (75%), Gaps = 6/442 (1%)
Query: 4 SSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPV 63
SS L+FTVRR +PEL+ PA PTP E K LSDIDDQEGLRF +PVI +R + SM KDPV
Sbjct: 6 SSELVFTVRRQEPELIAPAKPTPRETKFLSDIDDQEGLRFQIPVINFYRKDSSMGGKDPV 65
Query: 64 KVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPF 123
+V+K A++ TLV+YYP AGR+REG RKLMVDCTGEGVMF+EA AD+TL++FGD LQPPF
Sbjct: 66 EVIKKAIAETLVFYYPFAGRLREGNDRKLMVDCTGEGVMFVEANADVTLEEFGDELQPPF 125
Query: 124 PCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEI 183
PC +E+L DVPGS ++ P+ LIQVTRL+CGGFI AL LNHTM D GL QF++A E+
Sbjct: 126 PCLEELLYDVPGSAGVLHCPLLLIQVTRLRCGGFIFALRLNHTMSDAPGLVQFMTAVGEM 185
Query: 184 ARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTS 243
ARGA PS PVW RE+L AR+PP +TC H EYE++ P+T +VH+SFFF
Sbjct: 186 ARGATAPSTLPVWCRELLNARNPPQVTCTHHEYEEV--PDTKGTLIPLDDMVHRSFFFGP 243
Query: 244 AHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNR 303
++A+RR VP HL C+ ++++TA W CRT +++ +P+E+VR++CIVNARSRF+
Sbjct: 244 TEVSALRRFVPPHLHNCSTFEVLTAALWRCRTISIKPDPEEEVRVLCIVNARSRFNPQLP 303
Query: 304 SSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERC 363
S GYYGN AF AVTTA++LC N LGYA+EL++K K+ VTE+YM S+AD MVIK R
Sbjct: 304 S---GYYGNAFAFPVAVTTAEKLCKNPLGYALELVKKTKSDVTEEYMKSVADLMVIKGRP 360
Query: 364 LFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG-GTFLVTHKNAKGEEGL 422
FT RT +VSD TRA F EV+FGWG+AVYGG AKGG+G+ G +F + +N KGE G+
Sbjct: 361 HFTVVRTYLVSDVTRAGFGEVDFGWGKAVYGGPAKGGVGAIPGVASFYIPFRNKKGENGI 420
Query: 423 ILPICLPPEDMKRFVKELDDML 444
++PICLP M++FVKELD ML
Sbjct: 421 VVPICLPGFAMEKFVKELDSML 442
>emb|CAA64636.1| hsr201 [Nicotiana tabacum] gi|7489142|pir||T03274 hsr201 protein,
hypersensitivity-related - common tobacco
Length = 460
Score = 525 bits (1353), Expect = e-148
Identities = 261/443 (58%), Positives = 331/443 (73%), Gaps = 8/443 (1%)
Query: 4 SSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPV 63
SS L+FTVRR +PEL+ PA PTP E K LSDIDDQEGLRF +PVI + + SM KDPV
Sbjct: 6 SSELVFTVRRQKPELIAPAKPTPRETKFLSDIDDQEGLRFQIPVIQFYHKDSSMGRKDPV 65
Query: 64 KVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPF 123
KV+K A++ TLV+YYP AGR+REG GRKLMVDCTGEG+MF+EA+AD+TL+QFGD LQPPF
Sbjct: 66 KVIKKAIAETLVFYYPFAGRLREGNGRKLMVDCTGEGIMFVEADADVTLEQFGDELQPPF 125
Query: 124 PCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEI 183
PC +E+L DVP S +++ P+ LIQVTRL+CGGFI AL LNHTM D GL QF++A E+
Sbjct: 126 PCLEELLYDVPDSAGVLNCPLLLIQVTRLRCGGFIFALRLNHTMSDAPGLVQFMTAVGEM 185
Query: 184 ARGANQPSIQPVWNREILMARDPPYITCNHREYEQIL-PPNTYIKEEDTTIVVHQSFFFT 242
ARG + PSI PVW RE+L AR+PP +TC H EY+++ T I +D +VH+SFFF
Sbjct: 186 ARGGSAPSILPVWCRELLNARNPPQVTCTHHEYDEVRDTKGTIIPLDD---MVHKSFFFG 242
Query: 243 SAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANN 302
+ ++A+RR VP HL +C+ ++L+TA W CRT +L+ +P+E+VR +CIVNARSRF N
Sbjct: 243 PSEVSALRRFVPHHLRKCSTFELLTAVLWRCRTMSLKPDPEEEVRALCIVNARSRF---N 299
Query: 303 RSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKER 362
GYYGN AF AVTTA +L N LGYA+EL++K K+ VTE+YM S+AD MV+K R
Sbjct: 300 PPLPTGYYGNAFAFPVAVTTAAKLSKNPLGYALELVKKTKSDVTEEYMKSVADLMVLKGR 359
Query: 363 CLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG-GTFLVTHKNAKGEEG 421
FT RT +VSD TR F EV+FGWG+AVYGG AKGG+G+ G +F + KN KGE G
Sbjct: 360 PHFTVVRTFLVSDVTRGGFGEVDFGWGKAVYGGPAKGGVGAIPGVASFYIPFKNKKGENG 419
Query: 422 LILPICLPPEDMKRFVKELDDML 444
+++PICLP M+ FVKELD ML
Sbjct: 420 IVVPICLPGFAMETFVKELDGML 442
>gb|AAN09796.1| benzoyl coenzyme A: benzyl alcohol benzoyl transferase [Clarkia
breweri]
Length = 456
Score = 497 bits (1279), Expect = e-139
Identities = 257/450 (57%), Positives = 323/450 (71%), Gaps = 10/450 (2%)
Query: 1 MTSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHE-PSMKE 59
M L F V R +PEL+ PA TPHE K LSD++DQEGLRF +PVI ++H SM+E
Sbjct: 1 MAHDQSLSFEVCRRKPELIRPAKQTPHEFKKLSDVEDQEGLRFQIPVIQFYKHNNESMQE 60
Query: 60 KDPVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDAL 119
+DPV+V++ ++R LVYYYP AGR+RE GRKL+V+CTGEGVMFIEA+AD+TL+QFGDAL
Sbjct: 61 RDPVQVIREGIARALVYYYPFAGRLREVDGRKLVVECTGEGVMFIEADADVTLEQFGDAL 120
Query: 120 QPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSA 179
