

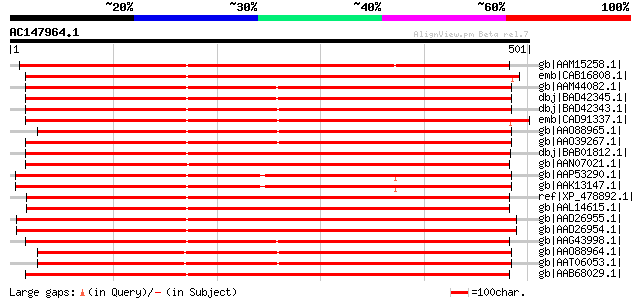
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.1 + phase: 0
(501 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM15258.1| putative sugar transporter [Arabidopsis thaliana]... 657 0.0
emb|CAB16808.1| sugar transporter like protein [Arabidopsis thal... 647 0.0
gb|AAM44082.1| putative sorbitol transporter [Prunus cerasus] 596 e-169
dbj|BAD42345.1| sorbitol transporter [Malus x domestica] 593 e-168
dbj|BAD42343.1| sorbitol transporter [Malus x domestica] 583 e-165
emb|CAD91337.1| sorbitol-like transporter [Glycine max] 580 e-164
gb|AAO88965.1| sorbitol transporter [Malus x domestica] 573 e-162
gb|AAO39267.1| sorbitol transporter [Prunus cerasus] 571 e-161
dbj|BAB01812.1| sugar transporter protein [Arabidopsis thaliana]... 571 e-161
gb|AAN07021.1| putative mannitol transporter [Orobanche ramosa] 563 e-159
gb|AAP53290.1| putative mannitol transporter protein [Oryza sati... 561 e-158
gb|AAK13147.1| Putative sugar transporter [Oryza sativa] 561 e-158
ref|XP_478892.1| putative sorbitol transporter [Oryza sativa (ja... 557 e-157
gb|AAL14615.1| putative sugar transporter [Oryza sativa] 557 e-157
gb|AAD26955.1| putative sugar transporter [Arabidopsis thaliana]... 554 e-156
gb|AAD26954.1| putative sugar transporter [Arabidopsis thaliana]... 554 e-156
gb|AAG43998.1| mannitol transporter [Apium graveolens var. dulce] 552 e-156
gb|AAO88964.1| sorbitol transporter [Malus x domestica] 550 e-155
gb|AAT06053.1| sorbitol transporter [Malus x domestica] 550 e-155
gb|AAB68029.1| putative sugar transporter; member of major facil... 549 e-155
>gb|AAM15258.1| putative sugar transporter [Arabidopsis thaliana]
gi|4218010|gb|AAD12218.1| putative sugar transporter
[Arabidopsis thaliana] gi|15224183|ref|NP_179438.1|
mannitol transporter, putative [Arabidopsis thaliana]
gi|25308961|pir||G84564 probable sugar transporter
[imported] - Arabidopsis thaliana
Length = 508
Score = 657 bits (1696), Expect = 0.0
Identities = 333/473 (70%), Positives = 398/473 (83%), Gaps = 2/473 (0%)
Query: 10 DQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCA 69
D NK+AF CAIVAS++SI+ GYDTGVMSGA +FI++DL I+DTQ EVLAGILNLCA
Sbjct: 13 DPNPHMNKFAFGCAIVASIISIIFGYDTGVMSGAQIFIRDDLKINDTQIEVLAGILNLCA 72
Query: 70 LVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMI 129
LVGSLTAG+TSD IGRRYTI L++++F++G+ LMGYGPNY +LMVGRC+ GVGVGFALMI
Sbjct: 73 LVGSLTAGKTSDVIGRRYTIALSAVIFLVGSVLMGYGPNYPVLMVGRCIAGVGVGFALMI 132
Query: 130 APVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPS 189
APVYSAEISSAS RGFLTSLPELCI +GILLGY+SNY GK L+LKLGWRLMLGIAA PS
Sbjct: 133 APVYSAEISSASHRGFLTSLPELCISLGILLGYVSNYCFGK-LTLKLGWRLMLGIAAFPS 191
Query: 190 FVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDE 249
++AF I MPESPRWLVMQG+L +AKK+++ VSNT +EAE R +DI AA +D E
Sbjct: 192 LILAFGITRMPESPRWLVMQGRLEEAKKIMVLVSNTEEEAEERFRDILTAAEVDVTEIKE 251
Query: 250 TVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAG 309
+K + G+ VW+EL+++P P+VR +LIAAVGIHFFEHATGIEAV+LYSPRIF+KAG
Sbjct: 252 VGGGVKKKNHGKSVWRELVIKPRPAVRLILIAAVGIHFFEHATGIEAVVLYSPRIFKKAG 311
Query: 310 ITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVD 369
+ SK+KLLLAT+GVGLTK F++IA FLLDK+GRR+LL STGGM+ LT L +SLT+V
Sbjct: 312 VVSKDKLLLATVGVGLTKAFFIIIATFLLDKVGRRKLLLTSTGGMVFALTSLAVSLTMVQ 371
Query: 370 KSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMN 429
+ G + WAL LSIV+TYA+VAFF+IGLGPITWVYSSEIFPL+LRAQGASIGVAVNR MN
Sbjct: 372 RF-GRLAWALSLSIVSTYAFVAFFSIGLGPITWVYSSEIFPLRLRAQGASIGVAVNRIMN 430
Query: 430 AVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
A VSM+F+S+ KAIT GG FF+FAGI+V AW FF+F LPETKG LEEME +F
Sbjct: 431 ATVSMSFLSMTKAITTGGVFFVFAGIAVAAWWFFFFMLPETKGLPLEEMEKLF 483
>emb|CAB16808.1| sugar transporter like protein [Arabidopsis thaliana]
gi|7270615|emb|CAB80333.1| sugar transporter like
protein [Arabidopsis thaliana]
gi|15234491|ref|NP_195385.1| mannitol transporter,
putative [Arabidopsis thaliana] gi|25308953|pir||A85433
sugar transporter like protein [imported] - Arabidopsis
thaliana
Length = 493
Score = 647 bits (1670), Expect = 0.0
Identities = 323/480 (67%), Positives = 392/480 (81%), Gaps = 4/480 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
N++A CAIVAS+VSI+ GYDTGVMSGAM+FI+EDL +D Q EVL GILNLCALVGSL
Sbjct: 14 NRFALQCAIVASIVSIIFGYDTGVMSGAMVFIEEDLKTNDVQIEVLTGILNLCALVGSLL 73
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD IGRRYTI LASILF+LG+ LMG+GPNY +L+ GRC G+GVGFALM+APVYSA
Sbjct: 74 AGRTSDIIGRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYSA 133
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
EI++AS RG L SLP LCI IGILLGYI NY K L + +GWRLMLGIAA+PS V+AF
Sbjct: 134 EIATASHRGLLASLPHLCISIGILLGYIVNYFFSK-LPMHIGWRLMLGIAAVPSLVLAFG 192
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
IL MPESPRWL+MQG+L + K++L VSN+ +EAELR +DIK AAG+D C D+ VK+
Sbjct: 193 ILKMPESPRWLIMQGRLKEGKEILELVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEG 252
