

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.12 - phase: 0
(214 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
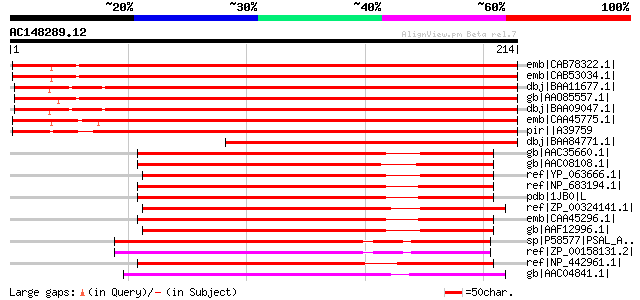
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB78322.1| probable photosystem I chain XI precursor [Arabi... 355 4e-97
emb|CAB53034.1| photosystem I subunit XI precursor [Arabidopsis ... 352 5e-96
dbj|BAA11677.1| subunit of photosystem I [Cucumis sativus] 350 2e-95
gb|AAO85557.1| photosystem I subunit XI [Nicotiana attenuata] 350 2e-95
dbj|BAA09047.1| PsaL [Cucumis sativus] gi|1363479|pir||JC4281 ph... 349 3e-95
emb|CAA45775.1| subunit XI of photosystem I reaction center [Spi... 344 1e-93
pir||A39759 photosystem I 18K protein precursor - barley gi|1312... 300 1e-80
dbj|BAA84771.1| photosystem I subunit PSI-L [Arabidopsis thaliana] 239 5e-62
gb|AAC35660.1| PSI reaction centre subunit XI [Guillardia theta]... 151 1e-35
gb|AAC08108.1| Photosystem I reaction centre subunit XI [Porphyr... 150 2e-35
ref|YP_063666.1| photosystem I reaction center subunit XI [Graci... 149 7e-35
ref|NP_683194.1| photosystem I subunit XI [Thermosynechococcus e... 148 9e-35
pdb|1JB0|L Chain L, Crystal Structure Of Photosystem I: A Photos... 148 9e-35
ref|ZP_00324141.1| COG0304: 3-oxoacyl-(acyl-carrier-protein) syn... 145 6e-34
emb|CAA45296.1| photosystem I subunit XI [Synechococcus sp.] gi|... 145 6e-34
gb|AAF12996.1| unknown; Photosystem I reaction centre subunit XI... 145 6e-34
sp|P58577|PSAL_ANASP Photosystem I reaction center subunit XI (P... 145 1e-33
ref|ZP_00158131.2| hypothetical protein Avar03006008 [Anabaena v... 144 2e-33
ref|NP_442961.1| photosystem I subunit XI [Synechocystis sp. PCC... 143 3e-33
gb|AAC04841.1| photosystem I subunit XI [Fischerella sp. PCC 760... 142 5e-33
>emb|CAB78322.1| probable photosystem I chain XI precursor [Arabidopsis thaliana]
gi|4586256|emb|CAB40997.1| probable photosystem I chain
XI precursor [Arabidopsis thaliana]
gi|21592939|gb|AAM64889.1| probable photosystem I chain
XI precursor [Arabidopsis thaliana]
gi|20453409|gb|AAM19943.1| AT4g12800/T20K18_150
[Arabidopsis thaliana] gi|17933283|gb|AAL48225.1|
AT4g12800/T20K18_150 [Arabidopsis thaliana]
gi|16649159|gb|AAL24431.1| probable photosystem I chain
XI precursor [Arabidopsis thaliana]
gi|16226622|gb|AAL16216.1| AT4g12800/T20K18_150
[Arabidopsis thaliana] gi|15235490|ref|NP_193016.1|
photosystem I reaction center subunit XI, chloroplast
(PSI-L) / PSI subunit V [Arabidopsis thaliana]
gi|7488011|pir||T06638 photosystem I chain XI precursor
- Arabidopsis thaliana gi|18203449|sp|Q9SUI4|PSAL_ARATH
Photosystem I reaction center subunit XI, chloroplast
precursor (PSI-L) (PSI subunit V)