QPPFPCF ++L DVPGS I+D P+ LIQVTRLKCG FI AL LNHTM D +G+ F+ A
Sbjct: 121 QPPFPCFDQLLFDVPGSGGILDSPLLLIQVTRLKCGSFIFALRLNHTMADAAGIVLFMKA 180
Query: 180 WAEIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSF 239
E+ARGA PS PVW+R IL AR PP +T NHREYE++ T D + H+SF
Sbjct: 181 VGEMARGAATPSTLPVWDRHILNARVPPQVTFNHREYEEV--KGTIFTPFDD--LAHRSF 236
Query: 240 FFTSAHIAAIRRLVPLHLSRC-TAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRF 298
FF S I+A+R+ +P HL C T +++TAC W CRT A++ PDE+VRM+CIVNARS+F
Sbjct: 237 FFGSTEISAMRKQIPPHLRSCSTTIEVLTACLWRCRTLAIKPNPDEEVRMICIVNARSKF 296
Query: 299 SANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMV 358
N GYYGN A AAVTTA +LC N LG+A+ELIRKAK +VTE+YMHS+AD MV
Sbjct: 297 ---NPPLPDGYYGNAFAIPAAVTTAGKLCNNPLGFALELIRKAKREVTEEYMHSVADLMV 353
Query: 359 IKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGT-FLVTHKNAK 417
R FT T +VSD TRA F EV+FGWGEAVYGG AKGG+G G T F + +N +
Sbjct: 354 ATGRPHFTVVNTYLVSDVTRAGFGEVDFGWGEAVYGGPAKGGVGVIPGVTSFYIPLRNRQ 413
Query: 418 GEEGLILPICLPPEDMKRFVKELDDMLGNQ 447
GE+G++LPICLP M+ F + L++ L +
Sbjct: 414 GEKGIVLPICLPSAAMEIFAEALNNTLNGK 443
>gb|AAP54496.1| putative hypersensitivity-related (hsr)protein [Oryza sativa
(japonica cultivar-group)] gi|37535814|ref|NP_922209.1|
putative hypersensitivity-related (hsr)protein [Oryza
sativa (japonica cultivar-group)]
gi|10140797|gb|AAG13627.1| putative
hypersensitivity-related (hsr)protein [Oryza sativa
(japonica cultivar-group)]
Length = 553
Score = 449 bits (1154), Expect = e-124
Identities = 232/449 (51%), Positives = 309/449 (68%), Gaps = 8/449 (1%)
Query: 2 TSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
++++ L FTVRR ELV PA PTP E+K LSDIDDQ+GLRF++PVI +R +M +D
Sbjct: 4 STAAALKFTVRRKPAELVAPAGPTPRELKKLSDIDDQDGLRFHIPVIQFYRRSAAMGGRD 63
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQP 121
P V++ A++R LV YYP AGR+RE GRKL VDCTGEGV+FIEA+AD+ L+ FG ALQP
Sbjct: 64 PAPVIRAAVARALVSYYPFAGRLRELEGRKLAVDCTGEGVLFIEADADVRLEHFGGALQP 123
Query: 122 PFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWA 181
PFPC +E++ DVPGS ++ P+ L QVTRL CGGFILA+ L+HTM D GL QF+ A A
Sbjct: 124 PFPCLEELVFDVPGSSEVLGSPLLLFQVTRLACGGFILAVRLHHTMADAQGLVQFLGAVA 183
Query: 182 EIARG--ANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSF 239
E+ARG A PS+ PVW RE+L AR PP HREY+++ P+T + H+SF
Sbjct: 184 EMARGGAAAAPSVAPVWGREMLEARSPPRPAFAHREYDEV--PDTKGTIIPLDDMAHRSF 241
Query: 240 FFTSAHIAAIR-RLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRF 298
FF + +AA+R L P R T ++++T C W CRT AL + DE +RM+CIVNAR
Sbjct: 242 FFGAREVAAVRSHLAPGIRERATTFEVLTGCLWRCRTAALAPDDDEVMRMICIVNARGGG 301
Query: 299 SANNRSSLI--GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADF 356
+ + +I GYYGN AF AV TA EL LGYAVEL+R AK +V+ +YM S+AD
Sbjct: 302 KSGGGAGMIPEGYYGNAFAFPVAVATAGELRARPLGYAVELVRAAKGEVSVEYMRSVADL 361
Query: 357 MVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG-GTFLVTHKN 415
MV + R FT R +VSD T+A F +++FGWG+ YGG AKGG+G+ G +FL+ KN
Sbjct: 362 MVQRGRPHFTVVRAYLVSDVTKAGFGDLDFGWGKPAYGGPAKGGVGAIPGVASFLIPFKN 421
Query: 416 AKGEEGLILPICLPPEDMKRFVKELDDML 444
AKGE+G+++P+CLP M +FV+E+ ++
Sbjct: 422 AKGEDGIVVPMCLPGPAMDKFVEEMGKLM 450
>dbj|BAD89275.1| (-)-13alpha-hydroxymultiflorine/(+)-13alpha- hydroxylupanine
O-tigloyltransferase [Lupinus albus]
Length = 453
Score = 442 bits (1136), Expect = e-122
Identities = 231/440 (52%), Positives = 307/440 (69%), Gaps = 11/440 (2%)
Query: 7 LLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVL 66
L+F VRR+ ELV PA PTP E KLLSDIDDQ LR P++ ++R+ PSM+ KDPV+++
Sbjct: 8 LVFKVRRNPQELVTPAKPTPKEFKLLSDIDDQTSLRSLTPLVTIYRNNPSMEGKDPVEII 67
Query: 67 KHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCF 126
+ ALS+TLV+YYP AGR+R G KLMVDCTGEGV+FIEA+AD+TLDQFG L PPFPCF
Sbjct: 68 REALSKTLVFYYPFAGRLRNGPNGKLMVDCTGEGVIFIEADADVTLDQFGIDLHPPFPCF 127
Query: 127 QEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARG 186
++L DVPGS+ I+D P+ LIQVTRLKCGGFI A+ LNH M D G+ QF+ AEIARG
Sbjct: 128 DQLLYDVPGSDGILDSPLLLIQVTRLKCGGFIFAVRLNHAMCDAIGMSQFMKGLAEIARG 187
Query: 187 ANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHI 246
