

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.1 - phase: 1 /pseudo
(451 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
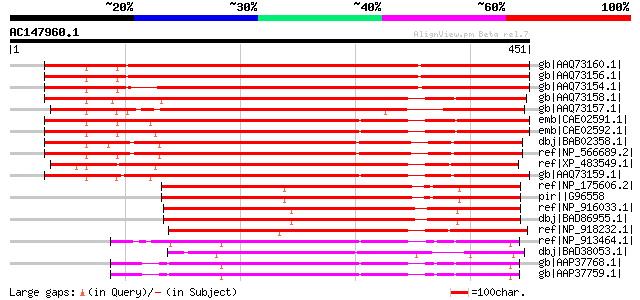
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ73160.1| LysM domain-containing receptor-like kinase 4 [Me... 622 e-177
gb|AAQ73156.1| LysM domain-containing receptor-like kinase 4 [Me... 622 e-177
gb|AAQ73154.1| LysM domain-containing receptor-like kinase 1 [Me... 579 e-164
gb|AAQ73158.1| LysM domain-containing receptor-like kinase 7 [Me... 432 e-120
gb|AAQ73157.1| LysM domain-containing receptor-like kinase 6 [Me... 428 e-118
emb|CAE02591.1| Nod-facor receptor 1a [Lotus corniculatus var. j... 412 e-114
emb|CAE02592.1| Nod-facor receptor 1b [Lotus corniculatus var. j... 411 e-113
dbj|BAB02358.1| unnamed protein product [Arabidopsis thaliana] 399 e-110
ref|NP_566689.2| protein kinase family protein [Arabidopsis thal... 398 e-109
ref|XP_483549.1| receptor protein kinase PERK1-like protein [Ory... 394 e-108
gb|AAQ73159.1| LysM domain-containing receptor-like kinase 3 [Me... 393 e-108
ref|NP_175606.2| protein kinase family protein / peptidoglycan-b... 240 8e-62
pir||G96558 probable protein kinase [imported] - Arabidopsis tha... 240 8e-62
ref|NP_916033.1| putative receptor protein kinase tmk1 precursor... 239 1e-61
dbj|BAD86955.1| putative Nod-factor receptor 1b [Oryza sativa (j... 239 1e-61
ref|NP_918232.1| protein kinase-like [Oryza sativa (japonica cul... 236 1e-60
ref|NP_913464.1| putative receptor protein kinase PERK1 [Oryza s... 214 4e-54
dbj|BAD38053.1| putative light repressible receptor protein kina... 213 1e-53
gb|AAP37768.1| At3g24600 [Arabidopsis thaliana] gi|13877617|gb|A... 213 1e-53
gb|AAP37759.1| At3g24550 [Arabidopsis thaliana] gi|22136228|gb|A... 213 1e-53
>gb|AAQ73160.1| LysM domain-containing receptor-like kinase 4 [Medicago truncatula]
Length = 496
Score = 622 bits (1603), Expect = e-177
Identities = 333/455 (73%), Positives = 365/455 (80%), Gaps = 37/455 (8%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
++F F+ P TL IA ++ LL+ +N NFS+GSG+V+I G+D NG+YVP
Sbjct: 33 KDFGLFVTYPLRSDDTLAKIATKADLDEGLLQNFNQDANFSKGSGIVFIPGRDENGVYVP 92
Query: 89 LPPK--------------------------------YFRKKNGEEESKFPPEDSMTPSTK 116
LP + YFRKKNGEE SK PPEDSM+PSTK
Sbjct: 93 LPSRKAGHLARSLVAAGICIRGVCMVLLLAICIYVRYFRKKNGEE-SKLPPEDSMSPSTK 151
Query: 117 DVDKDTNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIA 176
D DKD+ D SKYI VDKSP+FSY+ LANAT+NFSLAKKIGQGGFGEVYYG L G+K+A
Sbjct: 152 DGDKDSYSDTRSKYILVDKSPKFSYKVLANATENFSLAKKIGQGGFGEVYYGVLGGKKVA 211
Query: 177 IKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEK 236
IKKMK QATREFLSELKVLTSV H NLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEK
Sbjct: 212 IKKMKTQATREFLSELKVLTSVRHLNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEK 271
Query: 237 EPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTD 296
E MTLS RMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLN+NF GK+ADFGLTKLT+
Sbjct: 272 ELMTLSRRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNKNFNGKIADFGLTKLTN 331
Query: 297 AANSADNTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEF 356
ANS DNT H+AGTFGYMPPENAYGRISRK+DVYAFGVVLYELISAKAAVI IDK EFE
Sbjct: 332 IANSTDNTNHMAGTFGYMPPENAYGRISRKMDVYAFGVVLYELISAKAAVIMIDKNEFE- 390
Query: 357 KSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAK 416
S EIKTNES DEYKSLVALFDEVM+Q GD I+ LRKLVDPRLG NYSIDSISKMAKLAK
Sbjct: 391 -SHEIKTNESTDEYKSLVALFDEVMDQKGDPIEGLRKLVDPRLGDNYSIDSISKMAKLAK 449
Query: 417 ACINRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
ACINRDPKQRP MRDVVVSLM+L S+ID+++ T S
Sbjct: 450 ACINRDPKQRPKMRDVVVSLMKLISTIDDESRTDS 484
>gb|AAQ73156.1| LysM domain-containing receptor-like kinase 4 [Medicago truncatula]
Length = 624
Score = 622 bits (1603), Expect = e-177
Identities = 333/455 (73%), Positives = 365/455 (80%), Gaps = 37/455 (8%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
++F F+ P TL IA ++ LL+ +N NFS+GSG+V+I G+D NG+YVP
Sbjct: 161 KDFGLFVTYPLRSDDTLAKIATKADLDEGLLQNFNQDANFSKGSGIVFIPGRDENGVYVP 220
Query: 89 LPPK--------------------------------YFRKKNGEEESKFPPEDSMTPSTK 116
LP + YFRKKNGEE SK PPEDSM+PSTK
Sbjct: 221 LPSRKAGHLARSLVAAGICIRGVCMVLLLAICIYVRYFRKKNGEE-SKLPPEDSMSPSTK 279
Query: 117 DVDKDTNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIA 176
D DKD+ D SKYI VDKSP+FSY+ LANAT+NFSLAKKIGQGGFGEVYYG L G+K+A
Sbjct: 280 DGDKDSYSDTRSKYILVDKSPKFSYKVLANATENFSLAKKIGQGGFGEVYYGVLGGKKVA 339
Query: 177 IKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEK 236
