

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.7 + phase: 0
(248 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
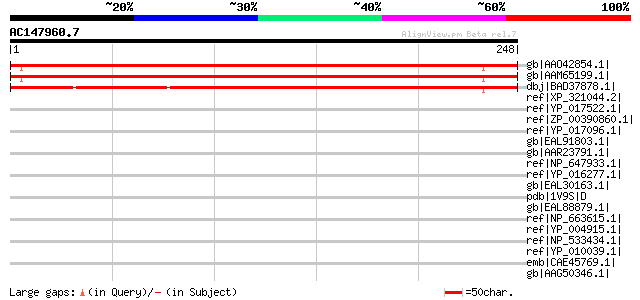
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO42854.1| At1g05060 [Arabidopsis thaliana] gi|18390475|ref|... 288 1e-76
gb|AAM65199.1| unknown [Arabidopsis thaliana] 286 5e-76
dbj|BAD37878.1| hypothetical protein [Oryza sativa (japonica cul... 229 7e-59
ref|XP_321044.2| ENSANGP00000007849 [Anopheles gambiae str. PEST... 40 0.077
ref|YP_017522.1| s-layer protein ea1 [Bacillus anthracis str. 'A... 38 0.22
ref|ZP_00390860.1| COG5280: Phage-related minor tail protein [Ba... 35 1.5
ref|YP_017096.1| prophage lambdaba04, tape measure protein, puta... 35 1.5
gb|EAL91803.1| serine/threonine protein kinase, putative [Asperg... 34 3.2
gb|AAR23791.1| S-layer protein [Bacillus thuringiensis] 33 5.5
ref|NP_647933.1| CG13708-PA [Drosophila melanogaster] gi|7292542... 33 5.5
ref|YP_016277.1| pyruvate dehydrogenase E3 component dihydrolipo... 33 5.5
gb|EAL30163.1| GA12474-PA [Drosophila pseudoobscura] 33 5.5
pdb|1V9S|D Chain D, Crystal Structure Of Tt0130 Protein From The... 33 5.5
gb|EAL88879.1| receptor L domain protein [Aspergillus fumigatus ... 33 7.2
ref|NP_663615.1| apolipoprotein L3 isoform 1 [Homo sapiens] 33 9.4
ref|YP_004915.1| uracil phosphoribosyltransferase [Thermus therm... 33 9.4
ref|NP_533434.1| acetyl-CoA acetyltransferase [Agrobacterium tum... 33 9.4
ref|YP_010039.1| hypothetical protein DVU0818 [Desulfovibrio vul... 33 9.4
emb|CAE45769.1| Tbx2/3 protein [Pleurobrachia pileus] 33 9.4
gb|AAG50346.1| apolipoprotein L-III splice variant alpha/d [Homo... 33 9.4
>gb|AAO42854.1| At1g05060 [Arabidopsis thaliana] gi|18390475|ref|NP_563727.1|
expressed protein [Arabidopsis thaliana]
gi|4056427|gb|AAC98001.1| Contains similarity to
gb|AJ006354 zinc finger protein (ZAC) from Homo sapiens.