+P I PVW+RE+L AR+PP +T H EY++ PP+ + I+ H SFFF +
Sbjct: 188 EPKPFILPVWHRELLCARNPPKVTFIHNEYQK--PPHD--NNNNNFILQHSSFFFGPNEL 243
Query: 247 AAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVE-PDEDVRMMCIVNARSRFSANNRSS 305
AIRRL+P H S+ T D++TA W CRT ALQ E P+ + R++ I+NAR + N
Sbjct: 244 DAIRRLLPYHHSKSTTSDILTAFLWRCRTLALQPENPNHEFRLLYILNARYGRCSFNPPL 303
Query: 306 LIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKER--C 363
G+YGN AA++T ++LC N L YA+EL+++AK++ TE+Y+HS+AD MVIK R
Sbjct: 304 PEGFYGNAFVSPAAISTGEKLCNNPLEYALELMKEAKSKGTEEYVHSVADLMVIKGRPSY 363
Query: 364 LFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLI 423
+ VSD T+A+F +V+FGWG+AVYGGA +G S L V++ N+KG EG++
Sbjct: 364 FYNDVGYLEVSDLTKARFRDVDFGWGKAVYGGATQGYFSSIL----YVSYTNSKGVEGIM 419
Query: 424 LPICLPPEDMKRFVKELDDM 443
LP + M+RF KELDD+
Sbjct: 420 ALTSLPTKAMERFEKELDDL 439
>gb|AAS48090.1| alcohol acyl transferase [Pyrus communis]
Length = 442
Score = 441 bits (1134), Expect = e-122
Identities = 228/436 (52%), Positives = 304/436 (69%), Gaps = 13/436 (2%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSM-KEKDPVKVLKHA 69
V+R QPEL+ PA PTP E K LSDIDDQEGLRF LPVI ++ PS+ K ++P+KV+K A
Sbjct: 9 VKRLQPELITPAKPTPQETKFLSDIDDQEGLRFQLPVIMCYKDNPSLNKNRNPIKVIKEA 68
Query: 70 LSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEI 129
LSR LVYYYP AGR+REG RKLMV+C GEG++F+EA AD+TL+Q GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLREGPNRKLMVNCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 130 LCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
L + PGS+ II P+ L+QVT L CGGFILAL LNHTM D +GL F++A E+ RGA+
Sbjct: 129 LFNFPGSDGIIGCPLLLVQVTCLTCGGFILALRLNHTMCDATGLLMFLTAITEMGRGADA 188
Query: 190 PSIQPVWNREILMARDPPYITCNHREYEQIL--PPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW RE+L ARDPP ITC H EYE ++ +Y + +V +SF+F + +
Sbjct: 189 PSILPVWERELLFARDPPRITCAHYEYEDVIDHSDGSYAFSNQSN-MVQRSFYFGAKEMR 247
Query: 248 AIRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL 306
+R+ +P HL S C+ +DLITAC W CRT L++ P + VR+ CIVNAR + NN
Sbjct: 248 VLRKQIPPHLISTCSTFDLITACLWKCRTLVLKINPKQAVRVSCIVNARGKH--NNVHIP 305
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFT 366
+GYYGN AF AAV+ A+ LC N LGYA+EL++KAKA + E+Y+ S+AD +V++ R ++
Sbjct: 306 LGYYGNAFAFPAAVSKAEPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYS 365
Query: 367 K-GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILP 425
G +VSD TRA F +VNFGWG+ V+ G AK + +F V HKN E+G+++P
Sbjct: 366 STGSYLIVSDNTRAGFGDVNFGWGQPVFAGPAK----ALDLISFYVQHKN-NIEDGILVP 420
Query: 426 ICLPPEDMKRFVKELD 441
+CLP M+RF +EL+
Sbjct: 421 MCLPSSAMERFQQELE 436
>gb|AAR99826.1| alcohol acyl transferase [Malus x domestica]
Length = 470
Score = 438 bits (1127), Expect = e-121
Identities = 228/436 (52%), Positives = 302/436 (68%), Gaps = 13/436 (2%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSM-KEKDPVKVLKHA 69
VRR Q EL+ PA PTP E K LSDIDDQEGLRF +PVI ++ PS+ K +PVKV++ A
Sbjct: 9 VRRLQLELITPAKPTPQETKFLSDIDDQEGLRFQVPVIMCYKDNPSLNKNCNPVKVIREA 68
Query: 70 LSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEI 129
LSR LVYYYP AGR++EG RKLMVDC GEG++F+EA AD+TL+Q GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLKEGPNRKLMVDCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 130 LCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
L + PGS+ II P+ L+QVT L CGGFILAL NHTM D GL F++A AE+ARGA+
Sbjct: 129 LFNFPGSDGIIGCPLLLVQVTCLTCGGFILALRANHTMCDAPGLLLFLTAIAEMARGAHA 188
Query: 190 PSIQPVWNREILMARDPPYITCNHREYEQIL--PPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW RE+L ARDPP ITC H EYE ++ +Y + +V +SF+F + +
Sbjct: 189 PSILPVWERELLFARDPPRITCAHHEYEDVIGHSDGSYASSNQSN-MVQRSFYFGAKEMR 247
Query: 248 AIRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL 306
+R+ +P HL S C+ +DLITAC W CRT AL + P E VR+ CIVNAR + NN
Sbjct: 248 VLRKQIPPHLISTCSTFDLITACLWKCRTLALNINPKEAVRVSCIVNARGKH--NNVRLP 305
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFT 366
+GYYGN AF AAV+ A+ LC N +GYA+EL++KAKA + E+Y+ S+AD +V++ R ++
Sbjct: 306 LGYYGNAFAFPAAVSKAEPLCKNPMGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYS 365