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
K GEGVWKELILRPTP+VR +L+ A+GIHFF+HA+GIEAV+LY PRIF+KAGIT+K+K
Sbjct: 253 KKTHGEGVWKELILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGPRIFKKAGITTKDK 312
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
L L TIGVG+ K F+ A LLDK+GRR+LL S GGM+I LT+LG LT+ + G +
Sbjct: 313 LFLVTIGVGIMKTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTMLGFGLTMAQNAGGKL 372
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
WAL+LSIVA Y++VAFF+IGLGPITWVYSSE+FPLKLRAQGAS+GVAVNR MNA VSM+
Sbjct: 373 AWALVLSIVAAYSFVAFFSIGLGPITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMS 432
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK---KSSGKNVA 492
F+S+ AIT GG+FFMFAG++ +AW FF+F LPETKGK+LEE+E +F + K G+N A
Sbjct: 433 FLSLTSAITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEIEALFQRDGDKVRGENGA 492
>gb|AAM44082.1| putative sorbitol transporter [Prunus cerasus]
Length = 538
Score = 596 bits (1536), Expect = e-169
Identities = 292/469 (62%), Positives = 376/469 (79%), Gaps = 2/469 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACAI+ASM SI+ GYD GVMSGA+++IK+DL +SD + EVL GILNL +L+GS
Sbjct: 32 NKYAFACAILASMTSILLGYDIGVMSGAVIYIKKDLKVSDVEIEVLVGILNLYSLIGSAA 91
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI LA +F GA LMG+ PNYA LM GR V G+GVG+ALMIAPVY+A
Sbjct: 92 AGRTSDWIGRRYTIVLAGAIFFAGALLMGFAPNYAFLMFGRFVAGIGVGYALMIAPVYTA 151
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFLTS PE+ I GIL GY+SNY K L LGWRLMLG+ A+PS +A
Sbjct: 152 EVSPASSRGFLTSFPEVFINAGILFGYVSNYGFSK-LPTHLGWRLMLGVGAIPSIFLAIG 210
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG A+KVL + S++ +E++LRL +IK AAG+ E+CND+ V++ +
Sbjct: 211 VLAMPESPRWLVMQGRLGDARKVLDKTSDSLEESKLRLGEIKEAAGIPEHCNDDIVEVKK 270
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
+S QG+ VWK+L+LRPTP+VR +L+ AVG+HFF+ A+GI+AV+LYSPRIF KAGIT+ +
Sbjct: 271 RS-QGQEVWKQLLLRPTPAVRHILMCAVGLHFFQQASGIDAVVLYSPRIFEKAGITNPDH 329
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
+LL T+ VG K VF+++A F+LD++GRR LL S GM+ L LGL LT++D S +
Sbjct: 330 VLLCTVAVGFVKTVFILVATFMLDRIGRRPLLLTSVAGMVFTLACLGLGLTIIDHSGEKI 389
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
+WA+ LS+ AYVAFF+IG+GPITWVYSSEIFPL+LRAQG SIGVAVNR ++ V+SMT
Sbjct: 390 MWAIALSLTMVLAYVAFFSIGMGPITWVYSSEIFPLQLRAQGCSIGVAVNRVVSGVLSMT 449
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
FIS+YKAITIGG+FF+FA I+ + W FF+ LPET+G+ LE+ME++F K
Sbjct: 450 FISLYKAITIGGAFFLFAAIAAVGWTFFFTMLPETQGRTLEDMEVLFGK 498
>dbj|BAD42345.1| sorbitol transporter [Malus x domestica]
Length = 535
Score = 593 bits (1528), Expect = e-168
Identities = 291/469 (62%), Positives = 375/469 (79%), Gaps = 2/469 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACAI+ASM SI+ GYD GVMSGA ++IK+DL ISD + EVL GILNL +L+GS
Sbjct: 31 NKYAFACAILASMTSILLGYDIGVMSGAAIYIKDDLKISDVEVEVLLGILNLYSLIGSAA 90
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD++GRRYTI LA +F +GA LMG+ NY+ LM GR V G+GVG+ALMIAPVY+A
Sbjct: 91 AGRTSDWVGRRYTIVLAGAIFFVGALLMGFATNYSFLMFGRFVAGIGVGYALMIAPVYTA 150
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFLTS PE+ I GILLGY+SNY K L LGWRLMLG+ A+PS +A
Sbjct: 151 EVSPASSRGFLTSFPEVFINSGILLGYVSNYAFSK-LPTHLGWRLMLGVGAIPSIFLAVG 209
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG A +VL + S++ +E+ LRL DIK AAG+ E+C D+ V++P+
Sbjct: 210 VLAMPESPRWLVMQGRLGDATRVLDKTSDSKEESMLRLADIKEAAGIPEHCTDDVVQVPK 269
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
+S QG+ VWKEL+L PTP+VR +LI A+GIHFF+ A+GI+AV+LYSPRIF KAGIT+ +K
Sbjct: 270 RS-QGQDVWKELLLHPTPAVRHILICAIGIHFFQQASGIDAVVLYSPRIFEKAGITNSDK 328
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LL T+ VG K VF+++A F +DK+GRR LL S GMI+ LT LGL LT++D+++ +
Sbjct: 329 KLLCTVAVGFVKTVFILVATFFVDKVGRRPLLLASVAGMILSLTGLGLGLTIIDQNHERI 388
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
LWA +L + YVAFF+IG+GPITWVYSSEIFPLKLRAQG S+GVA+NR ++ V+SMT
Sbjct: 389 LWAAVLCLTMVLLYVAFFSIGMGPITWVYSSEIFPLKLRAQGCSLGVAMNRVVSGVLSMT 448
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
FIS+Y+AITIGG+FF++A I+ +AW+FF+ LPET G+ LE+ME++F K
Sbjct: 449 FISLYEAITIGGAFFLYAAIASVAWVFFFTMLPETHGRTLEDMEVLFGK 497
>dbj|BAD42343.1| sorbitol transporter [Malus x domestica]
Length = 526
Score = 583 bits (1503), Expect = e-165
Identities = 294/469 (62%), Positives = 364/469 (76%), Gaps = 2/469 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
+K+A ACA++A S++ GYD GVMSGA L+I+++L ISD Q EVLAG LN+ +L+GS
Sbjct: 32 SKFAIACALLACTTSVLLGYDIGVMSGASLYIQKNLKISDVQVEVLAGTLNIYSLLGSAF 91
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGR+YTI LA ++F++GA LMG+ NYA LMVGR V GVGVG+ +MIAPVY+A
Sbjct: 92 AGRTSDWIGRKYTIVLAGVIFLVGALLMGFATNYAFLMVGRFVAGVGVGYGMMIAPVYTA 151
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
EIS AS RGFLTS PE+ + +GILLGYI+NY K L L LGWR MLG+ +P+ +
Sbjct: 152 EISPASFRGFLTSFPEVFVNVGILLGYIANYAFSK-LPLHLGWRFMLGVGGVPAIFLTVG 210