IKKMK QATREFLSELKVLTSV H NLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEK
Sbjct: 340 IKKMKTQATREFLSELKVLTSVRHLNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEK 399
Query: 237 EPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTD 296
E MTLS RMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLN+NF GK+ADFGLTKLT+
Sbjct: 400 ELMTLSRRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNKNFNGKIADFGLTKLTN 459
Query: 297 AANSADNTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEF 356
ANS DNT H+AGTFGYMPPENAYGRISRK+DVYAFGVVLYELISAKAAVI IDK EFE
Sbjct: 460 IANSTDNTNHMAGTFGYMPPENAYGRISRKMDVYAFGVVLYELISAKAAVIMIDKNEFE- 518
Query: 357 KSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAK 416
S EIKTNES DEYKSLVALFDEVM+Q GD I+ LRKLVDPRLG NYSIDSISKMAKLAK
Sbjct: 519 -SHEIKTNESTDEYKSLVALFDEVMDQKGDPIEGLRKLVDPRLGDNYSIDSISKMAKLAK 577
Query: 417 ACINRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
ACINRDPKQRP MRDVVVSLM+L S+ID+++ T S
Sbjct: 578 ACINRDPKQRPKMRDVVVSLMKLISTIDDESRTDS 612
>gb|AAQ73154.1| LysM domain-containing receptor-like kinase 1 [Medicago truncatula]
Length = 590
Score = 579 bits (1493), Expect = e-164
Identities = 313/452 (69%), Positives = 351/452 (77%), Gaps = 56/452 (12%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
+++ F+ P TLESIAK T ++ ELL++Y VNFS+GSGLV+I GKD NG+YVP
Sbjct: 152 KDYGLFITYPLSSEDTLESIAKHTKVKPELLQKYTPGVNFSKGSGLVFIPGKDKNGVYVP 211
Query: 89 LPPK-----------------------------YFRKKNGEEESKFPPEDSMTPSTKDVD 119
LP YFR KN +E SK P
Sbjct: 212 LPHGKAGHLARSLATAVGGTCTVLLLAISIYAIYFRNKNAKE-SKLP------------- 257
Query: 120 KDTNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKK 179
SKYI VDKSP+FSYEELANATD FSLA KIGQGGFGEVYYGE RG+K AIKK
Sbjct: 258 --------SKYIVVDKSPKFSYEELANATDKFSLANKIGQGGFGEVYYGEPRGKKTAIKK 309
Query: 180 MKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEKEPM 239
MKMQATREFL+ELK+LT VHH NLVHLIGYCVEG LFLVYEY++NGNLSQ+LH+SE+ PM
Sbjct: 310 MKMQATREFLAELKILTRVHHCNLVHLIGYCVEGSLFLVYEYIDNGNLSQNLHDSERGPM 369
Query: 240 TLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAAN 299
T STRM+IALDVARGLEYIH+HSVPVYIHRDIKSDNILLNENFTGK+ADFGLT+LTD+AN
Sbjct: 370 TWSTRMQIALDVARGLEYIHEHSVPVYIHRDIKSDNILLNENFTGKIADFGLTRLTDSAN 429
Query: 300 SADNTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSL 359
S DNT+HVAGTFGYMPPEN YGRISRKIDVYAFGVVLYELISAK AVIKIDKTEFE
Sbjct: 430 STDNTLHVAGTFGYMPPENVYGRISRKIDVYAFGVVLYELISAKPAVIKIDKTEFE---S 486
Query: 360 EIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACI 419
EI+TNESIDEYKSLVALFDEV++Q GD I+ LR LVDPRL NYSIDSISKMAKLA+AC+
Sbjct: 487 EIRTNESIDEYKSLVALFDEVIDQKGDPIEGLRNLVDPRLEDNYSIDSISKMAKLARACL 546
Query: 420 NRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
NRDPK+RPTMR VVVSLM LNS+ID+ + + S
Sbjct: 547 NRDPKRRPTMRAVVVSLMTLNSTIDDGSRSAS 578
>gb|AAQ73158.1| LysM domain-containing receptor-like kinase 7 [Medicago truncatula]
Length = 620
Score = 432 bits (1112), Expect = e-120
Identities = 250/461 (54%), Positives = 305/461 (65%), Gaps = 57/461 (12%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
+++ F+ P P +LE I+ T I+ ELL++YN VNFSQGSGLVYI GKD N YVP
Sbjct: 160 KDYGLFITYPLRPEDSLELISNKTEIDAELLQKYNPGVNFSQGSGLVYIPGKDQNRNYVP 219
Query: 89 ------------------------------LPPKYFRKKNGEEESKFPPEDS-MTPSTKD 117
+ Y+RKK ++ E S + K+
Sbjct: 220 FHISTGGLSGGVITGISVGAVAGLILLSFCIYVTYYRKKKIRKQEFLSEESSAIFGQVKN 279
Query: 118 VDKDTNDDNGSKY---------IWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYG 168
+ N G+ I V+KS EFSYEELANAT+NF++A KIGQGGFGEVYY
Sbjct: 280 DEVSGNATYGTSDSASPANMIGIRVEKSGEFSYEELANATNNFNMANKIGQGGFGEVYYA 339
Query: 169 ELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLS 228
EL G+K AIKKM M+AT+EFL+ELKVLT VHH NLV LIGYCVEG LFLVYEY++NGNL
Sbjct: 340 ELNGEKAAIKKMDMKATKEFLAELKVLTRVHHVNLVRLIGYCVEGSLFLVYEYIDNGNLG 399
Query: 229 QHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVAD 288
QHL +S+ EP++ S R+KIALD ARGLEYIH+H+VP YIHRDIKS+NILL++NF KVAD
Sbjct: 400 QHLRSSDGEPLSWSIRVKIALDSARGLEYIHEHTVPTYIHRDIKSENILLDKNFCAKVAD 459
Query: 289 FGLTKLTDAANSADNTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIK 348
FGLTKL DA S+ TV++AGTFGYMPPE AYG +S KIDVYAFGVVLYELISAKAAV
Sbjct: 460 FGLTKLIDAGISSVPTVNMAGTFGYMPPEYAYGSVSSKIDVYAFGVVLYELISAKAAV-- 517
Query: 349 IDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSI 408
I +S + K LV LF+EV +Q I+ L+KLVDPRLG NY ID +
Sbjct: 518 ------------IMGEDSGADLKGLVVLFEEVFDQPHP-IEGLKKLVDPRLGDNYPIDHV 564
Query: 409 SKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDNKNST 449
KMA+LAK C N DP+QRP M VVV+L L S+ ++ + T
