

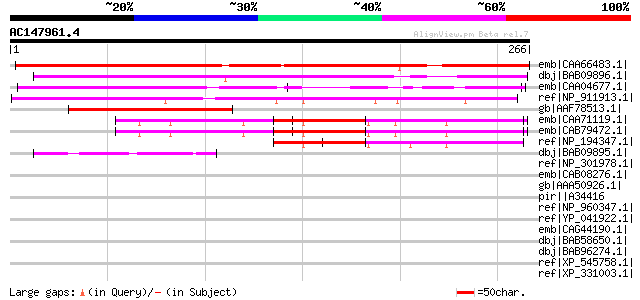
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147961.4 - phase: 0
(266 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA66483.1| transcripteion factor [Vicia faba] gi|7488868|pi... 327 2e-88
dbj|BAB09896.1| unnamed protein product [Arabidopsis thaliana] g... 167 4e-40
emb|CAA04677.1| putative transcription repressor HOTR [Hordeum v... 103 5e-21
ref|NP_911913.1| putative probable transcription repressor HOTR ... 97 5e-19
gb|AAF78513.1| putative transciption factor [Pyrus pyrifolia] 97 6e-19
emb|CAA71119.1| putative transcription factor [Arabidopsis thali... 85 2e-15
emb|CAB79472.1| putative transcription factor [Arabidopsis thali... 84 3e-15
ref|NP_194347.1| expressed protein [Arabidopsis thaliana] 71 4e-11
dbj|BAB09895.1| unnamed protein product [Arabidopsis thaliana] g... 68 3e-10
ref|NP_301978.1| possible pseudouridine synthase [Mycobacterium ... 37 0.57
emb|CAB08276.1| unnamed protein product [Mycobacterium leprae] g... 37 0.57
gb|AAA50926.1| u0247g [Mycobacterium leprae] gi|2145990|pir||S72... 36 0.97
pir||A34416 hydroxymethylglutaryl-CoA reductase (NADPH2) (EC 1.1... 35 2.8
ref|NP_960347.1| hypothetical protein MAP1413 [Mycobacterium avi... 34 3.7
ref|YP_041922.1| putative helicase [Staphylococcus aureus subsp.... 34 4.8
emb|CAG44190.1| putative helicase [Staphylococcus aureus subsp. ... 34 4.8
dbj|BAB58650.1| similar to putative helicase [Staphylococcus aur... 34 4.8
dbj|BAB96274.1| conserved hypothetical protein [Staphylococcus a... 34 4.8
ref|XP_545758.1| PREDICTED: similar to leukocyte differentiation... 33 6.3
ref|XP_331003.1| hypothetical protein [Neurospora crassa] gi|289... 33 6.3
>emb|CAA66483.1| transcripteion factor [Vicia faba] gi|7488868|pir||T12185
transcription factor - fava bean (fragment)
Length = 377
Score = 327 bits (838), Expect = 2e-88
Identities = 168/265 (63%), Positives = 205/265 (76%), Gaps = 13/265 (4%)
Query: 4 DSSLRTGRSLFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPD 63
+SS G SLF +IF Q FPIVYRWAPMQN+GDAL+TESQLLSTFDYAWNTI+NGTRRP
Sbjct: 118 ESSEEKGHSLFHDIFFQSFPIVYRWAPMQNKGDALQTESQLLSTFDYAWNTINNGTRRPA 177
Query: 64 DIHQMLNKLASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLTDDKLDEADSGSYNVLSR 123
DI QMLNK++SGTRTFS+VAK ++PFTQKKVGI IK+ KL +TD+K +D+ YN LSR
Sbjct: 178 DILQMLNKISSGTRTFSEVAKSLVPFTQKKVGILIKARKLPMTDNK---SDNDGYNFLSR 234
Query: 124 VFKFNRSRPRIVQDTTVGSATNENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINAS 183
VFKFNRSR ++ DT+ A +N KICGV+L DGSIC + PV KRVRC EHKGMRIN
Sbjct: 235 VFKFNRSRKVVIHDTS-DFAVEKNGKICGVILDDGSICSKMPVGKRVRCNEHKGMRINMV 293
Query: 184 TTKAIRSPKSELESI--VRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDG 241
TTKA+R KSE E++ + R ++ S V + V+E ITKT+ICGI+L+DG
Sbjct: 294 TTKAMRRSKSESENVFTAKEIRRSKSESEKVSE-------SLVDESITKTVICGIVLEDG 346