Query: 367 K-GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILP 425
G +VSD TRA F +VNFGWG+ ++ G AK + +F V HKN E+G+++P
Sbjct: 366 STGSYLIVSDNTRAGFGDVNFGWGQPIFAGPAK----ALDLISFYVQHKN-NSEDGILVP 420
Query: 426 ICLPPEDMKRFVKELD 441
+CLP M+RF +EL+
Sbjct: 421 MCLPFSAMERFQQELE 436
>gb|AAU14879.2| alcohol acyl transferase [Malus x domestica]
Length = 455
Score = 437 bits (1123), Expect = e-121
Identities = 225/436 (51%), Positives = 301/436 (68%), Gaps = 13/436 (2%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSM-KEKDPVKVLKHA 69
V+R QPEL+ PA TP E K LSDIDDQE LR +P+I ++ PS+ K ++PVK ++ A
Sbjct: 9 VKRLQPELITPAKSTPQETKFLSDIDDQESLRVQIPIIMCYKDNPSLNKNRNPVKAIREA 68
Query: 70 LSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEI 129
LSR LVYYYP AGR+REG RKL+VDC GEG++F+EA AD+TL+Q GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLREGPNRKLVVDCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 130 LCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
L + PGS+ IID P+ LIQVT L CGGFILAL LNHTM D +GL F++A AE+ARGA+
Sbjct: 129 LYNFPGSDGIIDCPLLLIQVTCLTCGGFILALRLNHTMCDAAGLLLFLTAIAEMARGAHA 188
Query: 190 PSIQPVWNREILMARDPPYITCNHREYEQIL--PPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW RE+L ARDPP ITC H EYE ++ +Y + +V +SF+F + +
Sbjct: 189 PSILPVWERELLFARDPPRITCAHHEYEDVIGHSDGSYASSNQSN-MVQRSFYFGAKEMR 247
Query: 248 AIRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL 306
+R+ +P HL S C+ +DLITAC W CRT AL + P E VR+ CIVNAR + NN
Sbjct: 248 VLRKQIPPHLISTCSTFDLITACLWKCRTLALNINPKEAVRVSCIVNARGKH--NNVRLP 305
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFT 366
+GYYGN AF AA++ A+ LC N LGYA+EL++KAKA + E+Y+ S+AD +V++ R ++
Sbjct: 306 LGYYGNAFAFPAAISKAEPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYS 365
Query: 367 K-GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILP 425
G +VSD TR F +VNFGWG+ V+ G K + +F V HKN E+G+++P
Sbjct: 366 STGSYLIVSDNTRVGFGDVNFGWGQPVFAGPVK----ALDLISFYVQHKN-NTEDGILVP 420
Query: 426 ICLPPEDMKRFVKELD 441
+CLP M+RF +EL+
Sbjct: 421 MCLPSSAMERFQQELE 436
>gb|AAS79797.1| alcohol acyl transferase [Malus x domestica]
Length = 459
Score = 434 bits (1116), Expect = e-120
Identities = 225/435 (51%), Positives = 303/435 (68%), Gaps = 11/435 (2%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSM-KEKDPVKVLKHA 69
V+R Q EL+ PA PT E K LSDIDDQEGLRF +PVI ++ PS+ K +PVKV++ A
Sbjct: 9 VKRLQLELITPAKPTLQEAKFLSDIDDQEGLRFQVPVIMCYKDNPSLNKNCNPVKVIREA 68
Query: 70 LSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEI 129
LSR LVYYYP AGR++EG RKLMVDC GEG++F+EA AD+TL+Q GD + PP P +E
Sbjct: 69 LSRALVYYYPLAGRLKEGPNRKLMVDCNGEGILFVEASADVTLEQLGDKILPPCPLLEEF 128
Query: 130 LCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
L + PGS+ II P+ L+QVT L CGGFILAL +NHTM D GL F++A AE+ARGA+
Sbjct: 129 LFNFPGSDGIIGCPLLLVQVTCLTCGGFILALRVNHTMCDAPGLLLFLTAIAEMARGAHA 188
Query: 190 PSIQPVWNREILMARDPPYITCNHREYEQILPPNTYI-KEEDTTIVVHQSFFFTSAHIAA 248
PSI PVW RE+L +RDPP ITC H EYE ++ + + + + +V +SF+F + +
Sbjct: 189 PSILPVWERELLFSRDPPRITCAHHEYEDVIDHSDGLYASSNQSNMVQRSFYFGAKEMRV 248
Query: 249 IRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+R+ +P HL S C+ +DLITAC W CRT AL + P E VR+ CIVNAR + NN +
Sbjct: 249 LRKQIPPHLISTCSTFDLITACLWKCRTLALNINPKEAVRVSCIVNARGKH--NNVRLPL 306
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
GYYGN AF AA++ A+ LC N LGYA+EL++KAKA + E+Y+ S+AD +V++ R ++
Sbjct: 307 GYYGNAFAFPAAISKAEPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYSS 366
Query: 368 -GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPI 426
G +VSD TRA F +VNFGWG+ V+ G AK + +F V HKN E+G+++P+
Sbjct: 367 TGSYLIVSDNTRAGFGDVNFGWGQPVFAGPAK----ALDLISFYVQHKN-NTEDGILVPM 421
Query: 427 CLPPEDMKRFVKELD 441
CLP M+RF +EL+
Sbjct: 422 CLPSSAMERFQQELE 436
>gb|AAL77060.1| putative acyltransferase [Cucumis melo]
Length = 461
Score = 433 bits (1113), Expect = e-120
Identities = 210/430 (48%), Positives = 299/430 (68%), Gaps = 9/430 (2%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F VR+ QPEL+ PA PTP+E K LSD+DDQ+ LRF LP++ ++ H PS++ +DPVKV+K