[Arabidopsis thaliana] gi|25373368|pir||F86184
hypothetical protein [imported] - Arabidopsis thaliana
Length = 253
Score = 288 bits (736), Expect = 1e-76
Identities = 146/252 (57%), Positives = 184/252 (72%), Gaps = 4/252 (1%)
Query: 1 MSFL-AGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRH 59
MSFL AGRLAGKEAAYFFQESK AV +LA+K+ K + DVLPE+LRH
Sbjct: 1 MSFLGAGRLAGKEAAYFFQESKHAVNRLAEKSPATGKKLPSSPPDPPEIQPDVLPEILRH 60
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWEL 119
SLPSK++ SS SKW L+SDP S+SPD +NPLR +VSLPQVTFG RRW+L
Sbjct: 61 SLPSKIYGRPPDPSSLSQFSKWALESDPNATVSISPDVLNPLRGYVSLPQVTFGRRRWDL 120
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
PE+++ V ASTANELR+DRY VNPEKLKAA EGL ++GKAFA AT ++FG A +V G
Sbjct: 121 PESENSVLASTANELRRDRYGTPVNPEKLKAAGEGLQHIGKAFAAATIIIFGSATLVFGT 180
Query: 180 VASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE---DVKQKA 236
ASKL++ N D++TKGKD+ +P+LE++K P++ WAENMS+KWH+E E +K+K
Sbjct: 181 AASKLDMRNADDIRTKGKDLFQPKLESMKEQVEPLRTWAENMSKKWHIENEGGNTIKEKP 240
Query: 237 IVKDLSKILGSK 248
I+K+LSKILG K
Sbjct: 241 ILKELSKILGPK 252
>gb|AAM65199.1| unknown [Arabidopsis thaliana]
Length = 253
Score = 286 bits (731), Expect = 5e-76
Identities = 145/252 (57%), Positives = 183/252 (72%), Gaps = 4/252 (1%)
Query: 1 MSFL-AGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRH 59
MSFL AGRL GKEAAYFFQESK AV +LA+K+ K + DVLPE+LRH
Sbjct: 1 MSFLGAGRLPGKEAAYFFQESKHAVNRLAEKSPATGKKLPSSPPDPPEIQPDVLPEILRH 60
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWEL 119
SLPSK++ SS SKW L+SDP S+SPD +NPLR +VSLPQVTFG RRW+L
Sbjct: 61 SLPSKIYGRPPDPSSLSQFSKWALESDPNATVSISPDVLNPLRGYVSLPQVTFGRRRWDL 120
Query: 120 PEAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
PE+++ V ASTANELR+DRY VNPEKLKAA EGL ++GKAFA AT ++FG A +V G
Sbjct: 121 PESENSVLASTANELRRDRYGTPVNPEKLKAAGEGLQHIGKAFAAATIIIFGSATLVFGT 180
Query: 180 VASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE---DVKQKA 236
ASKL++ N D++TKGKD+ +P+LE++K P++ WAENMS+KWH+E E +K+K
Sbjct: 181 AASKLDMRNADDIRTKGKDLFQPKLESMKEQVEPLRTWAENMSKKWHIENEGGNTIKEKP 240
Query: 237 IVKDLSKILGSK 248
I+K+LSKILG K
Sbjct: 241 ILKELSKILGPK 252
>dbj|BAD37878.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 251
Score = 229 bits (583), Expect = 7e-59
Identities = 120/249 (48%), Positives = 165/249 (66%), Gaps = 3/249 (1%)
Query: 1 MSFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHS 60
MSFLAGRLA KE AYF QESK A +LA+K P S + DVLPE+LRH+
Sbjct: 1 MSFLAGRLAAKEGAYFLQESKHAAGRLAEKL-PASAPAPAPAPGSTSPSPDVLPEILRHA 59
Query: 61 LPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELP 120
+P K S S S S+W + + +SPDA+NPLR++VSLPQ TFG +RW+LP
Sbjct: 60 VPIKATPPPGEPSLSAS-SRWAVPRGGAEAAGLSPDALNPLRSYVSLPQATFGPKRWQLP 118
Query: 121 EAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMV 180
+ S+STANE R+DR+ ++PEKLKA G + +GKAF AT +VFGG+ V+
Sbjct: 119 NEQPNYSSSTANERRRDRHPPPMDPEKLKAVIAGYSQIGKAFIAATILVFGGSTAVLLYT 178
Query: 181 ASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE-DVKQKAIVK 239