Query: 242 STCRRQPVKGRKRCQEHKGRRIRAN 266
STCR++PVKGRKRC EHKG+R+RA+
Sbjct: 347 STCRKEPVKGRKRCHEHKGKRVRAS 371
>dbj|BAB09896.1| unnamed protein product [Arabidopsis thaliana]
gi|15241932|ref|NP_200489.1| expressed protein
[Arabidopsis thaliana]
Length = 488
Score = 167 bits (422), Expect = 4e-40
Identities = 97/256 (37%), Positives = 141/256 (54%), Gaps = 26/256 (10%)
Query: 13 LFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKL 72
LF++IFS+G I+YRWAPM ++ +A TE LLSTFDYAWN SNG RR D+ + L
Sbjct: 142 LFEDIFSKGGSILYRWAPMGSKREAEATEGMLLSTFDYAWNKGSNGERRQLDLLKKLGDR 201
Query: 73 ASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLTDDK---LDEADSGSYNVLSRVFKFNR 129
++ S +++++ PF + +VGI IK K L +++ D + S N L+ + K R
Sbjct: 202 EFMSKRKSGISRMLFPFLRNQVGIRIKGEKHVLKEERKLTCDVDEEKSNNFLTSILKLTR 261
Query: 130 SRPRIVQDTTVGSATNENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTKAIR 189
SRP+ V D + + +CGV+L DG C R PV+ R RC EHKG R+ + +
Sbjct: 262 SRPQPVSDRFDEVDGSCSDIVCGVLLEDGGCCIRSPVKGRKRCIEHKGQRVCRVSPEKQT 321
Query: 190 SPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDGSTCRRQPV 249
PKSE+ + H+ +D ++CG+IL D C ++PV
Sbjct: 322 PPKSEIFT--------GQDHHNHKD---------------SDVVCGVILPDMEPCNKRPV 358
Query: 250 KGRKRCQEHKGRRIRA 265
GRKRC++HKG RI A
Sbjct: 359 PGRKRCEDHKGMRINA 374
>emb|CAA04677.1| putative transcription repressor HOTR [Hordeum vulgare subsp.
vulgare] gi|7489433|pir||T05738 probable transcription
repressor HOTR - barley
Length = 548
Score = 103 bits (257), Expect = 5e-21
Identities = 79/260 (30%), Positives = 121/260 (46%), Gaps = 49/260 (18%)
Query: 5 SSLRTGRSLFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDD 64
++L G LF+E+FS+G+ +++R A M ++ A +TE QLL FDYAWN + NG R ++
Sbjct: 123 NALTAGPGLFREVFSRGYSMMFRCALMGSKKAAEKTEGQLLGVFDYAWNKLQNGACRREE 182
Query: 65 IHQMLNKLASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLTDDKLDEADSGSYNVLSRV 124
I L + ++ S V L +K GI I SS ++ + S N+L RV
Sbjct: 183 ILLKLEQGSNRLSLLSRVRHLKQRVFGEKAGIKINSS------GSVEISSSSMKNMLPRV 236
Query: 125 FKFNRSRPRIVQDTTVGSATNENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINAST 184
F RPR+V G NE + I H+ A+T
Sbjct: 237 RTFVGFRPRLVNS---GDDLNEASDI------------------------HRKCTPQANT 269
Query: 185 TKAIRSPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDGSTC 244
+SE Y+ + ++ +RT E + CG++L+DGS+C
Sbjct: 270 AGKQAHRRSE-------GYKVKK----IDVIKRRTAPIREAEAV-----CGVMLEDGSSC 313
Query: 245 RRQPVKGRKRCQEHKGRRIR 264
P++GRKRC+ HKGRR+R
Sbjct: 314 LEDPMEGRKRCELHKGRRVR 333
Score = 58.5 bits (140), Expect = 2e-07
Identities = 38/120 (31%), Positives = 53/120 (43%), Gaps = 37/120 (30%)
Query: 143 ATNENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTKAIRSPKSELESIVRNA 202
A N+ + +CGV+ TD CK PV R RC EH+G+ + +++ A S +S L S
Sbjct: 456 ANNDASALCGVV-TDNGYCKLEPVIGRERCEEHRGIEVTGASS-APCSGRSVLPS----- 508