Length = 219
Score = 355 bits (912), Expect = 4e-97
Identities = 174/218 (79%), Positives = 197/218 (89%), Gaps = 6/218 (2%)
Query: 2 AAASPMASQLKSSFT-----RPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQV 56
A+ASPMASQL+SSF+ + L P+G+ G+ P P++R +FTV+A++S+K +QV
Sbjct: 3 ASASPMASQLRSSFSSASLSQRLAVPKGISGA-PFGVSPTKRVSSFTVRAVKSDKTTFQV 61
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
+QPINGDPFIGSLETPVTSSPL+AWYLSNLPGYRTAVNPLLRG+EVGLAHGFFLVGPFVK
Sbjct: 62 VQPINGDPFIGSLETPVTSSPLIAWYLSNLPGYRTAVNPLLRGVEVGLAHGFFLVGPFVK 121
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
AGPLRNT YAGSAGSLAAAGL+VILS+CLTIYG+SSF EG+PSIAPSLTLTGRKKQPD L
Sbjct: 122 AGPLRNTAYAGSAGSLAAAGLVVILSMCLTIYGISSFKEGEPSIAPSLTLTGRKKQPDQL 181
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
QTADGW+KFTGGFFFGGISGVTWAYFLLYVLDLPYFVK
Sbjct: 182 QTADGWAKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 219
>emb|CAB53034.1| photosystem I subunit XI precursor [Arabidopsis thaliana]
Length = 219
Score = 352 bits (902), Expect = 5e-96
Identities = 173/218 (79%), Positives = 195/218 (89%), Gaps = 6/218 (2%)
Query: 2 AAASPMASQLKSSFT-----RPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQV 56
A+ASPMASQL+SSF+ + L P+ G+ P P++R +FTV+A++S+K +QV
Sbjct: 3 ASASPMASQLRSSFSSASLSQRLAVPKQFSGA-PFGVSPTKRVSSFTVRAVKSDKTTFQV 61
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
+QPINGDPFIGSLETPVTSSPL+AWYLSNLPGYRTAVNPLLRG+EVGLAHGFFLVGPFVK
Sbjct: 62 VQPINGDPFIGSLETPVTSSPLIAWYLSNLPGYRTAVNPLLRGVEVGLAHGFFLVGPFVK 121
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
AGPLRNT YAGSAGSLAAAGL+VILS+CLTIYG+SSF EG+PSIAPSLTLTGRKKQPD L
Sbjct: 122 AGPLRNTAYAGSAGSLAAAGLVVILSMCLTIYGISSFKEGEPSIAPSLTLTGRKKQPDQL 181
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
QTADGW+KFTGGFFFGGISGVTWAYFLLYVLDLPYFVK
Sbjct: 182 QTADGWAKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 219
>dbj|BAA11677.1| subunit of photosystem I [Cucumis sativus]
Length = 217
Score = 350 bits (898), Expect = 2e-95
Identities = 172/216 (79%), Positives = 195/216 (89%), Gaps = 6/216 (2%)
Query: 3 AASPMASQLKSSF----TRPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQVIQ 58
A SP+ASQL SSF TR L++P+GL +SPL ++P+R +FT++AIQ++KP +QVIQ
Sbjct: 4 ATSPIASQLSSSFASSNTRALISPKGL-SASPLRRIPTRTH-SFTIRAIQADKPTFQVIQ 61
Query: 59 PINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVKAG 118
PINGDPFIGSLETPVTSSPL+AWYLSNLP YRTAV+PLLRGIEVGLAHGFFLVGPFVKAG
Sbjct: 62 PINGDPFIGSLETPVTSSPLIAWYLSNLPAYRTAVSPLLRGIEVGLAHGFFLVGPFVKAG 121
Query: 119 PLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPLQT 178
PLRNT YAG AGSLAA GLIVILS+CLT+YGV+SFNEG+PS APSLTLTGRKK PDPLQT
Sbjct: 122 PLRNTAYAGGAGSLAAGGLIVILSVCLTMYGVASFNEGEPSTAPSLTLTGRKKTPDPLQT 181