Sbjct: 565 FKMAQLAKVCTNSDPQQRPNMSSVVVALTTLTSTTEDWDIT 605
>gb|AAQ73157.1| LysM domain-containing receptor-like kinase 6 [Medicago truncatula]
Length = 574
Score = 428 bits (1101), Expect = e-118
Identities = 245/446 (54%), Positives = 297/446 (65%), Gaps = 72/446 (16%)
Query: 36 FLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVPLPP-- 91
F+ P TLESIAK IE ELL+RYN VNFS+GSGLV+I GKD NG Y+PL P
Sbjct: 150 FITYPLRSEDTLESIAKGAEIEAELLQRYNPGVNFSKGSGLVFIPGKDQNGSYLPLHPST 209
Query: 92 --------------------------KYFRKKNGEE--ESKFPPEDSMTPSTKDVDKDTN 123
KY+ KK ++ E +DS K TN
Sbjct: 210 VGTVAITGISVGVLAALLLLLFFVYIKYYLKKKNKKTWEKNLILDDS---KMKSAQIGTN 266
Query: 124 DDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQ 183
+ I V+KS EFSY+EL+ AT+NFS+A KIG+GGFGEV+Y ELRGQK AIKKMKM+
Sbjct: 267 IAS----IMVEKSEEFSYKELSIATNNFSMANKIGEGGFGEVFYAELRGQKAAIKKMKMK 322
Query: 184 ATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEKEPMTLST 243
A++EF +ELKVLT VHH NLV LIGYCVEGFLFLVYEY++NGNLSQ+LH+SE+EP++ ST
Sbjct: 323 ASKEFCAELKVLTLVHHLNLVGLIGYCVEGFLFLVYEYIDNGNLSQNLHDSEREPLSWST 382
Query: 244 RMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADN 303
RM+IALD ARGLEYIH+H+VPVYIHRDIKS+NILL+++F KVADFGL+KL D NS +
Sbjct: 383 RMQIALDSARGLEYIHEHTVPVYIHRDIKSENILLDKSFCAKVADFGLSKLADVGNSTSS 442
Query: 304 TVHVAGTFGYMPPENAYGRISR--KIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEI 361
T+ GTFGYMPPE A G +S K+DVYAFGVVLYELISAKAA
Sbjct: 443 TIVAEGTFGYMPPEYACGSVSSSPKVDVYAFGVVLYELISAKAA---------------- 486
Query: 362 KTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINR 421
FDEV D + ++ LVDPRLG NYSIDS+ KMA+LAKAC R
Sbjct: 487 ---------------FDEVFGYDQDPTEGIKNLVDPRLGDNYSIDSVCKMAQLAKACTMR 531
Query: 422 DPKQRPTMRDVVVSLMELNSSIDNKN 447
DP+ RP+MR +VV+LM L S+ ++ N
Sbjct: 532 DPQLRPSMRSIVVALMTLTSTTEDWN 557
>emb|CAE02591.1| Nod-facor receptor 1a [Lotus corniculatus var. japonicus]
gi|37651058|emb|CAE02589.1| Nod-factor receptor 1a
[Lotus corniculatus var. japonicus]
Length = 621
Score = 412 bits (1060), Expect = e-114
Identities = 239/464 (51%), Positives = 301/464 (64%), Gaps = 59/464 (12%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
+++ F+ P P TL+ IA + ++ L++ +N VNFS+ SG+ +I G+ NG+YVP
Sbjct: 161 KDYGLFITYPIRPGDTLQDIANQSSLDAGLIQSFNPSVNFSKDSGIAFIPGRYKNGVYVP 220
Query: 89 LPPK---------------------------YFR-KKNGEEESKFPPEDSMTPSTKDVDK 120
L + Y R +K EE++K P + SM ST+D
Sbjct: 221 LYHRTAGLASGAAVGISIAGTFVLLLLAFCMYVRYQKKEEEKAKLPTDISMALSTQDASS 280
Query: 121 D------------TNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYG 168
T G I V KS EFSY+ELA AT+NFSL KIGQGGFG VYY
Sbjct: 281 SAEYETSGSSGPGTASATGLTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVYYA 340
Query: 169 ELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLS 228
ELRG+K AIKKM +QA+ EFL ELKVLT VHH NLV LIGYCVEG LFLVYE+++NGNL
Sbjct: 341 ELRGKKTAIKKMDVQASTEFLCELKVLTHVHHLNLVRLIGYCVEGSLFLVYEHIDNGNLG 400
Query: 229 QHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVAD 288
Q+LH S KEP+ S+R++IALD ARGLEYIH+H+VPVYIHRD+KS NIL+++N GKVAD
Sbjct: 401 QYLHGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVAD 460
Query: 289 FGLTKLTDAANSADNTVHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVI 347
FGLTKL + NS T + GTFGYMPPE A YG IS KIDVYAFGVVL+ELISAK AV
Sbjct: 461 FGLTKLIEVGNSTLQT-RLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAV- 518
Query: 348 KIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDS 407
+KT E + E K LVALF+E +N++ C D LRKLVDPRLG NY IDS
Sbjct: 519 -------------LKTGELVAESKGLVALFEEALNKSDPC-DALRKLVDPRLGENYPIDS 564
Query: 408 ISKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
+ K+A+L +AC +P RP+MR +VV+LM L+S ++ + S
Sbjct: 565 VLKIAQLGRACTRDNPLLRPSMRSLVVALMTLSSLTEDCDDESS 608
>emb|CAE02592.1| Nod-facor receptor 1b [Lotus corniculatus var. japonicus]
gi|37651060|emb|CAE02590.1| Nod-factor receptor 1b
[Lotus corniculatus var. japonicus]
Length = 623
Score = 411 bits (1056), Expect = e-113
Identities = 238/466 (51%), Positives = 304/466 (65%), Gaps = 61/466 (13%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
+++ F+ P P TL+ IA + ++ L++ +N VNFS+ SG+ +I G+ NG+YVP
Sbjct: 161 KDYGLFITYPIRPGDTLQDIANQSSLDAGLIQSFNPSVNFSKDSGIAFIPGRYKNGVYVP 220
Query: 89 LPPK---------------------------YFR-KKNGEEESKFPPEDSMTPSTKDVDK 120
L + Y R +K EE++K P + SM ST+D +
Sbjct: 221 LYHRTAGLASGAAVGISIAGTFVLLLLAFCMYVRYQKKEEEKAKLPTDISMALSTQDGNA 280
Query: 121 DTNDD--------------NGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVY 166
++ + G I V KS EFSY+ELA AT+NFSL KIGQGGFG VY