Query: 203 YRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDGSTCRRQPVKGRKRCQEHKGRR 262
+CG DGS C+ QP+ RKRC HKG+R
Sbjct: 509 ------------------------------VCGARASDGSPCKNQPIARRKRCALHKGQR 538
Score = 40.8 bits (94), Expect = 0.039
Identities = 24/77 (31%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Query: 185 TKAIRSPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITK-TIICGIILDDGST 243
T S E E R + A P R C E + +CG++ D+G
Sbjct: 414 TSHAESQFHEDEPCGRKWFERLKAQKSANAPSSRGQGCQPREANNDASALCGVVTDNGY- 472
Query: 244 CRRQPVKGRKRCQEHKG 260
C+ +PV GR+RC+EH+G
Sbjct: 473 CKLEPVIGRERCEEHRG 489
Score = 40.8 bits (94), Expect = 0.039
Identities = 15/40 (37%), Positives = 24/40 (59%)
Query: 150 ICGVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTKAIR 189
+CG +DGS CK P+ +R RC HKG R ++ +++
Sbjct: 509 VCGARASDGSPCKNQPIARRKRCALHKGQRACCASAPSVK 548
>ref|NP_911913.1| putative probable transcription repressor HOTR [Oryza sativa
(japonica cultivar-group)] gi|23617230|dbj|BAC20898.1|
putative probable transcription repressor HOTR [Oryza
sativa (japonica cultivar-group)]
Length = 622
Score = 97.1 bits (240), Expect = 5e-19
Identities = 87/337 (25%), Positives = 138/337 (40%), Gaps = 84/337 (24%)
Query: 2 SDDSSLRTGRSLFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRR 61
++ ++ TG LF+E+F +G+ +V+R A M N+ +A +TE++LL FDYAWN + NG R
Sbjct: 127 AETNTRATGNGLFREVFVRGYSLVFRCALMGNKQEAEKTEARLLRVFDYAWNKLQNGGLR 186
Query: 62 PDDIHQMLNKLASGTRT--FSDVAKLILPFTQKKVGIPIKSSKLHLTDDKLDEADSGSYN 119
++I L + A R+ S V ++K GI I + +D + N
Sbjct: 187 REEILIKLEQGAVNNRSSLLSRVRHFKQEVFREKAGIKIS------RNGSVDVSSGIMKN 240
Query: 120 VLSRVFKFNRSRPRIV--------------QDTTVGSATNENAK---------------- 149
+L R+ F RP++V ++T+ G++ + A+
Sbjct: 241 MLPRIRTFVGFRPQLVNSGDNVDKEIGIRWKNTSEGNSYGKQARRSSEGYKVKRVNVIKR 300
Query: 150 ----------ICGVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTK------------- 186
+CGVML DGS C PV+ R RC HKG R+ T
Sbjct: 301 RTMPEQDSNDVCGVMLEDGSSCLDHPVQGRKRCELHKGRRLGRITVNPKGSSCSYSCQVE 360
Query: 187 ----AIRSPKSELES---------------IVRNAYRYQNASHDVEDPPQRTVKCHVEEG 227
SP +E ES + + + E +T + +E+G
Sbjct: 361 IPVVESISPLTENESESDQAQQTSELLSKFLPATVKKSSRPWYSFEAKEIKTGEAPIEDG 420
Query: 228 ITKTI----ICGIILDDGSTCRRQPVKGRKRCQEHKG 260
+T IC D S C + + G K+CQ H G
Sbjct: 421 KQETSEVIDICEAKKSDNSACTNKVISGSKKCQLHNG 457
Score = 45.4 bits (106), Expect = 0.002
Identities = 18/30 (60%), Positives = 21/30 (70%)
Query: 233 ICGIILDDGSTCRRQPVKGRKRCQEHKGRR 262
ICG DGS C+ QP+ GRKRC HKG+R
Sbjct: 585 ICGARASDGSPCKNQPIAGRKRCAMHKGQR 614
Score = 38.9 bits (89), Expect = 0.15
Identities = 16/30 (53%), Positives = 18/30 (59%)
Query: 150 ICGVMLTDGSICKRPPVEKRVRCPEHKGMR 179
ICG +DGS CK P+ R RC HKG R
Sbjct: 585 ICGARASDGSPCKNQPIAGRKRCAMHKGQR 614
>gb|AAF78513.1| putative transciption factor [Pyrus pyrifolia]