Sbjct: 11 FQVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRFQLPLVNIYHHNPSLEGRDPVKVIKE 70
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQ- 127
A+++ LV+YYP AGR+REG GRKL V+CTGEG++FIEA+AD++L+QF D L +
Sbjct: 71 AIAKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEQFRDTLPYSLSSMEN 130
Query: 128 EILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
I+ + S+ +++ P+ LIQVTRLKCGGFI + +HTM DG G+ QF+ A AEIARGA
Sbjct: 131 NIIHNSLNSDGVLNSPLLLIQVTRLKCGGFIFGIHFDHTMADGFGIAQFMKAIAEIARGA 190
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW R +L ARDPP IT H EY+Q++ + + + ++ + FFFT I+
Sbjct: 191 FAPSILPVWQRALLTARDPPRITVRHYEYDQVVDTKSTLIPANN--MIDRLFFFTQRQIS 248
Query: 248 AIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+R+ +P HL C++++++ A W RT A Q++P+E+VR +C+VN RS+ +
Sbjct: 249 TLRQTLPAHLHDCSSFEVLAAYVWRLRTIAFQLKPEEEVRFLCVVNLRSKIDIP-----L 303
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
G+YGN I F A +TT +LCGN LGYAV+LIRKAKA+ T++Y+ S+ DFMVIK R FT+
Sbjct: 304 GFYGNAIVFPAVITTVAKLCGNPLGYAVDLIRKAKAKATKEYIKSMVDFMVIKGRPRFTE 363
Query: 368 GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGG-TFLVTHKNAKGEEGLILPI 426
M+SD TR F V+FGWG+A++GG GG G G ++ + N GE+G+++P+
Sbjct: 364 IGPFMMSDITRIGFENVDFGWGKAIFGGPIIGGCGIIRGMISYSIAFMNRNGEKGIVVPL 423
Query: 427 CLPPEDMKRF 436
CLPP M+RF
Sbjct: 424 CLPPPAMERF 433
>dbj|BAB78588.1| alcohol acetyltransferase [Cucumis melo]
Length = 461
Score = 431 bits (1109), Expect = e-119
Identities = 211/430 (49%), Positives = 296/430 (68%), Gaps = 9/430 (2%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F VR+ QPEL+ PA PTP+E K LSD+DDQ+ LR LP + ++ H PS++ +DPVKV+K
Sbjct: 11 FHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSLEGRDPVKVIKE 70
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQ- 127
A+ + LV+YYP AGR+REG GRKL V+CTGEG++FIEA+AD++L++F D L Q
Sbjct: 71 AIGKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEEFWDTLPYSLSSMQN 130
Query: 128 EILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
I+ + S+ +++ P+ LIQVTRLKCGGFI L NHTM DG G+ QF+ A AEIARGA
Sbjct: 131 NIIHNALNSDEVLNSPLLLIQVTRLKCGGFIFGLCFNHTMADGFGIVQFMKATAEIARGA 190
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW R +L ARDPP IT H EY+Q++ + + + ++ + FFFT I+
Sbjct: 191 FAPSILPVWQRALLTARDPPRITVRHYEYDQVVDTKSTLIPANN--MIDRLFFFTQRQIS 248
Query: 248 AIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+R+ +P HL C++++++ A W RT A Q++P+E+VR +C+VN RS+ +
Sbjct: 249 TLRQTLPAHLHDCSSFEVLAAYVWRLRTIAFQLKPEEEVRFLCVVNLRSKIDIP-----L 303
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
G+YGN I F A +TT +LCGN LGYAV+LIRKAKA+ T++Y+ S+ DFMVIK R FT+
Sbjct: 304 GFYGNAIVFPAVITTIAKLCGNPLGYAVDLIRKAKAKATKEYIKSMVDFMVIKGRPRFTE 363
Query: 368 GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGG-TFLVTHKNAKGEEGLILPI 426
M+SD TR F V+FGWG+A++GG GG G G ++ + N GE+G+++P+
Sbjct: 364 IGPFMMSDITRIGFENVDFGWGKAIFGGPIIGGCGIIRGMISYSIAFMNRNGEKGIVVPL 423
Query: 427 CLPPEDMKRF 436
CLPP M+RF
Sbjct: 424 CLPPPAMERF 433
>emb|CAA94432.1| unknown [Cucumis melo] gi|7484744|pir||T09666 probable anthranilate
N-benzoyltransferase (EC 2.3.1.144) - muskmelon
(fragment)
Length = 455
Score = 416 bits (1068), Expect = e-114
Identities = 211/430 (49%), Positives = 287/430 (66%), Gaps = 9/430 (2%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F VR+ QPEL+ PA PTP+E K LSD+DDQ+ LR LP + ++ H PS++ +DPVKV+K
Sbjct: 4 FHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSLEGRDPVKVIKE 63
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQ- 127
A+ + LV+YYP AGR+REG GRKL V+CTGEG++FIEA+AD++L++F D L Q
Sbjct: 64 AIGKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEEFWDTLPYSLSSMQN 123
Query: 128 EILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
I+ + S+ +++ P+ LIQVTRLKCGGFI L NHTM DG G+ QF+ A AEIARGA
Sbjct: 124 NIIHNALNSDEVLNSPLLLIQVTRLKCGGFIFGLCFNHTMADGFGIVQFMKATAEIARGA 183
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW R +L ARDPP IT H EY+Q++ + + ++ I Q FFF+ I+
Sbjct: 184 FAPSILPVWQRALLTARDPPRITFRHYEYDQVVDMKSGLIPVNSKI--DQLFFFSQLQIS 241
Query: 248 AIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+R+ +P HL C +++++TA W RT ALQ +P+E+VR +C++N RS+ +