A KL+LH++ D++TKG+D V+P+ + IK P++ WAE MSRKWH E + D K+K+I++
Sbjct: 179 ADKLQLHSVDDVRTKGRDAVQPRADMIKEQIAPLRSWAEEMSRKWHFEGDKDAKEKSIIR 238
Query: 240 DLSKILGSK 248
+LS+ LGS+
Sbjct: 239 ELSRALGSR 247
>ref|XP_321044.2| ENSANGP00000007849 [Anopheles gambiae str. PEST]
gi|55233363|gb|EAA01243.2| ENSANGP00000007849 [Anopheles
gambiae str. PEST]
Length = 829
Score = 39.7 bits (91), Expect = 0.077
Identities = 18/46 (39%), Positives = 30/46 (65%)
Query: 53 LPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAI 98
LPE L L +L +ETA +S +A +W++Q+DP L++ V +A+
Sbjct: 710 LPEGLLQPLLQRLRLEETAHASKMTAKEWLMQADPSLKNIVGKEAL 755
>ref|YP_017522.1| s-layer protein ea1 [Bacillus anthracis str. 'Ames Ancestor']
gi|30261021|ref|NP_843398.1| S-layer protein EA1
[Bacillus anthracis str. Ames]
gi|49183866|ref|YP_027118.1| S-layer protein EA1
[Bacillus anthracis str. Sterne]
gi|30254635|gb|AAP24884.1| S-layer protein EA1 [Bacillus
anthracis str. Ames] gi|47501321|gb|AAT29997.1| S-layer
protein EA1 [Bacillus anthracis str. 'Ames Ancestor']
gi|65318302|ref|ZP_00391261.1| COG3210: Large
exoproteins involved in heme utilization or adhesion
[Bacillus anthracis str. A2012]
gi|49177793|gb|AAT53169.1| S-layer protein EA1 [Bacillus
anthracis str. Sterne] gi|1769919|emb|CAA68063.1| EA1
[Bacillus anthracis] gi|17368380|sp|P94217|SLAP2_BACAN
S-layer protein EA1 precursor
Length = 862
Score = 38.1 bits (87), Expect = 0.22
Identities = 37/126 (29%), Positives = 54/126 (42%), Gaps = 8/126 (6%)
Query: 103 AFVSLPQVTFGARRWELPEAKHGVSASTANELRQDRYD---VNVNPEKLKAASEGLANLG 159
AF + V+ + + P+ K ++ ST E + +Y V +PE L EG
Sbjct: 616 AFKNFELVSKVGQYGQSPDTKLDLNVSTTVEYQLSKYTSDRVYSDPENL----EGYEVES 671
Query: 160 KAFAIATAVVFGGAAMVIGMVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMK-VWA 218
K A+A A + G +V G K+++H + T GK VE E I V K V
Sbjct: 672 KNLAVADAKIVGNKVVVTGKTPGKVDIHLTKNGATAGKATVEIVQETIAIKSVNFKPVQT 731
Query: 219 ENMSRK 224
EN K
Sbjct: 732 ENFVEK 737
>ref|ZP_00390860.1| COG5280: Phage-related minor tail protein [Bacillus anthracis str.
A2012]
Length = 1311
Score = 35.4 bits (80), Expect = 1.5
Identities = 32/129 (24%), Positives = 50/129 (37%), Gaps = 8/129 (6%)
Query: 127 SASTANELRQDRYDVNVNPEKLKAASEGLANLGKAF--AIATAVVFGGAAMVIGMVASKL 184
S N ++ R D+N ++ + ASE + LG + V GG VIG
Sbjct: 197 SVDDGNSIQNIRNDLNQLSQEAEEASESVKGLGVELENVLGGIVAGGGIQEVIGQALDMS 256
Query: 185 ELHN----MGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE--DVKQKAIV 238
EL D+ K VE + + Y + E + R+W L ++ D I+
Sbjct: 257 ELKTKIDITFDVPESSKRSVEDAIRTVVAYGGDAEEALEGVRRQWSLNKDASDAANTEII 316
Query: 239 KDLSKILGS 247
K + I S
Sbjct: 317 KGAANIASS 325
>ref|YP_017096.1| prophage lambdaba04, tape measure protein, putative [Bacillus
anthracis str. 'Ames Ancestor']
gi|30260639|ref|NP_843016.1| prophage LambdaBa04, tape
measure protein, putative [Bacillus anthracis str. Ames]
gi|49183482|ref|YP_026734.1| prophage LambdaBa04, tape
measure protein, putative [Bacillus anthracis str.