Length = 104
Score = 96.7 bits (239), Expect = 6e-19
Identities = 47/84 (55%), Positives = 61/84 (71%)
Query: 31 MQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGTRTFSDVAKLILPFT 90
M+N+ DAL+TE+QLL TFDYAWNT NG RRPDD+ + L K++S T F++ A+ +LPF+
Sbjct: 1 MENKSDALKTETQLLDTFDYAWNTTINGARRPDDVLRKLKKISSSTTRFANFAEKLLPFS 60
Query: 91 QKKVGIPIKSSKLHLTDDKLDEAD 114
QKKVGI +SSK T DK D
Sbjct: 61 QKKVGIKNESSKPISTGDKFGAYD 84
>emb|CAA71119.1| putative transcription factor [Arabidopsis thaliana]
Length = 349
Score = 84.7 bits (208), Expect = 2e-15
Identities = 67/229 (29%), Positives = 105/229 (45%), Gaps = 46/229 (20%)
Query: 55 ISNGTRRPDDI--------HQMLNKLASGTRTFSD-------VAKLILPFTQKKVGIPIK 99
+S+ R DD+ + ++L S +R+F+ +++ ILP TQ K +
Sbjct: 35 LSHDHGRKDDVLVANLGQPESIRSRLRSYSRSFAHHDLLKQGLSQTILPTTQNKSDNQTE 94
Query: 100 SSKLHLTDDKLDEADSGSY--NVLSRVFKFNRSRPRIVQDTTVGSAT-NENAKICGVMLT 156
K +++ +D+ N L + + +RSRP+ V + +++A CGV+L
Sbjct: 95 EKKSDSEEEREVSSDAAEKESNSLPSILRLSRSRPQPVSEKHDDIVDESDSASACGVLLE 154
Query: 157 DGSICKRPPVEKRVRCPEHKGMRINASTTKAIRSPKSELESIVRNAYRYQNASHDVEDPP 216
DG+ C PV+ R RC EHKG R++ + I P E P
Sbjct: 155 DGTTCTTTPVKXRKRCTEHKGKRLSR-VSPGIHIP--------------------CEVPT 193
Query: 217 QRTVKCHVEEGITKTIICGIILDDGSTCRRQPVKGRKRCQEHKGRRIRA 265
R +C E I CG+IL D CR +PV RKRC++HKG R+ A
Sbjct: 194 VR--ECEETENI-----CGVILPDMIRCRSKPVSRRKRCEDHKGMRVNA 235
Score = 82.0 bits (201), Expect = 2e-14
Identities = 51/149 (34%), Positives = 69/149 (46%), Gaps = 31/149 (20%)
Query: 146 ENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA--------STTKAIRSPKSELE- 196
E ICGV+L D C+ PV +R RC +HKGMR+NA KA+ KS+ E
Sbjct: 199 ETENICGVILPDMIRCRSKPVSRRKRCEDHKGMRVNAFFFLLNPTERDKAVNEDKSKPET 258
Query: 197 --------SIVRNAYRYQNASHDVEDPPQRTVKC--------------HVEEGITKTIIC 234
S + +N P+ + +C +V+ +IC
Sbjct: 259 STGMNQEGSGLLCEATTKNGLPCTRSAPEGSKRCWQHKDKTLNHGSSENVQSATASQVIC 318
Query: 235 GIILDDGSTCRRQPVKGRKRCQEHKGRRI 263
G L +GS C + PVKGRKRC+EHKG RI
Sbjct: 319 GFKLYNGSVCEKSPVKGRKRCEEHKGMRI 347
Score = 49.3 bits (116), Expect = 1e-04
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 7/54 (12%)
Query: 136 QDTTVGSATNENAK-------ICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA 182
+D T+ ++EN + ICG L +GS+C++ PV+ R RC EHKGMRI +
Sbjct: 296 KDKTLNHGSSENVQSATASQVICGFKLYNGSVCEKSPVKGRKRCEEHKGMRITS 349
>emb|CAB79472.1| putative transcription factor [Arabidopsis thaliana]
gi|4538946|emb|CAB39682.1| putative transcription factor
[Arabidopsis thaliana] gi|25372096|pir||A85303 probable
transcription factor [imported] - Arabidopsis thaliana
gi|7485837|pir||T04272 hypothetical protein F20B18.280 -
Arabidopsis thaliana (fragment)
Length = 349
Score = 84.3 bits (207), Expect = 3e-15
Identities = 67/229 (29%), Positives = 105/229 (45%), Gaps = 46/229 (20%)
Query: 55 ISNGTRRPDDI--------HQMLNKLASGTRTFSD-------VAKLILPFTQKKVGIPIK 99