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG AKKVL + S + +E +LRL DIK AAG+ + ND+ V++ +
Sbjct: 211 VLFMPESPRWLVMQGRLGDAKKVLQRTSESKEECQLRLDDIKEAAGIPPHLNDDIVQVTK 270
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
SH GEGVWKELIL PTP+VR +LIAAVGIHFFE A+GI+ V+LYSPRIF KAGITS
Sbjct: 271 SSH-GEGVWKELILHPTPAVRHILIAAVGIHFFEQASGIDTVVLYSPRIFAKAGITSSNH 329
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLAT+ VG TK VF+++A F LDK GRR LL S GGM+ L LG+ LT+VD G+V
Sbjct: 330 KLLATVAVGFTKTVFILVATFFLDKFGRRPLLLTSVGGMVFSLMFLGVGLTIVDHHKGSV 389
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
WA+ L + Y VAFF+IGLGPITWVYSSEIFPLKLRAQG SIGVA NR + VVSMT
Sbjct: 390 PWAIGLCMAMVYFNVAFFSIGLGPITWVYSSEIFPLKLRAQGVSIGVACNRVTSGVVSMT 449
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
FIS+YKAITIGG+FF++AGIS AW+FFY LPET+G+ LE+ E++F K
Sbjct: 450 FISLYKAITIGGAFFLYAGISAAAWIFFYTMLPETQGRTLEDTEVLFGK 498
>emb|CAD91337.1| sorbitol-like transporter [Glycine max]
Length = 523
Score = 580 bits (1494), Expect = e-164
Identities = 292/490 (59%), Positives = 384/490 (77%), Gaps = 6/490 (1%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACA++ASM SI+ GYD GVMSGA ++IK DL +SD Q E+L GI+NL +L+GS
Sbjct: 28 NKYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDEQIEILLGIINLYSLIGSCL 87
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI L +F++G+ LMG+ P+Y+ LM GR V G+G+G+ALMIAPVY+A
Sbjct: 88 AGRTSDWIGRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYTA 147
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFLTS PE+ I GILLGYISNY K L+LK+GWR+MLG+ A+PS V+
Sbjct: 148 EVSPASSRGFLTSFPEVFINGGILLGYISNYGFSK-LTLKVGWRMMLGVGAIPSVVLTEG 206
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVM+G+LG+A+KVL + S++ +EA+LRL +IK AAG+ E+CND+ V++ +
Sbjct: 207 VLAMPESPRWLVMRGRLGEARKVLNKTSDSKEEAQLRLAEIKQAAGIPESCNDDVVQVNK 266
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
+S+ GEGVWKEL L PTP++R ++IAA+GIHFF+ A+G++AV+LYSPRIF KAGIT+
Sbjct: 267 QSN-GEGVWKELFLYPTPAIRHIVIAALGIHFFQQASGVDAVVLYSPRIFEKAGITNDTH 325
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLAT+ VG K VF++ A F LD++GRR LL S GGM++ L L +SLTV+D S +
Sbjct: 326 KLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTVIDHSERKL 385
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
+WA+ SI AYVA F+IG GPITWVYSSEIFPL+LRAQGA+ GVAVNRT +AVVSMT
Sbjct: 386 MWAVGSSIAMVLAYVATFSIGAGPITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVSMT 445
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF----TKKSSGKNV 491
F+S+ +AITIGG+FF++ GI+ + W+FFY LPET+GK LE+ME F +K ++ K V
Sbjct: 446 FLSLTRAITIGGAFFLYCGIATVGWIFFYTVLPETRGKTLEDMEGSFGTFRSKSNASKAV 505
Query: 492 AIEMDPIQKV 501
E + +V
Sbjct: 506 ENENGQVAQV 515
>gb|AAO88965.1| sorbitol transporter [Malus x domestica]
Length = 481
Score = 573 bits (1476), Expect = e-162
Identities = 283/457 (61%), Positives = 359/457 (77%), Gaps = 2/457 (0%)
Query: 28 MVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRTSDYIGRRY 87
M SI+ GYD GVMSGA LFIKE+L ISD Q E++ G LNL +L+GS AGRTSD+IGRRY
Sbjct: 1 MTSILLGYDIGVMSGASLFIKENLKISDVQVEIMNGTLNLYSLIGSALAGRTSDWIGRRY 60
Query: 88 TIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLT 147
TI LA +F +GA LMG+ PNYA LM GR V GVGVG+ALMIAPVY+AEIS AS RGFLT
Sbjct: 61 TIVLAGTIFFIGALLMGFAPNYAFLMFGRFVAGVGVGYALMIAPVYTAEISPASFRGFLT 120
Query: 148 SLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLV 207
S PE+ + IGILLGY+SNY K L + L WR+MLG+ A PS ++A +L MPESPRWLV
Sbjct: 121 SFPEVFVNIGILLGYVSNYAFSK-LPIHLNWRIMLGVGAFPSVILAVGVLAMPESPRWLV 179
Query: 208 MQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKEL 267
MQG+LG AK+VL + S + +E +LRL DIK AAG+ + ND+ V++ ++SH GEGVWKEL
Sbjct: 180 MQGRLGDAKRVLQKTSESIEECQLRLDDIKEAAGIPKESNDDVVQVSKRSH-GEGVWKEL 238
Query: 268 ILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTK 327
+L PTP+VR +LIAA+GIHFFE ++GI++V+LYSPRIF KAGITS + LLAT+ VG+ K
Sbjct: 239 LLHPTPAVRHILIAALGIHFFEQSSGIDSVVLYSPRIFEKAGITSYDHKLLATVAVGVVK 298
Query: 328 IVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATY 387
+ +++A LDK GRR LL S GM+ L+ LG SLT+VD+ +G ++WA++L I
Sbjct: 299 TICILVATVFLDKFGRRPLLLTSVAGMVFSLSCLGASLTIVDQQHGKIMWAIVLCITMVL 358
Query: 388 AYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGG 447
VAFF+IGLGPITWVYSSEIFPL+LRAQG S+GVAVNR + V+SMTFIS+YKAITIGG
Sbjct: 359 LNVAFFSIGLGPITWVYSSEIFPLQLRAQGCSMGVAVNRVTSGVISMTFISLYKAITIGG 418
Query: 448 SFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
+FF++AGI+ + W+FFY PET+G+ LE+ME++F K
Sbjct: 419 AFFLYAGIAAVGWVFFYMLYPETQGRTLEDMEVLFGK 455
>gb|AAO39267.1| sorbitol transporter [Prunus cerasus]
Length = 509
Score = 571 bits (1472), Expect = e-161
Identities = 282/469 (60%), Positives = 363/469 (77%), Gaps = 2/469 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
N YA CAI+ASM SI+ GYD GVMSGA ++I++DL ISD + E+L GILNL +L+GS
Sbjct: 24 NLYAIGCAILASMTSILLGYDIGVMSGASIYIQKDLKISDVEVEILIGILNLYSLIGSAA 83