Query: 179 ADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
ADGW+KF+GGFFFGGISGV WAYFLLYVLDLPY+VK
Sbjct: 182 ADGWAKFSGGFFFGGISGVIWAYFLLYVLDLPYYVK 217
>gb|AAO85557.1| photosystem I subunit XI [Nicotiana attenuata]
Length = 220
Score = 350 bits (897), Expect = 2e-95
Identities = 173/218 (79%), Positives = 190/218 (86%), Gaps = 7/218 (3%)
Query: 3 AASPMASQLKSSFTRPL------VTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQV 56
AASPMASQLKS F L VTP+G+ GS P PS R+ FT+KA+Q++KP YQV
Sbjct: 4 AASPMASQLKSKFASSLTRGSGLVTPKGISGS-PFKIFPSTRKSCFTIKAVQTDKPTYQV 62
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
IQP+NGDPFIGSLETPVTSSPL+AWYLSNLP YRTAVNPLLRG+EVGLAHGF LVGPFVK
Sbjct: 63 IQPLNGDPFIGSLETPVTSSPLIAWYLSNLPAYRTAVNPLLRGVEVGLAHGFLLVGPFVK 122
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
AGPLRNTEYAG AGSLAAAGL+VILS+CLTIYG+SSF EG+ S AP+LTLTGRKK PD L
Sbjct: 123 AGPLRNTEYAGGAGSLAAAGLVVILSMCLTIYGISSFKEGEASTAPALTLTGRKKVPDQL 182
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
QTA+GWSKFTGGFFFGGISGVTWAYFLLYVLDLPY+VK
Sbjct: 183 QTAEGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYYVK 220
>dbj|BAA09047.1| PsaL [Cucumis sativus] gi|1363479|pir||JC4281 photosystem I protein
psaL - cucumber gi|18202533|sp|Q39654|PSAL_CUCSA
Photosystem I reaction center subunit XI, chloroplast
precursor (PSI-L) (PSI subunit V)
Length = 217
Score = 349 bits (896), Expect = 3e-95
Identities = 172/216 (79%), Positives = 194/216 (89%), Gaps = 6/216 (2%)
Query: 3 AASPMASQLKSSF----TRPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQVIQ 58
A SP ASQL SSF TR L++P+GL +SPL ++P+R +FT++AIQ++KP +QVIQ
Sbjct: 4 ATSPTASQLSSSFASSNTRALISPKGL-SASPLRRIPTRTH-SFTIRAIQADKPTFQVIQ 61
Query: 59 PINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVKAG 118
PINGDPFIGSLETPVTSSPL+AWYLSNLP YRTAV+PLLRGIEVGLAHGFFLVGPFVKAG
Sbjct: 62 PINGDPFIGSLETPVTSSPLIAWYLSNLPAYRTAVSPLLRGIEVGLAHGFFLVGPFVKAG 121
Query: 119 PLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPLQT 178
PLRNT YAG AGSLAA GLIVILS+CLT+YGV+SFNEG+PS APSLTLTGRKK PDPLQT
Sbjct: 122 PLRNTAYAGGAGSLAAGGLIVILSVCLTMYGVASFNEGEPSTAPSLTLTGRKKTPDPLQT 181
Query: 179 ADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
ADGW+KF+GGFFFGGISGV WAYFLLYVLDLPY+VK
Sbjct: 182 ADGWAKFSGGFFFGGISGVIWAYFLLYVLDLPYYVK 217
>emb|CAA45775.1| subunit XI of photosystem I reaction center [Spinacia oleracea]
gi|479684|pir||S35151 photosystem I chain XI precursor -
spinach gi|3914473|sp|Q41385|PSAL_SPIOL Photosystem I
reaction center subunit XI, chloroplast precursor
(PSI-L) (PSI subunit V)
Length = 216
Score = 344 bits (882), Expect = 1e-93
Identities = 168/215 (78%), Positives = 192/215 (89%), Gaps = 3/215 (1%)