+S+ R DD+ + ++L S +R+F+ +++ ILP TQ K +
Sbjct: 35 LSHDHGRKDDVLVANLGQPESIRSRLRSYSRSFAHHDLLKQGLSQTILPTTQNKSDNQTE 94
Query: 100 SSKLHLTDDKLDEADSGSY--NVLSRVFKFNRSRPRIVQDTTVGSAT-NENAKICGVMLT 156
K +++ +D+ N L + + +RSRP+ V + +++A CGV+L
Sbjct: 95 EKKSDSEEEREVSSDAAEKESNSLPSILRLSRSRPQPVSEKHDDIVDESDSASACGVLLE 154
Query: 157 DGSICKRPPVEKRVRCPEHKGMRINASTTKAIRSPKSELESIVRNAYRYQNASHDVEDPP 216
DG+ C PV+ R RC EHKG R++ + I P E P
Sbjct: 155 DGTTCTTTPVKGRKRCTEHKGKRLSR-VSPGIHIP--------------------CEVPT 193
Query: 217 QRTVKCHVEEGITKTIICGIILDDGSTCRRQPVKGRKRCQEHKGRRIRA 265
R +C E I CG+IL D CR +PV RKRC++HKG R+ A
Sbjct: 194 VR--ECEETENI-----CGVILPDMIRCRSKPVSRRKRCEDHKGMRVNA 235
Score = 82.0 bits (201), Expect = 2e-14
Identities = 51/149 (34%), Positives = 69/149 (46%), Gaps = 31/149 (20%)
Query: 146 ENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA--------STTKAIRSPKSELE- 196
E ICGV+L D C+ PV +R RC +HKGMR+NA KA+ KS+ E
Sbjct: 199 ETENICGVILPDMIRCRSKPVSRRKRCEDHKGMRVNAFFFLLNPTERDKAVNEDKSKPET 258
Query: 197 --------SIVRNAYRYQNASHDVEDPPQRTVKC--------------HVEEGITKTIIC 234
S + +N P+ + +C +V+ +IC
Sbjct: 259 STGMNQEGSGLLCEATTKNGLPCTRSAPEGSKRCWQHKDKTLNHGSSENVQSATASQVIC 318
Query: 235 GIILDDGSTCRRQPVKGRKRCQEHKGRRI 263
G L +GS C + PVKGRKRC+EHKG RI
Sbjct: 319 GFKLYNGSVCEKSPVKGRKRCEEHKGMRI 347
Score = 49.3 bits (116), Expect = 1e-04
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 7/54 (12%)
Query: 136 QDTTVGSATNENAK-------ICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA 182
+D T+ ++EN + ICG L +GS+C++ PV+ R RC EHKGMRI +
Sbjct: 296 KDKTLNHGSSENVQSATASQVICGFKLYNGSVCEKSPVKGRKRCEEHKGMRITS 349
>ref|NP_194347.1| expressed protein [Arabidopsis thaliana]
Length = 139
Score = 70.9 bits (172), Expect = 4e-11
Identities = 43/134 (32%), Positives = 60/134 (44%), Gaps = 31/134 (23%)
Query: 161 CKRPPVEKRVRCPEHKGMRINA--------STTKAIRSPKSELESIVRNAYR-------- 204
C+ PV +R RC +HKGMR+NA KA+ KS+ E+
Sbjct: 4 CRSKPVSRRKRCEDHKGMRVNAFFFLLNPTERDKAVNEDKSKPETSTGMNQEGSGLLCEA 63
Query: 205 -YQNASHDVEDPPQRTVKC--------------HVEEGITKTIICGIILDDGSTCRRQPV 249
+N P+ + +C +V+ +ICG L +GS C + PV
Sbjct: 64 TTKNGLPCTRSAPEGSKRCWQHKDKTLNHGSSENVQSATASQVICGFKLYNGSVCEKSPV 123
Query: 250 KGRKRCQEHKGRRI 263
KGRKRC+EHKG RI
Sbjct: 124 KGRKRCEEHKGMRI 137
Score = 49.3 bits (116), Expect = 1e-04
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 7/54 (12%)
Query: 136 QDTTVGSATNENAK-------ICGVMLTDGSICKRPPVEKRVRCPEHKGMRINA 182
+D T+ ++EN + ICG L +GS+C++ PV+ R RC EHKGMRI +
Sbjct: 86 KDKTLNHGSSENVQSATASQVICGFKLYNGSVCEKSPVKGRKRCEEHKGMRITS 139
Score = 35.8 bits (81), Expect = 1.3
Identities = 13/22 (59%), Positives = 17/22 (77%)
Query: 244 CRRQPVKGRKRCQEHKGRRIRA 265
CR +PV RKRC++HKG R+ A
Sbjct: 4 CRSKPVSRRKRCEDHKGMRVNA 25
>dbj|BAB09895.1| unnamed protein product [Arabidopsis thaliana]
gi|15241930|ref|NP_200488.1| hypothetical protein