Sbjct: 281 SSSAEYETSGSSGPGTASATGLTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVY 340
Query: 167 YGELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGN 226
Y ELRG+K AIKKM +QA+ EFL ELKVLT VHH NLV LIGYCVEG LFLVYE+++NGN
Sbjct: 341 YAELRGKKTAIKKMDVQASTEFLCELKVLTHVHHLNLVRLIGYCVEGSLFLVYEHIDNGN 400
Query: 227 LSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKV 286
L Q+LH S KEP+ S+R++IALD ARGLEYIH+H+VPVYIHRD+KS NIL+++N GKV
Sbjct: 401 LGQYLHGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKV 460
Query: 287 ADFGLTKLTDAANSADNTVHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAA 345
ADFGLTKL + NS T + GTFGYMPPE A YG IS KIDVYAFGVVL+ELISAK A
Sbjct: 461 ADFGLTKLIEVGNSTLQT-RLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNA 519
Query: 346 VIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSI 405
V +KT E + E K LVALF+E +N++ C D LRKLVDPRLG NY I
Sbjct: 520 V--------------LKTGELVAESKGLVALFEEALNKSDPC-DALRKLVDPRLGENYPI 564
Query: 406 DSISKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
DS+ K+A+L +AC +P RP+MR +VV+LM L+S ++ + S
Sbjct: 565 DSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTLSSLTEDCDDESS 610
>dbj|BAB02358.1| unnamed protein product [Arabidopsis thaliana]
Length = 603
Score = 399 bits (1026), Expect = e-110
Identities = 227/443 (51%), Positives = 291/443 (65%), Gaps = 47/443 (10%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNG---- 84
++F F+ P P +L SIA+ + + ++L+RYN VNF+ G+G+VY+ G+DG G
Sbjct: 162 KDFGLFVTYPLRPEDSLSSIARSSGVSADILQRYNPGVNFNSGNGIVYVPGRDGVGAGVI 221
Query: 85 -----------LYVPLPPKYFRKKNGEEESKFPPEDSMTPSTK-DVDKDTNDDNGS---- 128
L + Y +KN + F S+ STK D T+ +G
Sbjct: 222 AGIVIGVIVALLLILFIVYYAYRKNKSKGDSF--SSSIPLSTKADHASSTSLQSGGLGGA 279
Query: 129 ------KYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKM 182
I VDKS EFS EELA ATDNF+L+ KIGQGGFG VYY ELRG+K AIKKM M
Sbjct: 280 GVSPGIAAISVDKSVEFSLEELAKATDNFNLSFKIGQGGFGAVYYAELRGEKAAIKKMDM 339
Query: 183 QATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEKEPMTLS 242
+A+++FL+ELKVLT VHH NLV LIGYCVEG LFLVYEY+ENGNL QHLH S +EP+ +
Sbjct: 340 EASKQFLAELKVLTRVHHVNLVRLIGYCVEGSLFLVYEYVENGNLGQHLHGSGREPLPWT 399
Query: 243 TRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSAD 302
R++IALD ARGLEYIH+H+VPVY+HRDIKS NIL+++ F KVADFGLTKLT+ SA
Sbjct: 400 KRVQIALDSARGLEYIHEHTVPVYVHRDIKSANILIDQKFRAKVADFGLTKLTEVGGSA- 458
Query: 303 NTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIK 362
T GTFGYM PE YG +S K+DVYAFGVVLYELISAK AV+K+
Sbjct: 459 -TRGAMGTFGYMAPETVYGEVSAKVDVYAFGVVLYELISAKGAVVKM------------- 504
Query: 363 TNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRD 422
E++ E++ LV +F+E +T D + LRK++DPRLG +Y DS+ KMA+L KAC +
Sbjct: 505 -TEAVGEFRGLVGVFEESFKET-DKEEALRKIIDPRLGDSYPFDSVYKMAELGKACTQEN 562
Query: 423 PKQRPTMRDVVVSLMELNSSIDN 445
+ RP+MR +VV+L L SS N
Sbjct: 563 AQLRPSMRYIVVALSTLFSSTGN 585
>ref|NP_566689.2| protein kinase family protein [Arabidopsis thaliana]
Length = 617
Score = 398 bits (1022), Expect = e-109
Identities = 227/457 (49%), Positives = 291/457 (63%), Gaps = 61/457 (13%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
++F F+ P P +L SIA+ + + ++L+RYN VNF+ G+G+VY+ G+D NG + P
Sbjct: 162 KDFGLFVTYPLRPEDSLSSIARSSGVSADILQRYNPGVNFNSGNGIVYVPGRDPNGAFPP 221
Query: 89 LPPK-----------------------------YFRKKNGEEESKFPPEDSMTPSTK-DV 118
Y +KN + F S+ STK D
Sbjct: 222 FKSSKQDGVGAGVIAGIVIGVIVALLLILFIVYYAYRKNKSKGDSF--SSSIPLSTKADH 279
Query: 119 DKDTNDDNGS----------KYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYG 168
T+ +G I VDKS EFS EELA ATDNF+L+ KIGQGGFG VYY
Sbjct: 280 ASSTSLQSGGLGGAGVSPGIAAISVDKSVEFSLEELAKATDNFNLSFKIGQGGFGAVYYA 339
Query: 169 ELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLS 228
ELRG+K AIKKM M+A+++FL+ELKVLT VHH NLV LIGYCVEG LFLVYEY+ENGNL
Sbjct: 340 ELRGEKAAIKKMDMEASKQFLAELKVLTRVHHVNLVRLIGYCVEGSLFLVYEYVENGNLG 399
Query: 229 QHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVAD 288
QHLH S +EP+ + R++IALD ARGLEYIH+H+VPVY+HRDIKS NIL+++ F KVAD
Sbjct: 400 QHLHGSGREPLPWTKRVQIALDSARGLEYIHEHTVPVYVHRDIKSANILIDQKFRAKVAD 459
Query: 289 FGLTKLTDAANSADNTVHVAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIK 348
FGLTKLT+ SA T GTFGYM PE YG +S K+DVYAFGVVLYELISAK AV+K
Sbjct: 460 FGLTKLTEVGGSA--TRGAMGTFGYMAPETVYGEVSAKVDVYAFGVVLYELISAKGAVVK 517
Query: 349 IDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSI 408
+ E++ E++ LV +F+E +T D + LRK++DPRLG +Y DS+
Sbjct: 518 M--------------TEAVGEFRGLVGVFEESFKET-DKEEALRKIIDPRLGDSYPFDSV 562
Query: 409 SKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDN 445