Sterne] gi|30254007|gb|AAP24502.1| prophage LambdaBa04,
tape measure protein, putative [Bacillus anthracis phage
lambda Ba01] gi|47500895|gb|AAT29571.1| prophage
LambdaBa04, tape measure protein, putative [Bacillus
anthracis str. 'Ames Ancestor']
gi|49177409|gb|AAT52785.1| prophage LambdaBa04, tape
measure protein, putative [Bacillus anthracis str.
Sterne]
Length = 1311
Score = 35.4 bits (80), Expect = 1.5
Identities = 32/129 (24%), Positives = 50/129 (37%), Gaps = 8/129 (6%)
Query: 127 SASTANELRQDRYDVNVNPEKLKAASEGLANLGKAF--AIATAVVFGGAAMVIGMVASKL 184
S N ++ R D+N ++ + ASE + LG + V GG VIG
Sbjct: 197 SVDDGNSIQNIRNDLNQLSQEAEEASESVKGLGVELENVLGGIVAGGGIQEVIGQALDMS 256
Query: 185 ELHN----MGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLERE--DVKQKAIV 238
EL D+ K VE + + Y + E + R+W L ++ D I+
Sbjct: 257 ELKTKIDITFDVPESSKRSVEDAIRTVVAYGGDAEEALEGVRRQWSLNKDASDAANTEII 316
Query: 239 KDLSKILGS 247
K + I S
Sbjct: 317 KGAANIASS 325
>gb|EAL91803.1| serine/threonine protein kinase, putative [Aspergillus fumigatus
Af293]
Length = 669
Score = 34.3 bits (77), Expect = 3.2
Identities = 21/61 (34%), Positives = 29/61 (47%)
Query: 40 VDQRHVVQDNADVLPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAIN 99
+D V + N DV + SLPSKLF+ +S S S P+LR +SP +
Sbjct: 482 LDLIQVRKTNGDVAQTHSQESLPSKLFKSAVPRASGSKESILEESSSPELRRDLSPTLSD 541
Query: 100 P 100
P
Sbjct: 542 P 542
>gb|AAR23791.1| S-layer protein [Bacillus thuringiensis]
Length = 862
Score = 33.5 bits (75), Expect = 5.5
Identities = 32/109 (29%), Positives = 46/109 (41%), Gaps = 8/109 (7%)
Query: 120 PEAKHGVSASTANELRQDRYD---VNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMV 176
P+ K ++ S E + +Y V +PE L EG K A+A A + G +V
Sbjct: 633 PDTKLDLNVSNKVEYQLSKYTSDRVYSDPENL----EGYVVESKNPAVAEAKIVGNKVVV 688
Query: 177 IGMVASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMK-VWAENMSRK 224
G K+++H + T GK +E E I V K V EN K
Sbjct: 689 TGKTPGKVDIHLTKNGATAGKATIEIVQETIAIESVNFKPVQTENFVEK 737
>ref|NP_647933.1| CG13708-PA [Drosophila melanogaster] gi|7292542|gb|AAF47944.1|
CG13708-PA [Drosophila melanogaster]
Length = 1301
Score = 33.5 bits (75), Expect = 5.5
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query: 53 LPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPK-LRSSVSPDAI 98
+PEV+ L ++L DET ++S S +W+L+ D K LR V +A+
Sbjct: 1162 MPEVMLQPLLARLRLDETCTASKLSPKEWLLRPDNKSLRLVVGKEAL 1208
>ref|YP_016277.1| pyruvate dehydrogenase E3 component dihydrolipoamide dehydrogenase
[Mycoplasma mobile 163K] gi|47458745|gb|AAT28066.1|
pyruvate dehydrogenase E3 component dihydrolipoamide
dehydrogenase [Mycoplasma mobile 163K]
Length = 600
Score = 33.5 bits (75), Expect = 5.5
Identities = 42/173 (24%), Positives = 72/173 (41%), Gaps = 18/173 (10%)