[Arabidopsis thaliana]
Length = 254
Score = 67.8 bits (164), Expect = 3e-10
Identities = 39/94 (41%), Positives = 55/94 (58%), Gaps = 10/94 (10%)
Query: 13 LFQEIFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKL 72
L+++IFS+G+ + YRWAP +A TE LLSTFDYAWNT SNG RR H L KL
Sbjct: 158 LYEDIFSEGYSVFYRWAP-----EAAATEGMLLSTFDYAWNTCSNGERR----HLELQKL 208
Query: 73 ASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLT 106
+++++P + +V + IK K + T
Sbjct: 209 GDPEFMSKRKSQVLVPSIRDQV-VTIKVEKSNYT 241
>ref|NP_301978.1| possible pseudouridine synthase [Mycobacterium leprae TN]
gi|13093266|emb|CAC31751.1| possible pseudouridine
synthase [Mycobacterium leprae] gi|25323975|pir||D87080
probable pseudouridine synthase [imported] -
Mycobacterium leprae
Length = 257
Score = 37.0 bits (84), Expect = 0.57
Identities = 42/140 (30%), Positives = 61/140 (43%), Gaps = 17/140 (12%)
Query: 134 IVQDTTVGSATNENAKICGVMLTD-GSICKRPPVEKRVRCPE---HKGMRINASTTKAIR 189
+V D+ V A N+ + M D G C +E+RVR + H G R++A T I
Sbjct: 72 VVDDSLVYLALNKPRGMYSTMSDDRGRPCVGDLIERRVRGNKKLFHVG-RLDADTEGLIL 130
Query: 190 SPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDG-------S 242
L + A+R + SH+V TVK V G+ K + G+ LDDG +
Sbjct: 131 -----LTNDGELAHRLMHPSHEVSKTYLATVKGAVPRGLGKKLSVGLELDDGPAHVDDFA 185
Query: 243 TCRRQPVKGRKRCQEHKGRR 262
P K R H+GR+
Sbjct: 186 VVDAIPGKTLVRLTLHEGRK 205
>emb|CAB08276.1| unnamed protein product [Mycobacterium leprae]
gi|3915542|sp|O05668|Y1370_MYCLE Hypothetical RNA
pseudouridine synthase ML1370 (RNA-uridine isomerase)
(RNA pseudouridylate synthase)
Length = 256
Score = 37.0 bits (84), Expect = 0.57
Identities = 42/140 (30%), Positives = 61/140 (43%), Gaps = 17/140 (12%)
Query: 134 IVQDTTVGSATNENAKICGVMLTD-GSICKRPPVEKRVRCPE---HKGMRINASTTKAIR 189
+V D+ V A N+ + M D G C +E+RVR + H G R++A T I
Sbjct: 71 VVDDSLVYLALNKPRGMYSTMSDDRGRPCVGDLIERRVRGNKKLFHVG-RLDADTEGLIL 129
Query: 190 SPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDG-------S 242
L + A+R + SH+V TVK V G+ K + G+ LDDG +
Sbjct: 130 -----LTNDGELAHRLMHPSHEVSKTYLATVKGAVPRGLGKKLSVGLELDDGPAHVDDFA 184
Query: 243 TCRRQPVKGRKRCQEHKGRR 262
P K R H+GR+
Sbjct: 185 VVDAIPGKTLVRLTLHEGRK 204
>gb|AAA50926.1| u0247g [Mycobacterium leprae] gi|2145990|pir||S72955 hypothetical
protein u0247g - Mycobacterium leprae
Length = 186
Score = 36.2 bits (82), Expect = 0.97
Identities = 42/140 (30%), Positives = 61/140 (43%), Gaps = 17/140 (12%)
Query: 134 IVQDTTVGSATNENAKICGVMLTD-GSICKRPPVEKRVRCPE---HKGMRINASTTKAIR 189
+V D+ V A N+ + M D G C +E+RVR + H G R++A T I
Sbjct: 1 MVDDSLVYLALNKPRGMYSTMSDDRGRPCVGDLIERRVRGNKKLFHVG-RLDADTEGLIL 59
Query: 190 SPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDG-------S 242
L + A+R + SH+V TVK V G+ K + G+ LDDG +
Sbjct: 60 -----LTNDGELAHRLMHPSHEVSKTYLATVKGAVPRGLGKKLSVGLELDDGPAHVDDFA 114