KMA+L KAC + + RP+MR +VV+L L SS N
Sbjct: 563 YKMAELGKACTQENAQLRPSMRYIVVALSTLFSSTGN 599
>ref|XP_483549.1| receptor protein kinase PERK1-like protein [Oryza sativa (japonica
cultivar-group)] gi|38175551|dbj|BAD01244.1| receptor
protein kinase PERK1-like protein [Oryza sativa
(japonica cultivar-group)] gi|50725672|dbj|BAD33138.1|
receptor protein kinase PERK1-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 594
Score = 394 bits (1012), Expect = e-108
Identities = 226/417 (54%), Positives = 283/417 (67%), Gaps = 26/417 (6%)
Query: 36 FLYLPFCP*ATLESIAKDTMI--EIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVPLPP 91
FL P TL S+A + ++++++RYN + + GSG+VYI KD NG Y+PL
Sbjct: 172 FLTYPLRAEDTLASVAATYGLSSQLDVVRRYNPGMESATGSGIVYIPVKDPNGSYLPLKS 231
Query: 92 KYFRKKNGEEESKFPPEDSMTPSTKDVDKDTNDD----NGSKYIWVDKSPEFSYEELANA 147
R+K + EDS T +DK T + I VDKS EFSYEEL+NA
Sbjct: 232 PG-RRKAKQATLLQSSEDSTQLGTISMDKVTPSTIVGPSPVAGITVDKSVEFSYEELSNA 290
Query: 148 TDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLI 207
T FS+ KIGQGGFG VYY ELRG+K AIKKM MQAT EFL+ELKVLT VHH NLV LI
Sbjct: 291 TQGFSIGNKIGQGGFGAVYYAELRGEKAAIKKMDMQATHEFLAELKVLTHVHHLNLVRLI 350
Query: 208 GYCVEGFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYI 267
GYC+E LFLVYE++ENGNLSQHL EP++ + R++IALD ARGLEYIH+H+VPVYI
Sbjct: 351 GYCIESSLFLVYEFIENGNLSQHLRGMGYEPLSWAARIQIALDSARGLEYIHEHTVPVYI 410
Query: 268 HRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNT-VHVAGTFGYMPPENA-YGRISR 325
HRDIKS NIL+++N+ KVADFGLTKLT+ ++ T V GTFGYMPPE A YG +S
Sbjct: 411 HRDIKSANILIDKNYRAKVADFGLTKLTEVGGTSMPTGTRVVGTFGYMPPEYARYGDVSP 470
Query: 326 KIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTG 385
K+DVYAFGVVLYELISAK A+ +++ ES + K LV LF+E +N +
Sbjct: 471 KVDVYAFGVVLYELISAKEAI--------------VRSTESSSDSKGLVYLFEEALN-SP 515
Query: 386 DCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVVVSLMELNSS 442
D + LR L+DP+LG +Y IDSI K+ +LAK C DPK RP+MR VVV+LM L+S+
Sbjct: 516 DPKEGLRTLIDPKLGEDYPIDSILKLTQLAKVCTQEDPKLRPSMRSVVVALMTLSST 572
>gb|AAQ73159.1| LysM domain-containing receptor-like kinase 3 [Medicago truncatula]
gi|34485516|gb|AAQ73155.1| LysM domain-containing
receptor-like kinase 3 [Medicago truncatula]
Length = 620
Score = 393 bits (1009), Expect = e-108
Identities = 233/465 (50%), Positives = 293/465 (62%), Gaps = 62/465 (13%)
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
+++ F+ P TL IA ++ L++ +N NFS GSG+V+I G+D NG + P
Sbjct: 161 KDYGLFVTYPLRSDDTLAKIATKAGLDEGLIQNFNQDANFSIGSGIVFIPGRDQNGHFFP 220
Query: 89 LPP-----------------------------KYFRKKNGEEESKFPPEDSMTPSTKDVD 119
L KYF+KK EEE P+ S ST+D
Sbjct: 221 LYSRTGIAKGSAVGIAMAGIFGLLLFVIYIYAKYFQKK--EEEKTKLPQTSRAFSTQDAS 278
Query: 120 KD------------TNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYY 167
T G I V KS EF+Y+ELA AT+NFSL KIGQGGFG VYY
Sbjct: 279 GSAEYETSGSSGHATGSAAGLTGIMVAKSTEFTYQELAKATNNFSLDNKIGQGGFGAVYY 338
Query: 168 GELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNL 227
ELRG+K AIKKM +QA+ EFL ELKVLT VHH NLV LIGYCVEG LFLVYE+++NGNL
Sbjct: 339 AELRGEKTAIKKMDVQASSEFLCELKVLTHVHHLNLVRLIGYCVEGSLFLVYEHIDNGNL 398
Query: 228 SQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVA 287
Q+LH EP+ S+R++IALD ARGLEYIH+H+VPVYIHRD+KS NIL+++N GKVA
Sbjct: 399 GQYLHGIGTEPLPWSSRVQIALDSARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVA 458
Query: 288 DFGLTKLTDAANSADNTVHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAV 346
DFGLTKL + NS +T + GTFGYMPPE A YG +S KIDVYAFGVVLYELI+AK AV
Sbjct: 459 DFGLTKLIEVGNSTLHT-RLVGTFGYMPPEYAQYGDVSPKIDVYAFGVVLYELITAKNAV 517
Query: 347 IKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSID 406
+KT ES+ E K LV LF+E +++ D ++ LRKLVDPRL NY ID
Sbjct: 518 --------------LKTGESVAESKGLVQLFEEALHRM-DPLEGLRKLVDPRLKENYPID 562
Query: 407 SISKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
S+ KMA+L +AC +P RP+MR +VV+LM L+S ++ + S
Sbjct: 563 SVLKMAQLGRACTRDNPLLRPSMRSIVVALMTLSSPTEDCDDDSS 607
>ref|NP_175606.2| protein kinase family protein / peptidoglycan-binding LysM
domain-containing protein [Arabidopsis thaliana]
Length = 651
Score = 240 bits (612), Expect = 8e-62
Identities = 139/321 (43%), Positives = 200/321 (62%), Gaps = 20/321 (6%)
Query: 133 VDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSEL 192
++K F+YEE+ ATD FS + +G G +G VY+G LR Q++A+K+M T+EF +E+
Sbjct: 323 IEKPMVFTYEEIRAATDEFSDSNLLGHGNYGSVYFGLLREQEVAVKRMTATKTKEFAAEM 382
Query: 193 KVLTSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKE---PMTLSTRMKIA 248
KVL VHH NLV LIGY LF+VYEY+ G L HLH+ + + P++ R +IA