Sbjct: 242 TLRQTLPAHLHDCPSFEVLTAYVWRLRTIALQFKPEEEVRFLCVMNLRSKIDIP-----L 296
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
GYYGN + A +TTA +LCGN LGYAV+LIRKAKA+ T +Y+ S D MVIK R FT
Sbjct: 297 GYYGNAVVVPAVITTAAKLCGNPLGYAVDLIRKAKAKATMEYIKSTVDLMVIKGRPYFTV 356
Query: 368 GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG-GTFLVTHKNAKGEEGLILPI 426
+ M+SD TR V+FGWG+A++GG G G +F V N GE+G L +
Sbjct: 357 VGSFMMSDLTRIGVENVDFGWGKAIFGGPTTTGARITRGLVSFCVPFMNRNGEKGTALSL 416
Query: 427 CLPPEDMKRF 436
CLPP M+RF
Sbjct: 417 CLPPPAMERF 426
>dbj|BAD53644.1| putative benzoyl coenzyme A, benzyl alcohol benzoyl transferase
[Oryza sativa (japonica cultivar-group)]
Length = 450
Score = 408 bits (1049), Expect = e-112
Identities = 219/444 (49%), Positives = 281/444 (62%), Gaps = 9/444 (2%)
Query: 3 SSSPL-LFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
+SSPL FTVRR +P LV PAAPTP EVK LSDIDD EG+RF I ++R+ P+ K +D
Sbjct: 2 ASSPLPAFTVRRGEPVLVTPAAPTPREVKALSDIDDGEGMRFYSSGIHLYRNNPAKKGQD 61
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQP 121
P V++ AL+R LV YYP AGR+RE AGRKL+V+C G+GVMF EA+AD+T D FGD P
Sbjct: 62 PAMVIREALARALVPYYPLAGRLREEAGRKLVVECAGQGVMFAEADADLTADDFGDVQSP 121
Query: 122 PFPCFQEILCD---VPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVS 178
PFPCF+ + + V G E ++ RP+ IQVTRL+CGGFI H + D G QF
Sbjct: 122 PFPCFERFILESTTVAGVEPVVGRPLLYIQVTRLRCGGFIFGQRFCHCVVDAPGGMQFEK 181
Query: 179 AWAEIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQS 238
A E+ARGA PS+ P W RE+ MARDPP + H EY + + +V
Sbjct: 182 AVCELARGAAAPSVSPSWGREMFMARDPPRPSYPHLEYREPAGGADRLLATPPEDMVRVP 241
Query: 239 FFFTSAHIAAIRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSR 297
FFF IA +R+ P + C+ ++L+ AC W RT AL P E+VR+ IVNAR R
Sbjct: 242 FFFGPREIAGLRQHAPASVRGACSRFELVAACIWRSRTAALGYAPGEEVRLSFIVNARGR 301
Query: 298 FSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFM 357
G+YGN A+S A TTA ELCG LGYA+ L++KAK+ VT +Y+ S+AD M
Sbjct: 302 ADVPLPE---GFYGNAFAYSVAATTAGELCGGDLGYALGLVKKAKSAVTYEYLQSVADLM 358
Query: 358 VIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGT-FLVTHKNA 416
V+ R LF RT +VSD + A F V+FGWGEAVYGG AKGG G LG T + KN
Sbjct: 359 VVAGRPLFALSRTYIVSDVSHAGFKSVDFGWGEAVYGGPAKGGEGPLLGVTNYFSRSKNG 418
Query: 417 KGEEGLILPICLPPEDMKRFVKEL 440
KGE+ +++PICLP + M +F E+
Sbjct: 419 KGEQSVVVPICLPKDAMDKFQLEV 442
>dbj|BAC58010.1| alcohol acyltransferase [Cucumis melo]
Length = 461
Score = 404 bits (1039), Expect = e-111
Identities = 201/430 (46%), Positives = 282/430 (64%), Gaps = 9/430 (2%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F VR+ QPEL+ PA PTP+E K LSD+DDQ+ LR LP + ++ H PS++ +DPVKV+K
Sbjct: 11 FHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSLEGRDPVKVIKE 70
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQ- 127
A+ + LV YYP AGR+REG GRKL V+CTGEG++FIEA+AD++L+QF D L +
Sbjct: 71 AIGKALVLYYPLAGRLREGPGRKLFVECTGEGILFIEADADVSLEQFRDTLPYSLSSMEN 130
Query: 128 EILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
I+ + S+ +++ + LIQVTRLKCGGFI + NHTM DG G+ QF+ A AEIARGA
Sbjct: 131 NIIHNALNSDGVLNSQLLLIQVTRLKCGGFIFGIRFNHTMADGFGIAQFMKAIAEIARGA 190
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
PSI PVW R +L AR IT H EY+Q++ + + + ++ + FFF+ I+
Sbjct: 191 FAPSILPVWQRALLTARYLSEITVRHYEYDQVVDTKSTLIPANN--MIDRLFFFSQLQIS 248
Query: 248 AIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+R+ +P H C++++++ C W RT Q++P+EDVR +C+VN RS+ +
Sbjct: 249 TLRQTLPAHRHDCSSFEVLADCVWRLRTIGFQLKPEEDVRFLCVVNLRSKIDIP-----L 303
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
GYYGN + F A +TTA +LCGN LGYAV+LIRKAKA+ T + DFMVIK R FT+
Sbjct: 304 GYYGNAVVFPAVITTAAKLCGNPLGYAVDLIRKAKAKATTHLTSPMVDFMVIKGRPRFTE 363
Query: 368 GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGG-TFLVTHKNAKGEEGLILPI 426
M+SD TR F V+FGWG+A++GG GG G G ++ + N + L++P+
Sbjct: 364 LMRFMMSDITRIGFENVDFGWGKAIFGGPIIGGCGIIRGMISYSIAFMNRHNKNSLVVPL 423
Query: 427 CLPPEDMKRF 436
CLPP DM+ F
Sbjct: 424 CLPPPDMESF 433
>emb|CAC01898.1| putative protein [Arabidopsis thaliana]
gi|15237985|ref|NP_197256.1| transferase family protein
[Arabidopsis thaliana] gi|11357934|pir||T51458