Query: 68 DETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELPEAKHGVS 127
D+ +SS S ++++ ++ + + VS ++FG + P + +
Sbjct: 76 DDGSSSQSIEVKPAEIKAEAPKKAGGGASVVGEVA--VSDEVMSFGKKTSSTPTSSTSIQ 133
Query: 128 ASTANELRQDRYDVNV---NPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMVASKL 184
++ N D+YDV V P AA E N GK I +GG + +G + +K
Sbjct: 134 PTSFNGKITDKYDVIVLGSGPGGYLAAEEAGKN-GKKTLIIEKEYWGGVCLNVGCIPTKA 192
Query: 185 ELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAI 237
L K +V E QL + +Y + + V M+ K ER KQK +
Sbjct: 193 LL--------KSTEVFE-QLSHASDYGLDIDVSKLKMNWKKMQER---KQKVV 233
>gb|EAL30163.1| GA12474-PA [Drosophila pseudoobscura]
Length = 1288
Score = 33.5 bits (75), Expect = 5.5
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query: 53 LPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPK-LRSSVSPDAI 98
+PEV+ L ++L DET ++S S +W+L+ D K LR V +A+
Sbjct: 1148 MPEVMLQPLLARLRLDETCTASKLSPKEWLLRPDNKSLRLVVGKEAL 1194
>pdb|1V9S|D Chain D, Crystal Structure Of Tt0130 Protein From Thermus
Thermophilus Hb8 gi|62738105|pdb|1V9S|C Chain C, Crystal
Structure Of Tt0130 Protein From Thermus Thermophilus
Hb8 gi|62738104|pdb|1V9S|B Chain B, Crystal Structure Of
Tt0130 Protein From Thermus Thermophilus Hb8
gi|62738103|pdb|1V9S|A Chain A, Crystal Structure Of
Tt0130 Protein From Thermus Thermophilus Hb8
Length = 208
Score = 33.5 bits (75), Expect = 5.5
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 1/67 (1%)
Query: 95 PDAINPLRAFVSLPQVTFGARRWEL-PEAKHGVSASTANELRQDRYDVNVNPEKLKAASE 153
P+++NP++ ++ LP R + L P G SAS A L ++R V + AA E
Sbjct: 105 PESLNPVQYYIKLPPDIAERRAFLLDPXLATGGSASLALSLLKERGATGVKLXAILAAPE 164
Query: 154 GLANLGK 160
GL + K
Sbjct: 165 GLERIAK 171
>gb|EAL88879.1| receptor L domain protein [Aspergillus fumigatus Af293]
Length = 406
Score = 33.1 bits (74), Expect = 7.2
Identities = 27/82 (32%), Positives = 38/82 (45%), Gaps = 3/82 (3%)
Query: 33 PISKTNVVDQRHVVQDNADVLPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKL-RS 91
P+S D+R V A +LP R S+P L S SS SAS + SDP L
Sbjct: 293 PVSPPPAADRRSSVP--ASLLPGSGRLSVPVSLLPGSNPSVSSASASHRRVTSDPSLFLH 350
Query: 92 SVSPDAINPLRAFVSLPQVTFG 113
++P A P + + +P + G
Sbjct: 351 HIAPSAPQPPPSEIDVPMLDSG 372
>ref|NP_663615.1| apolipoprotein L3 isoform 1 [Homo sapiens]
Length = 402
Score = 32.7 bits (73), Expect = 9.4
Identities = 45/215 (20%), Positives = 78/215 (35%), Gaps = 6/215 (2%)
Query: 2 SFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHSL 61
SFL + +EA +F+E V N + V + +D AD L E L+
Sbjct: 72 SFLEKKRFTEEATKYFRERVSPVHLQILLTNNEAWKRFVTAAELPRDEADALYEALKKLR 131
Query: 62 PSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVS-LPQVTFGARRWELP 120
DE +W L+ P+++ + ++I LRA + + +V G +