Query: 2 AAASPMASQLKSSFT-RPLVTPRGLCGSSPLHQLPS-RRQFNFTVKAIQSEKPNYQVIQP 59
A SPMASQLKS FT + LV P+G+ G + L PS RR +FTV+AI++EKP YQVIQP
Sbjct: 3 ATTSPMASQLKSGFTTKALVVPKGISGPA-LRGFPSPRRHTSFTVRAIKTEKPTYQVIQP 61
Query: 60 INGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVKAGP 119
+NGDPFIG LETPVTSSPL+AWYLSNLP YRTAVNPLLRG+EVGLAHGF LVGPFVKAGP
Sbjct: 62 LNGDPFIGGLETPVTSSPLIAWYLSNLPAYRTAVNPLLRGVEVGLAHGFLLVGPFVKAGP 121
Query: 120 LRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPLQTA 179
LRNTEYAG+AGSLAAAGL+VILS+CLT+YG++SF EG+PSIAP+LTLTGRKKQPD LQ+A
Sbjct: 122 LRNTEYAGAAGSLAAAGLVVILSMCLTMYGIASFKEGEPSIAPALTLTGRKKQPDQLQSA 181
Query: 180 DGWSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
DGW+KFTGGFFFGG+SGVTWA FL+YVLDLPY+ K
Sbjct: 182 DGWAKFTGGFFFGGVSGVTWACFLMYVLDLPYYFK 216
>pir||A39759 photosystem I 18K protein precursor - barley
gi|131225|sp|P23993|PSAL_HORVU Photosystem I reaction
center subunit XI, chloroplast precursor (PSI-L) (PSI
subunit V) gi|167087|gb|AAA62700.1| photosystem I
hydrophobic protein
Length = 209
Score = 300 bits (769), Expect = 1e-80
Identities = 150/213 (70%), Positives = 170/213 (79%), Gaps = 7/213 (3%)
Query: 2 AAASPMASQLKSSFTRPLVTPRGLCGSSPLHQLPSRRQFNFTVKAIQSEKPNYQVIQPIN 61
A A PMASQ+ S PRG+ G+S R+ F VKA++S+KP YQV+QPIN
Sbjct: 4 AYAPPMASQVMKSGLA-CSKPRGMSGAS------LTRRPRFVVKAVKSDKPTYQVVQPIN 56
Query: 62 GDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVKAGPLR 121
GDPFIGSLETPVTSSPLVAWYLSNLP YRTAV+PLLRGIEVGLAHG+ LVGPF GPLR
Sbjct: 57 GDPFIGSLETPVTSSPLVAWYLSNLPAYRTAVSPLLRGIEVGLAHGYLLVGPFALTGPLR 116
Query: 122 NTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPLQTADG 181
NT G AG+L A GL+ ILS+CLT+YGV+SFNEG+PS AP LTLTGRKK+ D LQTA+G
Sbjct: 117 NTPVHGQAGTLGAIGLVSILSVCLTMYGVASFNEGEPSTAPVLTLTGRKKEADKLQTAEG 176
Query: 182 WSKFTGGFFFGGISGVTWAYFLLYVLDLPYFVK 214
WS+FTGGFFFGG+SG WAYFLLYVLDLPYF K
Sbjct: 177 WSQFTGGFFFGGVSGAVWAYFLLYVLDLPYFFK 209
>dbj|BAA84771.1| photosystem I subunit PSI-L [Arabidopsis thaliana]
Length = 124
Score = 239 bits (609), Expect = 5e-62
Identities = 114/123 (92%), Positives = 120/123 (96%)
Query: 92 AVNPLLRGIEVGLAHGFFLVGPFVKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVS 151
AVNPLLRG+EVGLAHGFFLVGPFVKAGPLRNT YAGSAGSLAAAGL+VILS+CLTIYG+S
Sbjct: 2 AVNPLLRGVEVGLAHGFFLVGPFVKAGPLRNTAYAGSAGSLAAAGLVVILSMCLTIYGIS 61
Query: 152 SFNEGDPSIAPSLTLTGRKKQPDPLQTADGWSKFTGGFFFGGISGVTWAYFLLYVLDLPY 211
SF EG+PSIAPSLTLTGRKKQPD LQTADGW+KFTGGFFFGGISGVTWAYFLLYVLDLPY
Sbjct: 62 SFKEGEPSIAPSLTLTGRKKQPDQLQTADGWAKFTGGFFFGGISGVTWAYFLLYVLDLPY 121
Query: 212 FVK 214
FVK
Sbjct: 122 FVK 124
>gb|AAC35660.1| PSI reaction centre subunit XI [Guillardia theta]
gi|11467674|ref|NP_050726.1| photosystem I subunit XI