Sbjct: 383 KVLCKVHHSNLVELIGYAATVDELFVVYEYVRKGMLKSHLHDPQSKGNTPLSWIMRNQIA 442
Query: 249 LDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTV-HV 307
LD ARGLEYIH+H+ Y+HRDIK+ NILL+E F K++DFGL KL + + +V V
Sbjct: 443 LDAARGLEYIHEHTKTHYVHRDIKTSNILLDEAFRAKISDFGLAKLVEKTGEGEISVTKV 502
Query: 308 AGTFGYMPPEN-AYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNES 366
GT+GY+ PE + G + K D+YAFGVVL+E+IS + AVI+ + I T
Sbjct: 503 VGTYGYLAPEYLSDGLATSKSDIYAFGVVLFEIISGREAVIRTE---------AIGTKN- 552
Query: 367 IDEYKSLVALFDEVMNQTGDCID--DLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPK 424
E + L ++ V+ + D ++ L++ VDP + Y D + K+A LAK C++ DP
Sbjct: 553 -PERRPLASIMLAVLKNSPDSMNMSSLKEFVDPNMMDLYPHDCLFKIATLAKQCVDDDPI 611
Query: 425 QRPTMRDVVVSLME-LNSSID 444
RP M+ VV+SL + L SSI+
Sbjct: 612 LRPNMKQVVISLSQILLSSIE 632
>pir||G96558 probable protein kinase [imported] - Arabidopsis thaliana
gi|9802793|gb|AAF99862.1| Putative protein kinase
[Arabidopsis thaliana]
Length = 601
Score = 240 bits (612), Expect = 8e-62
Identities = 139/321 (43%), Positives = 200/321 (62%), Gaps = 20/321 (6%)
Query: 133 VDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSEL 192
++K F+YEE+ ATD FS + +G G +G VY+G LR Q++A+K+M T+EF +E+
Sbjct: 273 IEKPMVFTYEEIRAATDEFSDSNLLGHGNYGSVYFGLLREQEVAVKRMTATKTKEFAAEM 332
Query: 193 KVLTSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKE---PMTLSTRMKIA 248
KVL VHH NLV LIGY LF+VYEY+ G L HLH+ + + P++ R +IA
Sbjct: 333 KVLCKVHHSNLVELIGYAATVDELFVVYEYVRKGMLKSHLHDPQSKGNTPLSWIMRNQIA 392
Query: 249 LDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTV-HV 307
LD ARGLEYIH+H+ Y+HRDIK+ NILL+E F K++DFGL KL + + +V V
Sbjct: 393 LDAARGLEYIHEHTKTHYVHRDIKTSNILLDEAFRAKISDFGLAKLVEKTGEGEISVTKV 452
Query: 308 AGTFGYMPPEN-AYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNES 366
GT+GY+ PE + G + K D+YAFGVVL+E+IS + AVI+ + I T
Sbjct: 453 VGTYGYLAPEYLSDGLATSKSDIYAFGVVLFEIISGREAVIRTE---------AIGTKN- 502
Query: 367 IDEYKSLVALFDEVMNQTGDCID--DLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPK 424
E + L ++ V+ + D ++ L++ VDP + Y D + K+A LAK C++ DP
Sbjct: 503 -PERRPLASIMLAVLKNSPDSMNMSSLKEFVDPNMMDLYPHDCLFKIATLAKQCVDDDPI 561
Query: 425 QRPTMRDVVVSLME-LNSSID 444
RP M+ VV+SL + L SSI+
Sbjct: 562 LRPNMKQVVISLSQILLSSIE 582
>ref|NP_916033.1| putative receptor protein kinase tmk1 precursor [Oryza sativa
(japonica cultivar-group)]
Length = 612
Score = 239 bits (611), Expect = 1e-61
Identities = 140/320 (43%), Positives = 196/320 (60%), Gaps = 19/320 (5%)
Query: 134 DKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSELK 193
+K F+Y+E+ +TD+FS A +G G +G VYYG LR Q++AIK+M T+EF+ E+K
Sbjct: 284 EKPIVFTYQEILASTDSFSDANLLGHGTYGSVYYGVLRDQEVAIKRMTATKTKEFIVEMK 343
Query: 194 VLTSVHHWNLVHLIGYCV-EGFLFLVYEYMENGNLSQHLHNSEKEPMTLST---RMKIAL 249
VL VHH +LV LIGY + L+L+YEY + G+L HLH+ + + T + R++IAL
Sbjct: 344 VLCKVHHASLVELIGYAASKDELYLIYEYSQKGSLKNHLHDPQSKGYTSLSWIYRVQIAL 403
Query: 250 DVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTV-HVA 308
D ARGLEYIH+H+ Y+HRDIKS NILL+E+F K++DFGL KL + A+ +V V
Sbjct: 404 DAARGLEYIHEHTKDHYVHRDIKSSNILLDESFRAKISDFGLAKLVVKSTDAEASVTKVV 463
Query: 309 GTFGYMPPENAY-GRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESI 367
GTFGY+ PE G + K DVYAFGVVL+ELIS K A+ + D S
Sbjct: 464 GTFGYLAPEYLRDGLATTKNDVYAFGVVLFELISGKEAITRTDGL----------NEGSN 513
Query: 368 DEYKSLVALFDEVMNQTGDC--IDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQ 425
E +SL ++ + + + L+ +DP L Y D + KMA LAK C+ DP
Sbjct: 514 SERRSLASVMLSALKNCRNSMYMGSLKDCIDPNLMDLYPHDCVYKMAMLAKQCVEEDPVL 573
Query: 426 RPTMRDVVVSLME-LNSSID 444
RP M+ V++L + L SSI+
Sbjct: 574 RPDMKQAVITLSQILLSSIE 593
>dbj|BAD86955.1| putative Nod-factor receptor 1b [Oryza sativa (japonica
cultivar-group)]
Length = 420
Score = 239 bits (611), Expect = 1e-61
Identities = 140/320 (43%), Positives = 196/320 (60%), Gaps = 19/320 (5%)
Query: 134 DKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSELK 193
+K F+Y+E+ +TD+FS A +G G +G VYYG LR Q++AIK+M T+EF+ E+K
Sbjct: 92 EKPIVFTYQEILASTDSFSDANLLGHGTYGSVYYGVLRDQEVAIKRMTATKTKEFIVEMK 151
Query: 194 VLTSVHHWNLVHLIGYCV-EGFLFLVYEYMENGNLSQHLHNSEKEPMTLST---RMKIAL 249
VL VHH +LV LIGY + L+L+YEY + G+L HLH+ + + T + R++IAL
Sbjct: 152 VLCKVHHASLVELIGYAASKDELYLIYEYSQKGSLKNHLHDPQSKGYTSLSWIYRVQIAL 211
Query: 250 DVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTV-HVA 308
D ARGLEYIH+H+ Y+HRDIKS NILL+E+F K++DFGL KL + A+ +V V
Sbjct: 212 DAARGLEYIHEHTKDHYVHRDIKSSNILLDESFRAKISDFGLAKLVVKSTDAEASVTKVV 271
Query: 309 GTFGYMPPENAY-GRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESI 367