Sbjct: 132 TYAAIEDEYVQQKDEQFREWFLKEFPQVKRKIQ-ESIEKLRALANGIEEVHRGCTISNVV 190
Query: 121 EAKHGVSASTANELRQDRYDVNVNPE-KLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
+ G ++ + L AA GL I T++V
Sbjct: 191 SSSTGAASGIMSLAGLVLAPFTAGTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSAEA 250
Query: 180 VASKLELHNMGDLKT---KGKDVVEPQLENIKNYF 211
AS+L ++ LK +D+ L + NY+
Sbjct: 251 EASRLTATSIDRLKVFKEVMRDITPNLLSLLNNYY 285
>ref|YP_004915.1| uracil phosphoribosyltransferase [Thermus thermophilus HB27]
gi|55981281|ref|YP_144578.1| uracil
phosphoribosyltransferase [Thermus thermophilus HB8]
gi|46196873|gb|AAS81288.1| uracil
phosphoribosyltransferase [Thermus thermophilus HB27]
gi|61217001|sp|Q72J35|UPP_THET2 Uracil
phosphoribosyltransferase (UMP pyrophosphorylase)
(UPRTase) gi|55772694|dbj|BAD71135.1| uracil
phosphoribosyltransferase [Thermus thermophilus HB8]
Length = 208
Score = 32.7 bits (73), Expect = 9.4
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 1/67 (1%)
Query: 95 PDAINPLRAFVSLPQVTFGARRWEL-PEAKHGVSASTANELRQDRYDVNVNPEKLKAASE 153
P+++NP++ ++ LP R + L P G SAS A L ++R V + AA E
Sbjct: 105 PESLNPVQYYIKLPPDIAERRAFLLDPMLATGGSASLALSLLKERGATGVKLMAILAAPE 164
Query: 154 GLANLGK 160
GL + K
Sbjct: 165 GLERIAK 171
>ref|NP_533434.1| acetyl-CoA acetyltransferase [Agrobacterium tumefaciens str. C58]
gi|17741284|gb|AAL43750.1| acetyl-CoA acetyltransferase
[Agrobacterium tumefaciens str. C58]
gi|15157993|gb|AAK88484.1| AGR_C_5022p [Agrobacterium
tumefaciens str. C58] gi|15890018|ref|NP_355699.1|
hypothetical protein AGR_C_5022 [Agrobacterium
tumefaciens str. C58] gi|25286417|pir||AH2916 acetyl-CoA
acetyltransferase [imported] - Agrobacterium tumefaciens
(strain C58, Dupont) gi|25286413|pir||C97691 chain A,
unliganded biosynthetic thiolase [imported] -
Agrobacterium tumefaciens (strain C58, Cereon)
Length = 393
Score = 32.7 bits (73), Expect = 9.4
Identities = 35/159 (22%), Positives = 64/159 (40%), Gaps = 25/159 (15%)
Query: 104 FVSLPQVTFGARRWELPEAKHGVSASTANELRQDRYDV---NVNPEKLKAASEGLANLGK 160
F + P GA + GV+A+ +E+ + + NP + A G+
Sbjct: 23 FANTPAHELGATVISAVLERAGVAANEVDEVILGQVLMAGEGQNPARQAAMKAGIPQEAT 82
Query: 161 AFAI--------------ATAVVFGGAAMVIGMVASKLEL-----HNMGDLKTKGKDVVE 201
AF I +V G AA++I + + H G +K +++
Sbjct: 83 AFGINQLCGSGLRAVALGMQQIVTGDAAIIIAGGQESMSMAPHCAHLRGGVKMGDFKMID 142
Query: 202 PQLENIKN---YFVPMKVWAENMSRKWHLEREDVKQKAI 237
+++ Y PM + AEN++R+W L R++ Q A+
Sbjct: 143 TMMKDGLTDAFYGYPMGITAENIARQWQLSRDEQDQFAV 181
>ref|YP_010039.1| hypothetical protein DVU0818 [Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough] gi|46448645|gb|AAS95298.1|
conserved domain protein [Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough]
Length = 582