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI A +F GA LMG NYA LMVGR V G+GVG+ALMIAPVY+A
Sbjct: 84 AGRTSDWIGRRYTIVFAGAIFFTGALLMGLATNYAFLMVGRFVAGIGVGYALMIAPVYNA 143
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRG LTS PE+ + IGILLGY++NY L + LGWRLMLG+ PS ++A
Sbjct: 144 EVSPASSRGALTSFPEVFVNIGILLGYVANYAFSG-LPIDLGWRLMLGVGVFPSVILAVG 202
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L+MPESPRWLVMQG+LG+AK+VL + S++ +EA+LRL DIK AAG+ E+C ++ V++P+
Sbjct: 203 VLSMPESPRWLVMQGRLGEAKQVLDKTSDSLEEAQLRLADIKEAAGIPEHCVEDVVQVPK 262
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
SH GE VWKEL+L PTP VR +LIAA+G HFF+ +GI+A++LYSPRIF KAGIT
Sbjct: 263 HSH-GEEVWKELLLHPTPPVRHILIAAIGFHFFQQLSGIDALVLYSPRIFEKAGITDSST 321
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLLAT+ VG +K +F ++A+ LD++GRR LL S GMI L LG SLT+VD +
Sbjct: 322 LLLATVAVGFSKTIFTLVAIGFLDRVGRRPLLLTSVAGMIASLLCLGTSLTIVDHETEKM 381
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
+WA +L + AYV FF+IG+GPI WVYSSEIFPLKLRAQG S+G AVNR M+ V+SM+
Sbjct: 382 MWASVLCLTMVLAYVGFFSIGMGPIAWVYSSEIFPLKLRAQGCSMGTAVNRIMSGVLSMS 441
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
FIS+YKAIT+GG+FF++AGI+ + W+FFY LPET+G+ LE+ME++F K
Sbjct: 442 FISLYKAITMGGTFFLYAGIATVGWVFFYTMLPETQGRTLEDMEVLFGK 490
>dbj|BAB01812.1| sugar transporter protein [Arabidopsis thaliana]
gi|15230212|ref|NP_188513.1| mannitol transporter,
putative [Arabidopsis thaliana]
Length = 539
Score = 571 bits (1471), Expect = e-161
Identities = 274/468 (58%), Positives = 370/468 (78%), Gaps = 1/468 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
N YAFACAI+ASM SI+ GYD GVMSGAM++IK DL I+D Q +LAG LN+ +L+GS
Sbjct: 33 NNYAFACAILASMTSILLGYDIGVMSGAMIYIKRDLKINDLQIGILAGSLNIYSLIGSCA 92
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI LA +F GA LMG PNYA LM GR + G+GVG+ALMIAPVY+A
Sbjct: 93 AGRTSDWIGRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTA 152
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFL S PE+ I GI+LGY+SN L LK+GWRLMLGI A+PS ++A
Sbjct: 153 EVSPASSRGFLNSFPEVFINAGIMLGYVSNLAFSN-LPLKVGWRLMLGIGAVPSVILAIG 211
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG AK+VL + S++ EA LRL+DIK AAG+ +C+D+ V++ +
Sbjct: 212 VLAMPESPRWLVMQGRLGDAKRVLDKTSDSPTEATLRLEDIKHAAGIPADCHDDVVQVSR 271
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
++ GEGVW+EL++RPTP+VR ++IAA+GIHFF+ A+GI+AV+L+SPRIF+ AG+ + +
Sbjct: 272 RNSHGEGVWRELLIRPTPAVRRVMIAAIGIHFFQQASGIDAVVLFSPRIFKTAGLKTDHQ 331
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLAT+ VG+ K F+++A FLLD++GRR LL S GGM++ L LG SLT++D+S V
Sbjct: 332 QLLATVAVGVVKTSFILVATFLLDRIGRRPLLLTSVGGMVLSLAALGTSLTIIDQSEKKV 391
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
+WA++++I YVA F+IG GPITWVYSSEIFPL+LR+QG+S+GV VNR + V+S++
Sbjct: 392 MWAVVVAIATVMTYVATFSIGAGPITWVYSSEIFPLRLRSQGSSMGVVVNRVTSGVISIS 451
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT 483
F+ + KA+T GG+F++F GI+ +AW+FFY FLPET+G+ LE+M+ +F+
Sbjct: 452 FLPMSKAMTTGGAFYLFGGIATVAWVFFYTFLPETQGRMLEDMDELFS 499
>gb|AAN07021.1| putative mannitol transporter [Orobanche ramosa]
Length = 519
Score = 563 bits (1451), Expect = e-159
Identities = 271/468 (57%), Positives = 363/468 (76%), Gaps = 2/468 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
N YA +++ASM SI+ GYDTGVMSGA L+IK+DL I+D Q E+L G++N+ +L+GS
Sbjct: 29 NMYALLISVMASMTSILLGYDTGVMSGATLYIKKDLKITDVQVEILVGLINIYSLIGSAV 88
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSDY+GRR TI +AS++F +GA +MG NYA+LMVGR V G+GVG+ALMIAPVY+A
Sbjct: 89 AGRTSDYLGRRITIVIASVIFFVGAAVMGLANNYAVLMVGRFVAGLGVGYALMIAPVYAA 148
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E++ ASSRGFLTS PE+ I G+LLGY+SNY K+ SLKLGWRLMLG+ ALP+ +
Sbjct: 149 EVAPASSRGFLTSFPEVFINFGVLLGYLSNYAFAKF-SLKLGWRLMLGVGALPAIFIGLA 207
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
++ MPESPRWLVMQG+LG AKKVL + S++ QEA+LRL DI AAGL E+C+D+ V + +
Sbjct: 208 VIVMPESPRWLVMQGRLGDAKKVLDRTSDSPQEAQLRLADIMEAAGLPEDCHDDVVPVLK 267
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
+ G GVWKELI+ PT V + IAAVG FF+ A+GI+AV++YSPRI+ KAGITS EK
Sbjct: 268 QDRGGGGVWKELIVHPTKPVLHITIAAVGCQFFQQASGIDAVVMYSPRIYEKAGITSDEK 327
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLATI VGL K VF+++ F++D++GRR LL S GG+++ + L LTV+D +
Sbjct: 328 KLLATIAVGLCKTVFILVTTFMVDRIGRRVLLLTSCGGLVLSMLTLATGLTVIDHYGADR 387
Query: 376 L-WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSM 434
W ++L ++ TY+ VAFF++G+GPI WVYSSEIFPLKLRAQG +GVA+NR N V+ M
Sbjct: 388 FPWVVVLCVLTTYSSVAFFSMGMGPIAWVYSSEIFPLKLRAQGCGLGVAINRATNGVILM 447
Query: 435 TFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
+FIS+Y AITIGG+F++F+GI ++ W+FF+ LPET+G+ LE+ME++F
Sbjct: 448 SFISLYNAITIGGAFYLFSGIGIVTWIFFFTLLPETRGRTLEDMEVLF 495
>gb|AAP53290.1| putative mannitol transporter protein [Oryza sativa (japonica
cultivar-group)] gi|37533402|ref|NP_921003.1| putative
mannitol transporter protein [Oryza sativa (japonica
cultivar-group)] gi|18057108|gb|AAL58131.1| putative