Query: 243 TCRRQPVKGRKRCQEHKGRR 262
P K R H+GR+
Sbjct: 115 VVDAIPGKTLVRLTLHEGRK 134
>pir||A34416 hydroxymethylglutaryl-CoA reductase (NADPH2) (EC 1.1.1.34) - fluke
(Schistosoma mansoni) gi|123345|sp|P16237|HMDH_SCHMA
3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA
reductase) gi|161021|gb|AAA29896.1|
3-hydroxy-3-methylglutaryl coenzyme A reductase
Length = 948
Score = 34.7 bits (78), Expect = 2.8
Identities = 36/124 (29%), Positives = 54/124 (43%), Gaps = 28/124 (22%)
Query: 86 ILP-FTQKKVGIPIKSSK----LHLTDDKLDEADSGSYNVLSRVFKFNRSRPRIVQDTTV 140
+LP F++K IP++S K LH DD +D DS S + S K + RP V D +
Sbjct: 398 VLPKFSKKLNDIPLQSRKRIYCLHKDDDYIDRNDSSSVSTFSNTCKNSNERPSNVLD--L 455
Query: 141 GSATNENAKICGVMLTDGSICKRPPVEKRVRCPEHKGMRINASTTKAIRSPKSELESIVR 200
T + + G L+D I ++ H ++ ELES+VR
Sbjct: 456 DMLTEKIKQGLGHELSDTEI---------LQLLSHGRLKTR------------ELESVVR 494
Query: 201 NAYR 204
N +R
Sbjct: 495 NPFR 498
>ref|NP_960347.1| hypothetical protein MAP1413 [Mycobacterium avium subsp.
paratuberculosis str. k10] gi|41395864|gb|AAS03730.1|
hypothetical protein MAP1413 [Mycobacterium avium subsp.
paratuberculosis str. k10]
Length = 247
Score = 34.3 bits (77), Expect = 3.7
Identities = 34/112 (30%), Positives = 52/112 (46%), Gaps = 10/112 (8%)
Query: 134 IVQDTTVGSATNENAKICGVMLTD-GSICKRPPVEKRVRCPE---HKGMRINASTTKAIR 189
++ ++ V A N+ + M D G C +E+RVR + H G R++A T I
Sbjct: 62 VLDESQVYLALNKPRGVHSTMSDDRGRPCIGDLIERRVRGNKNLFHVG-RLDAETEGLIL 120
Query: 190 SPKSELESIVRNAYRYQNASHDVEDPPQRTVKCHVEEGITKTIICGIILDDG 241
L + A+R + SH+V TV + G+ KT+ GI LDDG
Sbjct: 121 -----LTNDGELAHRLMHPSHEVPKTYLATVTGSIPRGLGKTLRAGIELDDG 167
>ref|YP_041922.1| putative helicase [Staphylococcus aureus subsp. aureus MRSA252]
gi|49242827|emb|CAG41554.1| putative helicase
[Staphylococcus aureus subsp. aureus MRSA252]
Length = 953
Score = 33.9 bits (76), Expect = 4.8
Identities = 19/68 (27%), Positives = 34/68 (49%)
Query: 17 IFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGT 76
I SQ PI + ++ D+ + E + LS + W T SN T + ++ ++L+ A G
Sbjct: 833 IRSQEMPIFITYDKHEDISDSTKYEDEFLSQDELKWFTKSNRTLKSKEVQKILSHRAKGI 892
Query: 77 RTFSDVAK 84
+ + V K
Sbjct: 893 KMYIFVQK 900
>emb|CAG44190.1| putative helicase [Staphylococcus aureus subsp. aureus MSSA476]
gi|49487268|ref|YP_044489.1| putative helicase
[Staphylococcus aureus subsp. aureus MSSA476]
Length = 953
Score = 33.9 bits (76), Expect = 4.8
Identities = 19/68 (27%), Positives = 34/68 (49%)
Query: 17 IFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGT 76
I SQ PI + ++ D+ + E + LS + W T SN T + ++ ++L+ A G
Sbjct: 833 IRSQEMPIFITYDKHEDISDSTKYEDEFLSQDELKWFTKSNRTLKSKEVQKILSHRAKGI 892
Query: 77 RTFSDVAK 84
+ + V K
Sbjct: 893 KMYIFVQK 900
>dbj|BAB58650.1| similar to putative helicase [Staphylococcus aureus subsp. aureus
Mu50] gi|15928067|ref|NP_375600.1| hypothetical protein