[Guillardia theta] gi|6093806|sp|O78469|PSAL_GUITH
Photosystem I reaction center subunit XI (PSI-L) (PSI
subunit V)
Length = 152
Score = 151 bits (382), Expect = 1e-35
Identities = 73/150 (48%), Positives = 97/150 (64%), Gaps = 14/150 (9%)
Query: 55 QVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPF 114
Q ++P N DPF+G+L TP+T+S LSNLP YR ++PLLRG+E+G+ HG+FLVGPF
Sbjct: 3 QFVKPYNDDPFVGNLATPITTSSFTRTLLSNLPAYRAGLSPLLRGLEIGMTHGYFLVGPF 62
Query: 115 VKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
K GPLRN+E A AG +A GLI+I++ CL IYGV SFN D +
Sbjct: 63 YKLGPLRNSEVALLAGVFSALGLIIIMAACLAIYGVVSFNTNDGG--------------E 108
Query: 175 PLQTADGWSKFTGGFFFGGISGVTWAYFLL 204
LQ+A GW +FT G+ G I G ++AY L+
Sbjct: 109 QLQSAKGWRQFTSGWLVGSIGGASFAYILI 138
>gb|AAC08108.1| Photosystem I reaction centre subunit XI [Porphyra purpurea]
gi|11465688|ref|NP_053832.1| photosystem I subunit XI
[Porphyra purpurea] gi|1709823|sp|P51222|PSAL_PORPU
Photosystem I reaction center subunit XI (PSI-L) (PSI
subunit V) gi|2147924|pir||S73143 photosystem I chain XI
- red alga (Porphyra purpurea) chloroplast
Length = 153
Score = 150 bits (379), Expect = 2e-35
Identities = 73/150 (48%), Positives = 97/150 (64%), Gaps = 14/150 (9%)
Query: 55 QVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPF 114
+ I+P N DPF+G+L TPV++S L NLP YR ++PLLRG+E+G+AHG+FL+GPF
Sbjct: 3 EFIKPYNDDPFVGNLSTPVSTSSFSKGLLGNLPAYRRGLSPLLRGLEIGMAHGYFLIGPF 62
Query: 115 VKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
K GPLR T+ A AG L++ GLI+IL+ CL++YG SF D D
Sbjct: 63 DKLGPLRGTDVALLAGFLSSVGLIIILTTCLSMYGNVSFTRSD--------------SKD 108
Query: 175 PLQTADGWSKFTGGFFFGGISGVTWAYFLL 204
PLQTA+GW +FT GF G + G +AY LL
Sbjct: 109 PLQTAEGWGQFTAGFLVGAVGGSGFAYLLL 138
>ref|YP_063666.1| photosystem I reaction center subunit XI [Gracilaria tenuistipitata
var. liui] gi|50657756|gb|AAT79741.1| photosystem I
reaction center subunit XI [Gracilaria tenuistipitata
var. liui]
Length = 150
Score = 149 bits (375), Expect = 7e-35
Identities = 73/148 (49%), Positives = 98/148 (65%), Gaps = 14/148 (9%)
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
IQ N DPF+G+L TPV++S L+NLP YR ++PLLRG+E+G+AHG+FLVGPF K
Sbjct: 5 IQSYNNDPFLGNLSTPVSTSTFTKGLLNNLPAYRRGLSPLLRGLEIGMAHGYFLVGPFDK 64
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
GPLRNT+ A +G L+A GLI+IL++CL++YG SF++ D D L
Sbjct: 65 LGPLRNTDVALLSGFLSAVGLIIILTVCLSMYGNVSFDKDDAK--------------DLL 110
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLL 204
QT +GW +FT GF G + G +AY LL
Sbjct: 111 QTTEGWGQFTAGFLVGAVGGSGFAYLLL 138
>ref|NP_683194.1| photosystem I subunit XI [Thermosynechococcus elongatus BP-1]
gi|34222854|sp|Q8DGB4|PSAL_SYNEL Photosystem I reaction