mannitol transporter protein [Oryza sativa (japonica
cultivar-group)]
Length = 506
Score = 561 bits (1447), Expect = e-158
Identities = 282/481 (58%), Positives = 367/481 (75%), Gaps = 7/481 (1%)
Query: 6 GDKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGIL 65
G+++ NKYA C+I+ S++S++ GYDTGVMSGAMLFIKEDL +DTQ +VLAGIL
Sbjct: 2 GEEKQNDERKNKYAVGCSIIGSIISVLMGYDTGVMSGAMLFIKEDLKTNDTQVQVLAGIL 61
Query: 66 NLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGF 125
N+CALVGSLTAGR SD +GRR TI LA+ +F++G+ LMG PN+A L+ GRCV GVGVG+
Sbjct: 62 NVCALVGSLTAGRVSDCVGRRLTISLAACIFLVGSVLMGLAPNFATLLAGRCVAGVGVGY 121
Query: 126 ALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIA 185
ALMIAPVY+AEI+SA RG LTSLPE+CI GIL+GY++NY+L K L L GWR MLG+
Sbjct: 122 ALMIAPVYAAEIASADIRGSLTSLPEICISFGILIGYVANYLLAK-LPLVYGWRAMLGLG 180
Query: 186 ALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDEN 245
ALPS +A +L MPESPRWLV+QG+ +A VL +V + EA+ RL +IK AAGL
Sbjct: 181 ALPSAALALGVLAMPESPRWLVVQGRAEEALSVLRRVCDRPSEADARLAEIKAAAGL--- 237
Query: 246 CNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIF 305
D+ G+GVW+EL L PTP VR ++IAA+GIHFF+H TGIEAV+LYSPRIF
Sbjct: 238 -ADDDGAAANAGSGGKGVWRELFLHPTPPVRRIVIAALGIHFFQHLTGIEAVVLYSPRIF 296
Query: 306 RKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSL 365
+ AGI S+ +L ATIGVG+TK F++ A+ L+D++GRR L S G+I L LG+ L
Sbjct: 297 KAAGIASRNSVLAATIGVGVTKTAFILTAILLVDRIGRRPLYLSSLAGIIASLACLGMGL 356
Query: 366 TVVDKS--NGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVA 423
TV+++S + + WA++L+I + +VA F+IG+GPITW YSSE++PL+LRAQGAS+GVA
Sbjct: 357 TVIERSPPHHSPAWAVVLAIATVFTFVASFSIGVGPITWAYSSEVYPLRLRAQGASVGVA 416
Query: 424 VNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT 483
+NR MNA VSMTF+S+YKAITIGG+FF+FAG++V A FFY PET+GK LEE+E VF+
Sbjct: 417 INRVMNAGVSMTFVSLYKAITIGGAFFLFAGLAVAAATFFYLLCPETQGKPLEEIEEVFS 476
Query: 484 K 484
+
Sbjct: 477 Q 477
>gb|AAK13147.1| Putative sugar transporter [Oryza sativa]
Length = 574
Score = 561 bits (1447), Expect = e-158
Identities = 282/481 (58%), Positives = 367/481 (75%), Gaps = 7/481 (1%)
Query: 6 GDKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGIL 65
G+++ NKYA C+I+ S++S++ GYDTGVMSGAMLFIKEDL +DTQ +VLAGIL
Sbjct: 70 GEEKQNDERKNKYAVGCSIIGSIISVLMGYDTGVMSGAMLFIKEDLKTNDTQVQVLAGIL 129
Query: 66 NLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGF 125
N+CALVGSLTAGR SD +GRR TI LA+ +F++G+ LMG PN+A L+ GRCV GVGVG+
Sbjct: 130 NVCALVGSLTAGRVSDCVGRRLTISLAACIFLVGSVLMGLAPNFATLLAGRCVAGVGVGY 189
Query: 126 ALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIA 185
ALMIAPVY+AEI+SA RG LTSLPE+CI GIL+GY++NY+L K L L GWR MLG+
Sbjct: 190 ALMIAPVYAAEIASADIRGSLTSLPEICISFGILIGYVANYLLAK-LPLVYGWRAMLGLG 248
Query: 186 ALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDEN 245
ALPS +A +L MPESPRWLV+QG+ +A VL +V + EA+ RL +IK AAGL
Sbjct: 249 ALPSAALALGVLAMPESPRWLVVQGRAEEALSVLRRVCDRPSEADARLAEIKAAAGL--- 305
Query: 246 CNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIF 305
D+ G+GVW+EL L PTP VR ++IAA+GIHFF+H TGIEAV+LYSPRIF
Sbjct: 306 -ADDDGAAANAGSGGKGVWRELFLHPTPPVRRIVIAALGIHFFQHLTGIEAVVLYSPRIF 364
Query: 306 RKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSL 365
+ AGI S+ +L ATIGVG+TK F++ A+ L+D++GRR L S G+I L LG+ L
Sbjct: 365 KAAGIASRNSVLAATIGVGVTKTAFILTAILLVDRIGRRPLYLSSLAGIIASLACLGMGL 424
Query: 366 TVVDKS--NGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVA 423
TV+++S + + WA++L+I + +VA F+IG+GPITW YSSE++PL+LRAQGAS+GVA
Sbjct: 425 TVIERSPPHHSPAWAVVLAIATVFTFVASFSIGVGPITWAYSSEVYPLRLRAQGASVGVA 484
Query: 424 VNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT 483
+NR MNA VSMTF+S+YKAITIGG+FF+FAG++V A FFY PET+GK LEE+E VF+
Sbjct: 485 INRVMNAGVSMTFVSLYKAITIGGAFFLFAGLAVAAATFFYLLCPETQGKPLEEIEEVFS 544
Query: 484 K 484
+
Sbjct: 545 Q 545
>ref|XP_478892.1| putative sorbitol transporter [Oryza sativa (japonica
cultivar-group)] gi|51963504|ref|XP_506429.1| PREDICTED
OJ1301_C12.3 gene product [Oryza sativa (japonica
cultivar-group)] gi|34393630|dbj|BAC83310.1| putative
sorbitol transporter [Oryza sativa (japonica
cultivar-group)]
Length = 510
Score = 557 bits (1435), Expect = e-157
Identities = 281/468 (60%), Positives = 363/468 (77%), Gaps = 3/468 (0%)
Query: 17 KYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTA 76
++AFACAI+ASM SI+ GYD GVMSGA L+IK+D ISD + EVL GILNL +L+GS A
Sbjct: 19 RFAFACAILASMTSILLGYDIGVMSGASLYIKKDFNISDGKVEVLMGILNLYSLIGSFAA 78
Query: 77 GRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAE 136
GRTSD+IGRRYTI A+++F GA LMG+ NYA+LM GR V G+GVG+ALMIAPVY+AE
Sbjct: 79 GRTSDWIGRRYTIVFAAVIFFAGAFLMGFAVNYAMLMFGRFVAGIGVGYALMIAPVYTAE 138
Query: 137 ISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCI 196
+S AS+RGFLTS PE+ I GILLGY+SNY + L L LGWR+MLGI A PS ++A +
Sbjct: 139 VSPASARGFLTSFPEVFINFGILLGYVSNYAFSR-LPLNLGWRIMLGIGAAPSVLLALMV 197
Query: 197 LTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQK 256