hypothetical protein K10A8_20 - Arabidopsis thaliana
Length = 461
Score = 403 bits (1036), Expect = e-111
Identities = 219/466 (46%), Positives = 304/466 (64%), Gaps = 13/466 (2%)
Query: 4 SSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPV 63
S L F + R +PELV PA PTP E+K LSDIDDQEGLRF++P IF +RH P+ DPV
Sbjct: 2 SGSLTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPTTNS-DPV 60
Query: 64 KVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFG--DALQP 121
V++ AL+ TLVYYYP AGR+REG RKL VDCTGEGV+FIEA+AD+TL +F DAL+P
Sbjct: 61 AVIRRALAETLVYYYPFAGRLREGPNRKLAVDCTGEGVLFIEADADVTLVEFEEKDALKP 120
Query: 122 PFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWA 181
PFPCF+E+L +V GS +++ P+ L+QVTRLKCGGFI A+ +NH M D GL F+
Sbjct: 121 PFPCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMC 180
Query: 182 EIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFF 241
E RG + P++ PVW R +L AR +T HREY+++ T + + V +S FF
Sbjct: 181 EFVRGYHAPTVAPVWERHLLSARVLLRVTHAHREYDEMPAIGTELGSRRDNL-VGRSLFF 239
Query: 242 TSAHIAAIRRLVPLHL-SRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSA 300
++AIRRL+P +L + T +++T+ W RT AL+ + D+++R++ IVNARS+
Sbjct: 240 GPCEMSAIRRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNARSKL-- 297
Query: 301 NNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIK 360
N GYYGN AF A+ TA EL L +A+ LI++AK+ VTE+YM SLAD MVIK
Sbjct: 298 KNPPLPRGYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADLMVIK 357
Query: 361 ERCLFTKGRTCMVSDWTRAKFSEVNFG-WGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGE 419
R F+ +VSD F++++FG WG+ VYGG G+ G +F V+ + GE
Sbjct: 358 GRPSFSSDGAYLVSD--VRIFADIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFEKRNGE 415
Query: 420 EGLILPICLPPEDMKRFVKELDDMLGNQ---NYPTMSGPSFILSTL 462
G+++P+CLP + M+RFV+EL+ + Q N + S I+S+L
Sbjct: 416 IGIVVPVCLPEKAMQRFVEELEGVFNGQVVFNRGSKSSNKLIMSSL 461
>gb|AAF01587.1| putative hypersensitivity-related gene [Arabidopsis thaliana]
gi|22854878|gb|AAN09797.1| acetyl coenzyme A:
cis-3-hexen-1-ol acetyl transferase [Arabidopsis
thaliana] gi|15228568|ref|NP_186998.1| transferase
family protein [Arabidopsis thaliana]
Length = 454
Score = 373 bits (957), Expect = e-102
Identities = 204/447 (45%), Positives = 285/447 (63%), Gaps = 11/447 (2%)
Query: 2 TSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
++++ L F V R Q ELV PA PTP E+K LSDIDDQ+GLRF +PVIF +R S + D
Sbjct: 10 STTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLS-SDLD 68
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQF--GDAL 119
PV+V+K AL+ LVYYYP AGR+RE + RKL VDCTGEGV+FIEAEAD+ L + DAL
Sbjct: 69 PVQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADAL 128
Query: 120 QPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSA 179
PPFP +E+L DV GS +++ P+ L+QVTRLKC GFI AL NHTM DG+GL F+ +
Sbjct: 129 LPPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKS 188
Query: 180 WAEIARGANQPSIQPVWNREIL-MARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQS 238
E+A G + PS+ PVWNR +L ++ +T HREY+ + + +V +S
Sbjct: 189 LCELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGIDVVATGHP---LVSRS 245
Query: 239 FFFTSAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRF 298
FFF + I+AIR+L+P L T+++ +++ W CRT AL +P+ ++R+ CI+N+RS+
Sbjct: 246 FFFRAEEISAIRKLLPPDLHN-TSFEALSSFLWRCRTIALNPDPNTEMRLTCIINSRSKL 304
Query: 299 SANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMV 358
N GYYGN AA+ TA++L L +A+ LI++ K+ VTE Y+ S+ M
Sbjct: 305 --RNPPLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVRSVTALMA 362
Query: 359 IKERCLFTKGRTCMVSDWTRAKFSEVNFG-WGEAVYGGAAKGGIGSFLGGTFLVTHKNAK 417
+ R +F ++SD +++FG WG+ VYGG AK GI F G +F V KN K
Sbjct: 363 TRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKNKK 422
Query: 418 GEEGLILPICLPPEDMKRFVKELDDML 444
GE G ++ I LP M+ FV EL+ +L
Sbjct: 423 GETGTVVAISLPVRAMETFVAELNGVL 449
>gb|AAM61186.1| putative hypersensitivity-related gene [Arabidopsis thaliana]
Length = 454
Score = 371 bits (953), Expect = e-101
Identities = 203/447 (45%), Positives = 284/447 (63%), Gaps = 11/447 (2%)