center subunit XI (PSI-L) (PSI subunit V)
gi|22296132|dbj|BAC09956.1| photosystem I subunit XI
[Thermosynechococcus elongatus BP-1]
Length = 155
Score = 148 bits (374), Expect = 9e-35
Identities = 67/150 (44%), Positives = 100/150 (66%), Gaps = 13/150 (8%)
Query: 55 QVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPF 114
++++P NGDPF+G L TP++ S LV ++ NLP YR ++P+LRG+EVG+AHG+FL+GP+
Sbjct: 4 ELVKPYNGDPFVGHLSTPISDSGLVKTFIGNLPAYRQGLSPILRGLEVGMAHGYFLIGPW 63
Query: 115 VKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
VK GPLR+++ A G ++ LI++ + CL YG+ SF +G S D
Sbjct: 64 VKLGPLRDSDVANLGGLISGIALILVATACLAAYGLVSFQKGGSS-------------SD 110
Query: 175 PLQTADGWSKFTGGFFFGGISGVTWAYFLL 204
PL+T++GWS+FT GFF G + A+FLL
Sbjct: 111 PLKTSEGWSQFTAGFFVGAMGSAFVAFFLL 140
>pdb|1JB0|L Chain L, Crystal Structure Of Photosystem I: A Photosynthetic
Reaction Center And Core Antenna System From
Cyanobacteria
Length = 154
Score = 148 bits (374), Expect = 9e-35
Identities = 67/150 (44%), Positives = 100/150 (66%), Gaps = 13/150 (8%)
Query: 55 QVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPF 114
++++P NGDPF+G L TP++ S LV ++ NLP YR ++P+LRG+EVG+AHG+FL+GP+
Sbjct: 3 ELVKPYNGDPFVGHLSTPISDSGLVKTFIGNLPAYRQGLSPILRGLEVGMAHGYFLIGPW 62
Query: 115 VKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
VK GPLR+++ A G ++ LI++ + CL YG+ SF +G S D
Sbjct: 63 VKLGPLRDSDVANLGGLISGIALILVATACLAAYGLVSFQKGGSS-------------SD 109
Query: 175 PLQTADGWSKFTGGFFFGGISGVTWAYFLL 204
PL+T++GWS+FT GFF G + A+FLL
Sbjct: 110 PLKTSEGWSQFTAGFFVGAMGSAFVAFFLL 139
>ref|ZP_00324141.1| COG0304: 3-oxoacyl-(acyl-carrier-protein) synthase [Trichodesmium
erythraeum IMS101]
Length = 161
Score = 145 bits (367), Expect = 6e-34
Identities = 68/153 (44%), Positives = 98/153 (63%), Gaps = 12/153 (7%)
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
+QP GDPF+G L TP++ S ++ NLP YR ++P+LRG+EVG+AHG+F+VGP+ K
Sbjct: 10 VQPYKGDPFVGHLSTPISDSDFTRAFIGNLPIYRPGLSPILRGLEVGMAHGYFIVGPWTK 69
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
GPLR++ A G ++ L++I +ICL+ YG+ SF P A DPL
Sbjct: 70 LGPLRDSAVANLGGLISTIALVLIATICLSAYGLVSFQGKSPEGA------------DPL 117
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLLYVLDL 209
+T++GWS+FTGGFF G + G A+FLL +L
Sbjct: 118 KTSEGWSQFTGGFFIGAMGGAVVAFFLLENFEL 150
>emb|CAA45296.1| photosystem I subunit XI [Synechococcus sp.] gi|97606|pir||S22209
photosystem I chain XI - Synechococcus sp
gi|131226|sp|P25902|PSAL_SYNEN Photosystem I reaction
center subunit XI (PSI-L) (PSI subunit V)
Length = 149
Score = 145 bits (367), Expect = 6e-34
Identities = 66/150 (44%), Positives = 99/150 (66%), Gaps = 13/150 (8%)
Query: 55 QVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPF 114