L MPESPRWLVM+G+L AK VL + S+T +EA RL DIK AAG+ E + + V +P++
Sbjct: 198 LGMPESPRWLVMKGRLADAKVVLEKTSDTAEEAAERLADIKAAAGIPEELDGDVVTVPKR 257
Query: 257 SHQGE-GVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
E VWKELIL PTP++R +L++ +GIHFF+ A+GI++V+LYSPR+F+ AGIT +
Sbjct: 258 GSGNEKRVWKELILSPTPAMRRILLSGIGIHFFQQASGIDSVVLYSPRVFKSAGITDDKH 317
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDK-SNGN 374
LL T VG+TK +F+++A F LD++GRR LL STGGMI+ L LG LTVV + +
Sbjct: 318 LLGTTCAVGVTKTLFILVATFFLDRVGRRPLLLSSTGGMILSLIGLGAGLTVVGQHPDAK 377
Query: 375 VLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSM 434
+ WA+ LSI +T AYVAFF+IGLGPITWVYSSEIFPL++RA G S+GVA NR + V+SM
Sbjct: 378 IPWAIGLSIASTLAYVAFFSIGLGPITWVYSSEIFPLQVRALGCSLGVAANRVTSGVISM 437
Query: 435 TFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
TF+S+ KAITIGGSFF+++GI+ LAW+FFY +LPET+G+ LEEM +F
Sbjct: 438 TFLSLSKAITIGGSFFLYSGIAALAWVFFYTYLPETRGRTLEEMSKLF 485
>gb|AAL14615.1| putative sugar transporter [Oryza sativa]
Length = 577
Score = 557 bits (1435), Expect = e-157
Identities = 281/468 (60%), Positives = 363/468 (77%), Gaps = 3/468 (0%)
Query: 17 KYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTA 76
++AFACAI+ASM SI+ GYD GVMSGA L+IK+D ISD + EVL GILNL +L+GS A
Sbjct: 86 RFAFACAILASMTSILLGYDIGVMSGASLYIKKDFNISDGKVEVLMGILNLYSLIGSFAA 145
Query: 77 GRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAE 136
GRTSD+IGRRYTI A+++F GA LMG+ NYA+LM GR V G+GVG+ALMIAPVY+AE
Sbjct: 146 GRTSDWIGRRYTIVFAAVIFFAGAFLMGFAVNYAMLMFGRFVAGIGVGYALMIAPVYTAE 205
Query: 137 ISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCI 196
+S AS+RGFLTS PE+ I GILLGY+SNY + L L LGWR+MLGI A PS ++A +
Sbjct: 206 VSPASARGFLTSFPEVFINFGILLGYVSNYAFSR-LPLNLGWRIMLGIGAAPSVLLALMV 264
Query: 197 LTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQK 256
L MPESPRWLVM+G+L AK VL + S+T +EA RL DIK AAG+ E + + V +P++
Sbjct: 265 LGMPESPRWLVMKGRLADAKVVLEKTSDTAEEAAERLADIKAAAGIPEELDGDVVTVPKR 324
Query: 257 SHQGE-GVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
E VWKELIL PTP++R +L++ +GIHFF+ A+GI++V+LYSPR+F+ AGIT +
Sbjct: 325 GSGNEKRVWKELILSPTPAMRRILLSGIGIHFFQQASGIDSVVLYSPRVFKSAGITDDKH 384
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDK-SNGN 374
LL T VG+TK +F+++A F LD++GRR LL STGGMI+ L LG LTVV + +
Sbjct: 385 LLGTTCAVGVTKTLFILVATFFLDRVGRRPLLLSSTGGMILSLIGLGAGLTVVGQHPDAK 444
Query: 375 VLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSM 434
+ WA+ LSI +T AYVAFF+IGLGPITWVYSSEIFPL++RA G S+GVA NR + V+SM
Sbjct: 445 IPWAIGLSIASTLAYVAFFSIGLGPITWVYSSEIFPLQVRALGCSLGVAANRVTSGVISM 504
Query: 435 TFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
TF+S+ KAITIGGSFF+++GI+ LAW+FFY +LPET+G+ LEEM +F
Sbjct: 505 TFLSLSKAITIGGSFFLYSGIAALAWVFFYTYLPETRGRTLEEMSKLF 552
>gb|AAD26955.1| putative sugar transporter [Arabidopsis thaliana]
gi|15226696|ref|NP_179210.1| mannitol transporter,
putative [Arabidopsis thaliana] gi|25308959|pir||A84537
probable sugar transporter [imported] - Arabidopsis
thaliana
Length = 511
Score = 554 bits (1427), Expect = e-156
Identities = 273/484 (56%), Positives = 361/484 (74%), Gaps = 2/484 (0%)
Query: 7 DKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILN 66
+ E +++AFACAI+ASM SI+ GYD GVMSGA +FIK+DL +SD Q E+L GILN
Sbjct: 14 ESEPPRGNRSRFAFACAILASMTSIILGYDIGVMSGAAIFIKDDLKLSDVQLEILMGILN 73
Query: 67 LCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFA 126
+ +L+GS AGRTSD+IGRRYTI LA F GA LMG+ NY +MVGR V G+GVG+A
Sbjct: 74 IYSLIGSGAAGRTSDWIGRRYTIVLAGFFFFCGALLMGFATNYPFIMVGRFVAGIGVGYA 133
Query: 127 LMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAA 186
+MIAPVY+ E++ ASSRGFL+S PE+ I IGILLGY+SNY K L +GWR MLGI A
Sbjct: 134 MMIAPVYTTEVAPASSRGFLSSFPEIFINIGILLGYVSNYFFAK-LPEHIGWRFMLGIGA 192
Query: 187 LPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENC 246
+PS +A +L MPESPRWLVMQG+LG A KVL + SNT +EA RL DIK A G+ ++
Sbjct: 193 VPSVFLAIGVLAMPESPRWLVMQGRLGDAFKVLDKTSNTKEEAISRLNDIKRAVGIPDDM 252
Query: 247 NDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFR 306
D+ + +P K G+GVWK+L++RPTPSVR +LIA +GIHF + A+GI+AV+LYSP IF
Sbjct: 253 TDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFSQQASGIDAVVLYSPTIFS 312
Query: 307 KAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLT 366
+AG+ SK LLAT+ VG+ K +F+V+ L+D+ GRR LL S GGM LT LG SLT
Sbjct: 313 RAGLKSKNDQLLATVAVGVVKTLFIVVGTCLVDRFGRRALLLTSMGGMFFSLTALGTSLT 372
Query: 367 VVDKSNGNVL-WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVN 425
V+D++ G L WA+ L++ +VA F++G GP+TWVY+SEIFP++LRAQGAS+GV +N
Sbjct: 373 VIDRNPGQTLKWAIGLAVTTVMTFVATFSLGAGPVTWVYASEIFPVRLRAQGASLGVMLN 432
Query: 426 RTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKK 485
R M+ ++ MTF+S+ K +TIGG+F +FAG++V AW+FF+ FLPET+G LEE+E +F
Sbjct: 433 RLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAVAAWVFFFTFLPETRGVPLEEIESLFGSY 492
Query: 486 SSGK 489
S+ K
Sbjct: 493 SANK 496
>gb|AAD26954.1| putative sugar transporter [Arabidopsis thaliana]