SA2276 [Staphylococcus aureus subsp. aureus N315]
gi|13702438|dbj|BAB43579.1| conserved hypothetical
protein [Staphylococcus aureus subsp. aureus N315]
gi|25506073|pir||A99952 conserved hypothetical protein
SA2276 [imported] - Staphylococcus aureus (strain N315)
gi|15925478|ref|NP_373012.1| similar to DNA or RNA
helicases of superfamily II (truncated) [Staphylococcus
aureus subsp. aureus Mu50]
Length = 496
Score = 33.9 bits (76), Expect = 4.8
Identities = 19/68 (27%), Positives = 34/68 (49%)
Query: 17 IFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGT 76
I SQ PI + ++ D+ + E + LS + W T SN T + ++ ++L+ A G
Sbjct: 376 IRSQEMPIFITYDKHEDISDSTKYEDEFLSQDELKWFTKSNRTLKSKEVQKILSHRAKGI 435
Query: 77 RTFSDVAK 84
+ + V K
Sbjct: 436 KMYIFVQK 443
>dbj|BAB96274.1| conserved hypothetical protein [Staphylococcus aureus subsp. aureus
MW2] gi|21284138|ref|NP_647226.1| hypothetical protein
MW2409 [Staphylococcus aureus subsp. aureus MW2]
Length = 951
Score = 33.9 bits (76), Expect = 4.8
Identities = 19/68 (27%), Positives = 34/68 (49%)
Query: 17 IFSQGFPIVYRWAPMQNEGDALRTESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGT 76
I SQ PI + ++ D+ + E + LS + W T SN T + ++ ++L+ A G
Sbjct: 833 IRSQEMPIFITYDKHEDISDSTKYEDEFLSQDELKWFTKSNRTLKSKEVQKILSHRAKGI 892
Query: 77 RTFSDVAK 84
+ + V K
Sbjct: 893 KMYIFVQK 900
>ref|XP_545758.1| PREDICTED: similar to leukocyte differentiation antigen CD84 [Canis
familiaris]
Length = 641
Score = 33.5 bits (75), Expect = 6.3
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 5/59 (8%)
Query: 24 IVYRWAPMQNEGDALR----TESQLLSTFDYAWNTISNGTRRPDDIHQMLNKLASGTRT 78
+ Y W+P+ EG+A+R +SQ L+ AWN +SN + + Q+ A+G R+
Sbjct: 166 VTYSWSPLGEEGNAIRIFQTPDSQRLTYTCTAWNPVSNSSDSISGL-QLCADAATGPRS 223
>ref|XP_331003.1| hypothetical protein [Neurospora crassa] gi|28921086|gb|EAA30404.1|
hypothetical protein [Neurospora crassa]
Length = 452
Score = 33.5 bits (75), Expect = 6.3
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 8/77 (10%)
Query: 65 IHQMLNKLASGTRTFSDVAKLILPFTQKKVGIPIKSSKLHLTDDKL------DEADSGSY 118
+H+ L ++ S +RT +V K +L + + +G K+ L DK D+ G +
Sbjct: 65 LHKSLIEIPSISRTEQEVGKFLLDYLRNNLGYVAKAQ--FLESDKTSHGSDGDDHSQGRF 122
Query: 119 NVLSRVFKFNRSRPRIV 135
NVL+ N S PR++
Sbjct: 123 NVLAWPSSHNLSSPRVL 139
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 443,676,720
Number of Sequences: 2540612
Number of extensions: 17627217
Number of successful extensions: 36366
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 36304
Number of HSP's gapped (non-prelim): 54
length of query: 266
length of database: 863,360,394
effective HSP length: 126
effective length of query: 140
effective length of database: 543,243,282
effective search space: 76054059480
effective search space used: 76054059480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC147961.4