Score = 32.7 bits (73), Expect = 9.4
Identities = 16/51 (31%), Positives = 27/51 (52%)
Query: 149 KAASEGLANLGKAFAIATAVVFGGAAMVIGMVASKLELHNMGDLKTKGKDV 199
+A SE + +L +A ++FGG A + GMV + + D++T G V
Sbjct: 89 EAISERVPDLRAGAVVAAIILFGGPATLFGMVGPYVARLRLADMRTAGATV 139
>emb|CAE45769.1| Tbx2/3 protein [Pleurobrachia pileus]
Length = 560
Score = 32.7 bits (73), Expect = 9.4
Identities = 43/178 (24%), Positives = 75/178 (41%), Gaps = 20/178 (11%)
Query: 22 QAVTKLAQKNNPISK--------TNVVDQRHVVQDNADVLPEVLRHSLPSKLFR----DE 69
+ +T+L ++NP +K T ++RH +D V+ + + P +
Sbjct: 250 EKITQLKIEHNPFAKGFREPGTATQAAEERHESKDVNHVIIALEHYGAPPFIAAGGSIQH 309
Query: 70 TASSSSFSASKWVLQSDPKLRSSVS---PDAINPLRAFVSLPQVTFGARRWELPEAKHGV 126
T + + S+ K+ QS + S V P I P + PQ T A +P
Sbjct: 310 TQAPAGKSSCKYQ-QSQFDMNSPVQNGVPLHIAPYPEDLPTPQATSSA----VPLPMLLG 364
Query: 127 SASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMVASKL 184
S+++ ++L + YD++ P A S+GLAN ++ A A A +G S L
Sbjct: 365 SSTSPSQLSKACYDIDTYPYSATAWSDGLANAQIPYSAALADFPPAARESVGYRGSPL 422
>gb|AAG50346.1| apolipoprotein L-III splice variant alpha/d [Homo sapiens]
gi|13446777|gb|AAK26528.1| apolipoprotein L-III splice
variant alpha/d [Homo sapiens]
gi|17433280|sp|O95236|APOL3_HUMAN Apolipoprotein-L3
(Apolipoprotein L-III) (ApoL-III) (TNF-inducible protein
CG12-1) (CG12_1)
Length = 402
Score = 32.7 bits (73), Expect = 9.4
Identities = 45/215 (20%), Positives = 78/215 (35%), Gaps = 6/215 (2%)
Query: 2 SFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHSL 61
SFL + +EA +F+E V N + V + +D AD L E L+
Sbjct: 72 SFLEKKRFTEEATKYFRERVSPVHLQILLTNNEAWKRFVTAAELPRDEADALYEALKKLR 131
Query: 62 PSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVS-LPQVTFGARRWELP 120
DE +W L+ P+++ + ++I LRA + + +V G +
Sbjct: 132 TYAAIEDEYVQQKDEQFREWFLKEFPQVKRKIQ-ESIEKLRALANGIEEVHRGCTISNVV 190
Query: 121 EAKHGVSASTANELRQDRYDVNVNPE-KLKAASEGLANLGKAFAIATAVVFGGAAMVIGM 179
+ G ++ + L AA GL I T++V
Sbjct: 191 SSSTGAASGIMSLAGLVLAPFTAGTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSAEA 250
Query: 180 VASKLELHNMGDLKT---KGKDVVEPQLENIKNYF 211
AS+L ++ LK +D+ L + NY+
Sbjct: 251 EASRLTATSIDRLKVFKEVMRDITPNLLSLLNNYY 285
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 389,700,911
Number of Sequences: 2540612
Number of extensions: 14792581
Number of successful extensions: 34248
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 34233
Number of HSP's gapped (non-prelim): 20
length of query: 248
length of database: 863,360,394
effective HSP length: 125
effective length of query: 123
effective length of database: 545,783,894
effective search space: 67131418962
effective search space used: 67131418962
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC147960.7