gi|15226682|ref|NP_179209.1| mannitol transporter,
putative [Arabidopsis thaliana] gi|25308957|pir||H84536
probable sugar transporter [imported] - Arabidopsis
thaliana
Length = 511
Score = 554 bits (1427), Expect = e-156
Identities = 274/484 (56%), Positives = 361/484 (73%), Gaps = 2/484 (0%)
Query: 7 DKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILN 66
+ E ++YAFACAI+ASM SI+ GYD GVMSGA +FIK+DL +SD Q E+L GILN
Sbjct: 14 ESEPPRGNRSRYAFACAILASMTSIILGYDIGVMSGASIFIKDDLKLSDVQLEILMGILN 73
Query: 67 LCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFA 126
+ +LVGS AGRTSD++GRRYTI LA F GA LMG+ NY +MVGR V G+GVG+A
Sbjct: 74 IYSLVGSGAAGRTSDWLGRRYTIVLAGAFFFCGALLMGFATNYPFIMVGRFVAGIGVGYA 133
Query: 127 LMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAA 186
+MIAPVY+AE++ ASSRGFLTS PE+ I IGILLGY+SNY K L LGWR MLG+ A
Sbjct: 134 MMIAPVYTAEVAPASSRGFLTSFPEIFINIGILLGYVSNYFFSK-LPEHLGWRFMLGVGA 192
Query: 187 LPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENC 246
+PS +A +L MPESPRWLV+QG+LG A KVL + SNT +EA RL DIK A G+ ++
Sbjct: 193 VPSVFLAIGVLAMPESPRWLVLQGRLGDAFKVLDKTSNTKEEAISRLDDIKRAVGIPDDM 252
Query: 247 NDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFR 306
D+ + +P K G+GVWK+L++RPTPSVR +LIA +GIHF + A+GI+AV+LYSP IF
Sbjct: 253 TDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFAQQASGIDAVVLYSPTIFS 312
Query: 307 KAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLT 366
KAG+ SK LLAT+ VG+ K +F+V+ ++D+ GRR LL S GGM + LT LG SLT
Sbjct: 313 KAGLKSKNDQLLATVAVGVVKTLFIVVGTCVVDRFGRRALLLTSMGGMFLSLTALGTSLT 372
Query: 367 VVDKSNGNVL-WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVN 425
V++++ G L WA+ L++ +VA F+IG GP+TWVY SEIFP++LRAQGAS+GV +N
Sbjct: 373 VINRNPGQTLKWAIGLAVTTVMTFVATFSIGAGPVTWVYCSEIFPVRLRAQGASLGVMLN 432
Query: 426 RTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKK 485
R M+ ++ MTF+S+ K +TIGG+F +FAG++ AW+FF+ FLPET+G LEEME +F
Sbjct: 433 RLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAAAAWVFFFTFLPETRGIPLEEMETLFGSY 492
Query: 486 SSGK 489
++ K
Sbjct: 493 TANK 496
>gb|AAG43998.1| mannitol transporter [Apium graveolens var. dulce]
Length = 513
Score = 552 bits (1423), Expect = e-156
Identities = 275/467 (58%), Positives = 359/467 (75%), Gaps = 2/467 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACA++ASM SI+ GYDTGV+SGA ++IKEDL SD Q E++ GI+N+ +L+GS
Sbjct: 21 NKYAFACALLASMNSILLGYDTGVLSGASIYIKEDLHFSDVQIEIIIGIINIYSLLGSAI 80
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYT+ LA I+F LGA MG N+A LM GR V G+GVG+A+MIAPVY+A
Sbjct: 81 AGRTSDWIGRRYTMVLAGIIFFLGAIFMGLATNFAFLMFGRFVAGIGVGYAMMIAPVYTA 140
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E++ +SSRGFLTS PE+ I G+LLGY+SN+ K L LGWR+MLGI A PS +A
Sbjct: 141 EVAPSSSRGFLTSFPEVFINSGVLLGYVSNFAFAK-CPLWLGWRIMLGIGAFPSVALAII 199
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG+A+ VL + S + +EA RL DIK AAG+D++CND+ V++P+
Sbjct: 200 VLYMPESPRWLVMQGRLGEARTVLEKTSTSKEEAHQRLSDIKEAAGIDKDCNDDVVQVPK 259
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
++ + E VWKELIL PT VR I +GIHFF+ A GI+AV+LYSPRIF KAGI S K
Sbjct: 260 RT-KDEAVWKELILHPTKPVRHAAITGIGIHFFQQACGIDAVVLYSPRIFEKAGIKSNSK 318
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLATI VG+ K VF++I+ F LDK+GRR L+ S GGM+I L +L SLTV++KS+
Sbjct: 319 KLLATIAVGVCKTVFILISTFQLDKIGRRPLMLTSMGGMVIALFVLAGSLTVINKSHHTG 378
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
WA L+I YA+V+ F+ G+GPI WVYSSE+FPL+LRAQG SIGVAVNR M+ ++ MT
Sbjct: 379 HWAGGLAIFTVYAFVSIFSSGMGPIAWVYSSEVFPLRLRAQGCSIGVAVNRGMSGIIGMT 438
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
FIS+YKA+TIGG+F +FA ++ + W+F Y PET+G+ LEE+E++F
Sbjct: 439 FISMYKAMTIGGAFLLFAVVASIGWVFMYTMFPETQGRNLEEIELLF 485
>gb|AAO88964.1| sorbitol transporter [Malus x domestica]
Length = 491
Score = 550 bits (1418), Expect = e-155
Identities = 267/458 (58%), Positives = 356/458 (77%), Gaps = 3/458 (0%)
Query: 28 MVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRTSDYIGRRY 87
M SI+ GYD GVMSGA ++I++DL ++DTQ E++ G++ + +L+GS AG+TSD++GRRY
Sbjct: 1 MTSILMGYDIGVMSGASIYIEKDLKVTDTQIEIMIGVIEIYSLIGSAMAGKTSDWVGRRY 60
Query: 88 TIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLT 147
TI ++ +F +GA LMG+ NY LM GR V G+GVG+AL IAPVYSAE+S SSRGFLT
Sbjct: 61 TIVISGAIFFIGAILMGFSTNYTFLMCGRFVAGIGVGYALTIAPVYSAEVSPTSSRGFLT 120
Query: 148 SLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLV 207
S PE+ + IGILLGY+SNY + L LGWRLMLG+ A+PS +A +L MPESPRWLV
Sbjct: 121 SFPEVFVNIGILLGYLSNYAF-SFCPLDLGWRLMLGVGAIPSVGLAVGVLAMPESPRWLV 179
Query: 208 MQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKEL 267
MQG+LG+AK+VL + S++ +E+ LRL DIK AAG+ E CND+ V++ SH GEGVWKEL
Sbjct: 180 MQGRLGEAKRVLDRTSDSKEESMLRLADIKEAAGIPEECNDDIVQVSGHSH-GEGVWKEL 238
Query: 268 ILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTK 327
++ PTP+VR +LIAA+G HFF+ A+GI+A++LYSPR+F KAGITS +LLL T+GVGL+K
Sbjct: 239 LVHPTPTVRHILIAAIGFHFFQQASGIDALVLYSPRVFAKAGITSTNQLLLCTVGVGLSK 298