GTFGY+ PE G + K DVYAFGVVL+ELIS K A+ + D S
Sbjct: 272 GTFGYLAPEYLRDGLATTKNDVYAFGVVLFELISGKEAITRTDGL----------NEGSN 321
Query: 368 DEYKSLVALFDEVMNQTGDC--IDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQ 425
E +SL ++ + + + L+ +DP L Y D + KMA LAK C+ DP
Sbjct: 322 SERRSLASVMLSALKNCRNSMYMGSLKDCIDPNLMDLYPHDCVYKMAMLAKQCVEEDPVL 381
Query: 426 RPTMRDVVVSLME-LNSSID 444
RP M+ V++L + L SSI+
Sbjct: 382 RPDMKQAVITLSQILLSSIE 401
>ref|NP_918232.1| protein kinase-like [Oryza sativa (japonica cultivar-group)]
Length = 599
Score = 236 bits (602), Expect = 1e-60
Identities = 131/319 (41%), Positives = 198/319 (62%), Gaps = 22/319 (6%)
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSELKVLTSV 198
FS + +AT NF +KIG+GG+G VY G + +IA+KKMK ++EF +ELKVL +
Sbjct: 283 FSLRAIEDATSNFDEKRKIGEGGYGSVYLGFIGTHEIAVKKMKASKSKEFFAELKVLCKI 342
Query: 199 HHWNLVHLIGYCV-EGFLFLVYEYMENGNLSQHLHN---SEKEPMTLSTRMKIALDVARG 254
HH N+V LIGY + L+LVYEY++NG+LS+HLH+ +P++ + R +IA+D ARG
Sbjct: 343 HHINVVELIGYAAGDDHLYLVYEYVQNGSLSEHLHDPLLKGHQPLSWTARTQIAMDSARG 402
Query: 255 LEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSAD-NTVHVAGTFGY 313
+EYIHDH+ Y+HRDIK+ NILL+ KVADFGL KL ++ + + GT GY
Sbjct: 403 IEYIHDHTKTCYVHRDIKTSNILLDNGLRAKVADFGLVKLVQRSDEDECLATRLVGTPGY 462
Query: 314 MPPENAYG-RISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKS 372
+PPE+ ++ K DVYAFGVVL ELI+ A+ ++ N+ ++ KS
Sbjct: 463 LPPESVLELHMTTKSDVYAFGVVLAELITGLRAL--------------VRDNKEANKTKS 508
Query: 373 LVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDV 432
L+++ + + D L +VDP L NY I+ + K+A ++ C++ DP RP MR+V
Sbjct: 509 LISIMRKAF-KPEDLESSLETIVDPYLKDNYPIEEVCKLANISMWCLSEDPLHRPEMREV 567
Query: 433 VVSLMELN-SSIDNKNSTG 450
+ L +++ +SI+ + S G
Sbjct: 568 MPILAQIHMASIEWEASLG 586
>ref|NP_913464.1| putative receptor protein kinase PERK1 [Oryza sativa (japonica
cultivar-group)] gi|17385728|dbj|BAB78668.1| putative
brassinosteroid insensitive 1-associated receptor kinase
1 [Oryza sativa (japonica cultivar-group)]
Length = 597
Score = 214 bits (545), Expect = 4e-54
Identities = 149/383 (38%), Positives = 211/383 (54%), Gaps = 54/383 (14%)
Query: 88 PLPPKYFRKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPE--------- 138
P PP + KK S PP P+ +V + +GS Y D S
Sbjct: 154 PPPPDHVLKKVPSHPSPPPP-----PAPLNVH---SGGSGSNYSGGDNSQPLVSPGAALG 205
Query: 139 -----FSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFL 189
F+YE+L+ ATD FS A +GQGGFG V+ G L G ++A+K+++ Q REF
Sbjct: 206 FSRCTFTYEDLSAATDGFSDANLLGQGGFGYVHKGVLPNGTEVAVKQLRDGSGQGEREFQ 265
Query: 190 SELKVLTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIA 248
+E+++++ VHH +LV L+GYC+ G LVYEY+ N L HLH + M TR++IA
Sbjct: 266 AEVEIISRVHHKHLVTLVGYCISGGKRLLVYEYVPNNTLELHLHGRGRPTMEWPTRLRIA 325
Query: 249 LDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVA 308
L A+GL Y+H+ P IHRDIKS NILL+ F KVADFGL KLT N+ +T V
Sbjct: 326 LGAAKGLAYLHEDCHPKIIHRDIKSANILLDARFEAKVADFGLAKLTSDNNTHVST-RVM 384
Query: 309 GTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNES- 366
GTFGY+ PE A G+++ K DV++FGV+L ELI+ + V ++N+S
Sbjct: 385 GTFGYLAPEYASSGQLTEKSDVFSFGVMLLELITGRRPV---------------RSNQSQ 429
Query: 367 IDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQR 426
+D+ SLV +M + D + LVDPRLG Y+ + +++M A AC+ ++R
Sbjct: 430 MDD--SLVDWARPLMMRASD-DGNYDALVDPRLGQEYNGNEMARMIACAAACVRHSARRR 486
Query: 427 PTMRDVV------VSLMELNSSI 443
P M VV VSL +LN +
Sbjct: 487 PRMSQVVRALEGDVSLDDLNEGV 509
>dbj|BAD38053.1| putative light repressible receptor protein kinase [Oryza sativa
(japonica cultivar-group)]
Length = 899
Score = 213 bits (542), Expect = 1e-53
Identities = 141/327 (43%), Positives = 185/327 (56%), Gaps = 47/327 (14%)
Query: 138 EFSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIK---KMKMQATREFLSELK 193
EFSY EL + T+NFS +++G+GGFG V+ G L G +A+K + Q +EFL+E +
Sbjct: 565 EFSYRELKHITNNFS--QQVGKGGFGAVFLGYLENGNPVAVKVRSESSSQGGKEFLAEAQ 622
Query: 194 VLTSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVA 252
LT +HH NLV LIGYC + L LVYEYM GNL HL + +P+T R+ IALD A
Sbjct: 623 HLTRIHHKNLVSLIGYCKDKNHLALVYEYMPEGNLQDHLRATTNKPLTWEQRLHIALDAA 682
Query: 253 RGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFG 312
+GLEY+H P IHRD+KS NILL N K+ADFGLTK+ + + T AGTFG
Sbjct: 683 QGLEYLHVACKPALIHRDVKSRNILLTTNLGAKIADFGLTKVFSESRT-HMTTEPAGTFG 741
Query: 313 YMPPENAYG-RISRKIDVYAFGVVLYELISAKAAVIKIDKT------EFEFKSLEIKTNE 365
Y+ PE IS K DVY+FGVVL ELI+ + VI ID++ EF +SL+ + E
Sbjct: 742 YLDPEYYRNYHISEKSDVYSFGVVLLELITGRPPVIPIDESVSIHIGEFVHQSLDHGSIE 801