Query: 2 TSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
++++ L F V R Q ELV PA PTP E+K LSDIDDQ+GLRF +PVIF +R S + D
Sbjct: 10 STTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLS-SDLD 68
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQF--GDAL 119
PV+V+K AL+ LVYYYP AGR+RE + RKL VDCTGEGV+FIEAEAD+ L + DAL
Sbjct: 69 PVQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADAL 128
Query: 120 QPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSA 179
PPFP +E+L DV GS +++ P+ L+QVTRLKC GFI AL NHTM DG+GL F+ +
Sbjct: 129 LPPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKS 188
Query: 180 WAEIARGANQPSIQPVWNREIL-MARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQS 238
E+A G + PS+ PVWNR +L ++ +T HREY+ + + +V +S
Sbjct: 189 LCELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGIDVVATGHP---LVSRS 245
Query: 239 FFFTSAHIAAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRF 298
FFF + I+AIR+L+P L T+++ +++ W CRT AL +P+ ++R+ CI+N+RS+
Sbjct: 246 FFFRAEEISAIRKLLPPDLHN-TSFEALSSFLWRCRTIALNPDPNTEMRLTCIINSRSKL 304
Query: 299 SANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMV 358
N GYYGN AA+ TA++L L + + LI++ K+ VTE Y+ S+ M
Sbjct: 305 --RNPPLEPGYYGNVFVIPAAIATARDLIEKPLEFVLRLIQETKSSVTEDYVRSVTALMA 362
Query: 359 IKERCLFTKGRTCMVSDWTRAKFSEVNFG-WGEAVYGGAAKGGIGSFLGGTFLVTHKNAK 417
+ R +F ++SD +++FG WG+ VYGG AK GI F G +F V KN K
Sbjct: 363 TRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKNKK 422
Query: 418 GEEGLILPICLPPEDMKRFVKELDDML 444
GE G ++ I LP M+ FV EL+ +L
Sbjct: 423 GETGTVVAISLPVRAMETFVAELNGVL 449
>ref|XP_475094.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|46576045|gb|AAT01406.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 464
Score = 361 bits (927), Expect = 2e-98
Identities = 199/446 (44%), Positives = 282/446 (62%), Gaps = 12/446 (2%)
Query: 1 MTSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEK 60
M + L F+VRR + ELV PA PTP+E K+LSDIDDQ+ LRFN I +RH PS
Sbjct: 1 MAGAPTLAFSVRRRERELVAPAKPTPYEFKMLSDIDDQDILRFNRSGILFYRHSPSKDGL 60
Query: 61 DPVKVLKHALSRTLVYYYPGAGRIRE-GAGRKLMVDCTGEGVMFIEAEADITLDQFGDAL 119
DPVKV+K A+S TLV++YP AGR RE RKL+V+CTGEGV+F+EA+A+ +D+ G +L
Sbjct: 61 DPVKVIKAAISETLVHFYPVAGRFRELRPTRKLVVECTGEGVVFVEADANFRMDELGTSL 120
Query: 120 QPPFPCFQEILCDVPG-SEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVS 178
PP PC+ +LC+ + ++DRP+ IQVTRL CGGF+ + + H M DGSG+ QF++
Sbjct: 121 APPVPCYDMLLCEPESPTADVVDRPLLFIQVTRLACGGFVFGMHICHCMADGSGIVQFLT 180
Query: 179 AWAEIARGAN-QPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQ 237
A E ARG + P+++PVW RE+L AR PP +T +H EY + P + T H
Sbjct: 181 ALTEFARGVHGAPTVRPVWEREVLTARWPPTVTRDHVEYTPLPNPGKDVL-SPTDAYAHH 239
Query: 238 SFFFTSAHIAAIRRLVPLHLSRCTA-YDLITACYWCCRTKALQVEPDEDVRMMCIVNARS 296
FFF ++ IAA+R P L ++ +DL+ A W CRT AL+ +P + VR+ VNAR
Sbjct: 240 VFFFGASEIAALRSQAPPDLRAVSSRFDLVGAFMWRCRTAALRYDPGDVVRLHMFVNARV 299
Query: 297 RFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEK-YMHSLAD 355
R + + R GYYGN I F+AA A EL GYA+ L+ +AKA+ +E+ Y+ S+A+
Sbjct: 300 R-NRSKRPVPRGYYGNAIVFAAASVPAGELWRRPFGYALRLLMQAKARASEEGYVQSVAN 358
Query: 356 FMVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKN 415
F R F K RT ++SD T+A ++FGWG+ VYGG A + + TF + +N
Sbjct: 359 FNAAHRRPPFPKARTYLISDMTQAGLMAIDFGWGKPVYGGPA-----TTMLATFHLEGRN 413
Query: 416 AKGEEGLILPICLPPEDMKRFVKELD 441
GE G+I+PI LP ++R ++E++
Sbjct: 414 EVGEAGVIVPIRLPNPVIERLIQEVN 439
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 823,140,981
Number of Sequences: 2540612
Number of extensions: 35558218
Number of successful extensions: 69016
Number of sequences better than 10.0: 330
Number of HSP's better than 10.0 without gapping: 221
Number of HSP's successfully gapped in prelim test: 109
Number of HSP's that attempted gapping in prelim test: 67921
Number of HSP's gapped (non-prelim): 394
length of query: 462
length of database: 863,360,394
effective HSP length: 132
effective length of query: 330
effective length of database: 527,999,610
effective search space: 174239871300
effective search space used: 174239871300
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148345.12