Query: 328 IVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNG-NVLWALILSIVAT 386
VF ++A F LD++GRR LL S GM+ L LG SLT+VD+ G + WA+IL +
Sbjct: 299 TVFTLVATFFLDRVGRRPLLLTSMAGMVGALVCLGTSLTIVDQHEGVRMTWAVILCLCCV 358
Query: 387 YAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIG 446
AYV FF+ G+GPI WVYSSEIFPL+LRAQG +GVAVNR M+ ++SMTFIS+YKAIT+G
Sbjct: 359 LAYVGFFSSGIGPIAWVYSSEIFPLRLRAQGCGMGVAVNRLMSGILSMTFISLYKAITMG 418
Query: 447 GSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
G+FF++A I + W+FF+ LPET+G+ LE+ME++F K
Sbjct: 419 GTFFLYAAIGTVGWIFFFTMLPETQGRTLEDMEVLFGK 456
>gb|AAT06053.1| sorbitol transporter [Malus x domestica]
Length = 491
Score = 550 bits (1416), Expect = e-155
Identities = 267/458 (58%), Positives = 356/458 (77%), Gaps = 3/458 (0%)
Query: 28 MVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRTSDYIGRRY 87
M SI+ GYD GVMSGA ++I++DL ++DTQ E++ G++ + +L+GS AG+TSD++GRRY
Sbjct: 1 MTSILMGYDIGVMSGASIYIEKDLKVTDTQIEIMIGVIEIYSLIGSAMAGKTSDWVGRRY 60
Query: 88 TIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLT 147
TI ++ +F +GA LMG+ NY LM GR V G+GVG+AL IAPVYSAE+S SSRGFLT
Sbjct: 61 TIVISGAIFFIGAILMGFSTNYTFLMCGRFVAGIGVGYALTIAPVYSAEVSPTSSRGFLT 120
Query: 148 SLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLV 207
S PE+ + IGILLGY+SNY + L LGWRLMLG+ A+PS +A +L MPESPRWLV
Sbjct: 121 SFPEVFVNIGILLGYLSNYAF-SFCPLDLGWRLMLGVGAIPSVGLAVGVLAMPESPRWLV 179
Query: 208 MQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKEL 267
MQG+LG+AK+VL + S++ +E+ LRL DIK AAG+ E CND+ V++ SH GEGVWKEL
Sbjct: 180 MQGRLGEAKRVLDRTSDSKEESMLRLADIKEAAGIPEECNDDIVQVSGHSH-GEGVWKEL 238
Query: 268 ILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTK 327
++ PTP+VR +LIAA+G HFF+ A+GI+A++LYSPR+F KAGITS +LLL T+GVGL+K
Sbjct: 239 LVHPTPTVRHILIAAIGFHFFQQASGIDALVLYSPRVFAKAGITSTNQLLLCTVGVGLSK 298
Query: 328 IVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNG-NVLWALILSIVAT 386
VF ++A F LD++GRR LL S GM+ L LG SLT+VD+ G + WA+IL +
Sbjct: 299 TVFTLVATFFLDRVGRRPLLLTSMAGMVGALVCLGTSLTMVDQHEGVRMTWAVILCLCCV 358
Query: 387 YAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIG 446
AYV FF+ G+GPI WVYSSEIFPL+LRAQG +GVAVNR M+ ++SMTFIS+YKAIT+G
Sbjct: 359 LAYVGFFSSGIGPIAWVYSSEIFPLRLRAQGCGMGVAVNRLMSGILSMTFISLYKAITMG 418
Query: 447 GSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
G+FF++A I + W+FF+ LPET+G+ LE+ME++F K
Sbjct: 419 GTFFLYAAIGTVGWIFFFTMLPETQGRTLEDMEVLFGK 456
>gb|AAB68029.1| putative sugar transporter; member of major facilitative
superfamily; integral membrane protein [Beta vulgaris]
Length = 545
Score = 549 bits (1414), Expect = e-155
Identities = 276/468 (58%), Positives = 350/468 (73%), Gaps = 2/468 (0%)
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NK+AFACA +ASM S++ GYD GVMSGA++++KED ISDTQ VL GILN+ L GS
Sbjct: 34 NKFAFACATLASMTSVLLGYDIGVMSGAIIYLKEDWHISDTQIGVLVGILNIYCLFGSFA 93
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI LA +F +GA LMG+ NYA LMVGR V G+GVG+ALMIAPVY+A
Sbjct: 94 AGRTSDWIGRRYTIVLAGAIFFVGALLMGFATNYAFLMVGRFVTGIGVGYALMIAPVYTA 153
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFLTS PE+ I GILLGYISN L L WR MLGI A+PS +A
Sbjct: 154 EVSPASSRGFLTSFPEVFINAGILLGYISNLAFSS-LPTHLSWRFMLGIGAIPSIFLAIG 212
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVMQG+LG AKKVL ++S++ +EA+LRL +IK AG+ C+++ K+ +
Sbjct: 213 VLAMPESPRWLVMQGRLGDAKKVLNRISDSPEEAQLRLSEIKQTAGIPAECDEDIYKVEK 272
Query: 256 -KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKE 314
K G VWKEL PTP+VR +IA +GIHFF+ A+GI+AV+LYSPRIF+ AGIT+
Sbjct: 273 TKIKSGNAVWKELFFNPTPAVRRAVIAGIGIHFFQQASGIDAVVLYSPRIFQSAGITNAR 332
Query: 315 KLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGN 374
K LLAT+ VG+ K +F+++A F LDK GRR LL S GGMII + L +SLTV+D S+
Sbjct: 333 KQLLATVAVGVVKTLFILVATFQLDKYGRRPLLLTSVGGMIIAILTLAMSLTVIDHSHHK 392
Query: 375 VLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSM 434
+ WA+ L I A VA F+IGLGPITWVYSSE+FPL+LRAQG S+GVAVNR ++ V+S+
Sbjct: 393 ITWAIALCITMVCAVVASFSIGLGPITWVYSSEVFPLRLRAQGTSMGVAVNRVVSGVISI 452
Query: 435 TFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
F+ + IT GG+FF+F GI+++AW FF FLPET+G+ LE M +F
Sbjct: 453 FFLPLSHKITTGGAFFLFGGIAIIAWFFFLTFLPETRGRTLENMHELF 500
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 806,946,394
Number of Sequences: 2540612
Number of extensions: 33779521
Number of successful extensions: 136868
Number of sequences better than 10.0: 6304
Number of HSP's better than 10.0 without gapping: 3272
Number of HSP's successfully gapped in prelim test: 3034
Number of HSP's that attempted gapping in prelim test: 120912
Number of HSP's gapped (non-prelim): 9743
length of query: 501
length of database: 863,360,394
effective HSP length: 132
effective length of query: 369
effective length of database: 527,999,610
effective search space: 194831856090
effective search space used: 194831856090
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC147964.1