++++P NGDPF+G L TP++ S LV ++ NLP YR ++P+L G+EVG+AHG+FL+GP+
Sbjct: 4 ELVKPYNGDPFVGHLSTPISDSGLVKTFIGNLPAYRQGLSPILPGLEVGMAHGYFLIGPW 63
Query: 115 VKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
VK GPLR+++ A G ++ LI++ + CL YG+ SF +G S D
Sbjct: 64 VKLGPLRDSDVANLGGLISGIALILVATACLAAYGLVSFQKGGSS-------------SD 110
Query: 175 PLQTADGWSKFTGGFFFGGISGVTWAYFLL 204
PL+T++GWS+FT GFF G + A+FLL
Sbjct: 111 PLKTSEGWSQFTAGFFVGAMGSAFVAFFLL 140
>gb|AAF12996.1| unknown; Photosystem I reaction centre subunit XI [Cyanidium
caldarium] gi|11465558|ref|NP_045050.1| photosystem I
subunit XI [Cyanidium caldarium]
gi|14423881|sp|Q9TM17|PSAL_CYACA Photosystem I reaction
center subunit XI (PSI-L) (PSI subunit V)
Length = 152
Score = 145 bits (367), Expect = 6e-34
Identities = 68/148 (45%), Positives = 96/148 (63%), Gaps = 14/148 (9%)
Query: 57 IQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPFVK 116
I+P +PF+G+L TPV SS + WYL NLP YR ++PLLRG+E+G+AHG+F++GPF K
Sbjct: 5 IKPYGSNPFVGNLSTPVNSSKVTIWYLKNLPIYRRGLSPLLRGLEIGMAHGYFIIGPFYK 64
Query: 117 AGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPDPL 176
GPLRNT+ + +G +AA GLI+I SI + IYG+ +F+ + + D L
Sbjct: 65 LGPLRNTDLSLLSGLIAAIGLIIISSIAMIIYGIVTFDNSENN--------------DKL 110
Query: 177 QTADGWSKFTGGFFFGGISGVTWAYFLL 204
QTA+GW + GF G + G +AY LL
Sbjct: 111 QTANGWRQLASGFLLGAVGGAGFAYILL 138
>sp|P58577|PSAL_ANASP Photosystem I reaction center subunit XI (PSI-L) (PSI subunit V)
gi|17135085|dbj|BAB77631.1| photosystem I subunit XI
[Nostoc sp. PCC 7120] gi|17227603|ref|NP_484151.1|
photosystem I subunit XI [Nostoc sp. PCC 7120]
Length = 172
Score = 145 bits (365), Expect = 1e-33
Identities = 71/159 (44%), Positives = 97/159 (60%), Gaps = 7/159 (4%)
Query: 45 KAIQSEKPNYQVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGL 104
K + S+ N +V+ P DP G+LETPV +SPLV W+++NLP YR + P RG+EVG+
Sbjct: 9 KNLPSDPRNREVVFPAGRDPQWGNLETPVNASPLVKWFINNLPAYRPGLTPFRRGLEVGM 68
Query: 105 AHGFFLVGPFVKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSL 164
AHG+FL GPF K GPLR+ A AG L A GL+V+ ++ L++Y N P+ S+
Sbjct: 69 AHGYFLFGPFAKLGPLRDAANANLAGLLGAIGLVVLFTLALSLYA----NSNPPTALASV 124
Query: 165 TLTGRKKQPDPLQTADGWSKFTGGFFFGGISGVTWAYFL 203
T+ PD Q+ +GW+ F F GGI G AYFL
Sbjct: 125 TV---PNPPDAFQSKEGWNNFASAFLIGGIGGAVVAYFL 160
>ref|ZP_00158131.2| hypothetical protein Avar03006008 [Anabaena variabilis ATCC 29413]
Length = 172
Score = 144 bits (363), Expect = 2e-33
Identities = 71/159 (44%), Positives = 96/159 (59%), Gaps = 7/159 (4%)
Query: 45 KAIQSEKPNYQVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGL 104
K + S+ N +V+ P DP G+LETPV +SPLV W+++NLP YR + P RG+EVG+
Sbjct: 9 KNLPSDPRNREVVFPAGRDPQWGNLETPVNASPLVKWFINNLPAYRPGLTPFRRGLEVGM 68