Query: 366 SIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRL--GYNYSIDSISKMAKLAKACINRDP 423
SI VD R+ G Y I+S+ K+A LA C
Sbjct: 802 SI---------------------------VDARMGGGGGYDINSVWKVADLALHCKREVS 834
Query: 424 KQRPTMRDVVVSL---MELNSSIDNKN 447
++RPTM +VV L +EL S D K+
Sbjct: 835 RERPTMTEVVAQLKESLELESHGDRKH 861
>gb|AAP37768.1| At3g24600 [Arabidopsis thaliana] gi|13877617|gb|AAK43886.1| protein
kinase-like protein [Arabidopsis thaliana]
Length = 652
Score = 213 bits (541), Expect = 1e-53
Identities = 138/368 (37%), Positives = 200/368 (53%), Gaps = 45/368 (12%)
Query: 88 PLPPKYFRKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPEFSYEELANA 147
P PP + G + S P +P + KS F+YEEL+ A
Sbjct: 232 PPPPAFMSSSGGSDYSDLPVLPPPSPGL--------------VLGFSKST-FTYEELSRA 276
Query: 148 TDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKVLTSVHHWNL 203
T+ FS A +GQGGFG V+ G L G+++A+K++K Q REF +E+++++ VHH +L
Sbjct: 277 TNGFSEANLLGQGGFGYVHKGILPSGKEVAVKQLKAGSGQGEREFQAEVEIISRVHHRHL 336
Query: 204 VHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHS 262
V LIGYC+ G LVYE++ N NL HLH + M STR+KIAL A+GL Y+H+
Sbjct: 337 VSLIGYCMAGVQRLLVYEFVPNNNLEFHLHGKGRPTMEWSTRLKIALGSAKGLSYLHEDC 396
Query: 263 VPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPE-NAYG 321
P IHRDIK+ NIL++ F KVADFGL K+ N+ +T V GTFGY+ PE A G
Sbjct: 397 NPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVST-RVMGTFGYLAPEYAASG 455
Query: 322 RISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVM 381
+++ K DV++FGVVL ELI+ + V N +D+ SLV ++
Sbjct: 456 KLTEKSDVFSFGVVLLELITGRRPV--------------DANNVYVDD--SLVDWARPLL 499
Query: 382 NQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV------VS 435
N+ + D L D ++G Y + +++M A AC+ ++RP M +V VS
Sbjct: 500 NRASE-EGDFEGLADSKMGNEYDREEMARMVACAAACVRHSARRRPRMSQIVRALEGNVS 558
Query: 436 LMELNSSI 443
L +LN +
Sbjct: 559 LSDLNEGM 566
>gb|AAP37759.1| At3g24550 [Arabidopsis thaliana] gi|22136228|gb|AAM91192.1| protein
kinase-like protein [Arabidopsis thaliana]
gi|9294050|dbj|BAB02007.1| protein kinase-like protein
[Arabidopsis thaliana] gi|20260332|gb|AAM13064.1|
unknown protein [Arabidopsis thaliana]
gi|16649063|gb|AAL24383.1| protein kinase-like protein
[Arabidopsis thaliana] gi|15983765|gb|AAL10479.1|
AT3g24550/MOB24_8 [Arabidopsis thaliana]
gi|15230130|ref|NP_189098.1| protein kinase family
protein [Arabidopsis thaliana]
Length = 652
Score = 213 bits (541), Expect = 1e-53
Identities = 138/368 (37%), Positives = 200/368 (53%), Gaps = 45/368 (12%)
Query: 88 PLPPKYFRKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPEFSYEELANA 147
P PP + G + S P +P + KS F+YEEL+ A
Sbjct: 232 PPPPAFMSSSGGSDYSDLPVLPPPSPGL--------------VLGFSKST-FTYEELSRA 276
Query: 148 TDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKVLTSVHHWNL 203
T+ FS A +GQGGFG V+ G L G+++A+K++K Q REF +E+++++ VHH +L
Sbjct: 277 TNGFSEANLLGQGGFGYVHKGILPSGKEVAVKQLKAGSGQGEREFQAEVEIISRVHHRHL 336
Query: 204 VHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHS 262
V LIGYC+ G LVYE++ N NL HLH + M STR+KIAL A+GL Y+H+
Sbjct: 337 VSLIGYCMAGVQRLLVYEFVPNNNLEFHLHGKGRPTMEWSTRLKIALGSAKGLSYLHEDC 396
Query: 263 VPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPE-NAYG 321
P IHRDIK+ NIL++ F KVADFGL K+ N+ +T V GTFGY+ PE A G
Sbjct: 397 NPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVST-RVMGTFGYLAPEYAASG 455
Query: 322 RISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVM 381
+++ K DV++FGVVL ELI+ + V N +D+ SLV ++
Sbjct: 456 KLTEKSDVFSFGVVLLELITGRRPV--------------DANNVYVDD--SLVDWARPLL 499
Query: 382 NQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV------VS 435
N+ + D L D ++G Y + +++M A AC+ ++RP M +V VS
Sbjct: 500 NRASE-EGDFEGLADSKMGNEYDREEMARMVACAAACVRHSARRRPRMSQIVRALEGNVS 558
Query: 436 LMELNSSI 443
L +LN +
Sbjct: 559 LSDLNEGM 566
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.140 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 749,958,430
Number of Sequences: 2540612
Number of extensions: 33248777
Number of successful extensions: 144721
Number of sequences better than 10.0: 20350
Number of HSP's better than 10.0 without gapping: 7439
Number of HSP's successfully gapped in prelim test: 12914
Number of HSP's that attempted gapping in prelim test: 97689
Number of HSP's gapped (non-prelim): 25666
length of query: 451
length of database: 863,360,394
effective HSP length: 131
effective length of query: 320
effective length of database: 530,540,222
effective search space: 169772871040
effective search space used: 169772871040
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC147960.1