Query: 105 AHGFFLVGPFVKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSL 164
AHG+FL GPF K GPLR+ A AG L A GL+V+ ++ L++Y N P S+
Sbjct: 69 AHGYFLFGPFAKLGPLRDAANANLAGLLGAIGLVVLFTLSLSLYA----NSNPPKALASV 124
Query: 165 TLTGRKKQPDPLQTADGWSKFTGGFFFGGISGVTWAYFL 203
T+ PD Q+ +GW+ F F GGI G AYFL
Sbjct: 125 TV---PNPPDAFQSKEGWNNFASAFLIGGIGGAVVAYFL 160
>ref|NP_442961.1| photosystem I subunit XI [Synechocystis sp. PCC 6803]
gi|1653863|dbj|BAA18773.1| photosystem I subunit XI
[Synechocystis sp. PCC 6803]
gi|585737|sp|P37277|PSAL_SYNY3 Photosystem I reaction
center subunit XI (PSI-L) (PSI subunit V)
gi|310724|gb|AAA27296.1| photosystem I subunit
Length = 157
Score = 143 bits (361), Expect = 3e-33
Identities = 68/150 (45%), Positives = 94/150 (62%), Gaps = 13/150 (8%)
Query: 55 QVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGFFLVGPF 114
QV+Q NGDPF+G L TP++ S ++ NLP YR ++P+LRG+EVG+AHG+FL+GP+
Sbjct: 6 QVVQAYNGDPFVGHLSTPISDSAFTRTFIGNLPAYRKGLSPILRGLEVGMAHGYFLIGPW 65
Query: 115 VKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTGRKKQPD 174
GPLR++EY G + A LI++ + L+ YG+ +T G + D
Sbjct: 66 TLLGPLRDSEYQYIGGLIGALALILVATAALSSYGL-------------VTFQGEQGSGD 112
Query: 175 PLQTADGWSKFTGGFFFGGISGVTWAYFLL 204
LQTADGWS+F GFF GG+ G AYFLL
Sbjct: 113 TLQTADGWSQFAAGFFVGGMGGAFVAYFLL 142
>gb|AAC04841.1| photosystem I subunit XI [Fischerella sp. PCC 7605]
gi|3334302|sp|O31126|PSAL_MASLA Photosystem I reaction
center subunit XI (PSI-L) (PSI subunit V)
Length = 173
Score = 142 bits (359), Expect = 5e-33
Identities = 73/161 (45%), Positives = 97/161 (59%), Gaps = 7/161 (4%)
Query: 49 SEKPNYQVIQPINGDPFIGSLETPVTSSPLVAWYLSNLPGYRTAVNPLLRGIEVGLAHGF 108
S+ N +V+ P DP IG LETP+ SSPLV W++ NLP YR + RG+EVG+AHG+
Sbjct: 13 SDPRNREVVFPAYRDPQIGDLETPINSSPLVKWFIGNLPAYRPGITTFRRGLEVGMAHGY 72
Query: 109 FLVGPFVKAGPLRNTEYAGSAGSLAAAGLIVILSICLTIYGVSSFNEGDPSIAPSLTLTG 168
++ GPF K GPLRNT A AG L+A GLI+IL+ LT+Y S + S+A
Sbjct: 73 WIFGPFAKLGPLRNTVNANLAGLLSALGLIIILTGALTLYANSKPPKPVKSVA------- 125
Query: 169 RKKQPDPLQTADGWSKFTGGFFFGGISGVTWAYFLLYVLDL 209
P+ Q+++GW+ F F GGI G AYFL L+L
Sbjct: 126 TPNPPEAFQSSEGWNNFASAFLIGGIGGAVVAYFLTSNLEL 166
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 407,142,636
Number of Sequences: 2540612
Number of extensions: 18839346
Number of successful extensions: 36472
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 36389
Number of HSP's gapped (non-prelim): 58
length of query: 214
length of database: 863,360,394
effective HSP length: 123
effective length of query: 91
effective length of database: 550,865,118
effective search space: 50128725738
effective search space used: 50128725738
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC148289.12


