

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.1 + phase: 0
(371 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
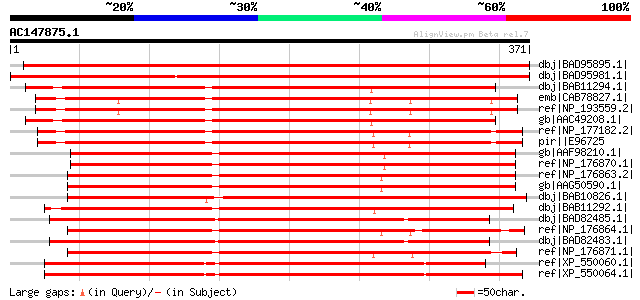
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD95895.1| Ser/Thr protein kinase [Lotus corniculatus var. ... 516 e-145
dbj|BAD95981.1| Ser/Thr protein kinase [Lotus corniculatus var. ... 503 e-141
dbj|BAB11294.1| receptor serine/threonine kinase [Arabidopsis th... 385 e-105
emb|CAB78827.1| receptor serine/threonine kinase-like protein [A... 384 e-105
ref|NP_193559.2| receptor serine/threonine kinase, putative [Ara... 384 e-105
gb|AAC49208.1| receptor serine/threonine kinase PR5K gi|1589714|... 381 e-104
ref|NP_177182.2| receptor serine/threonine kinase, putative [Ara... 378 e-103
pir||E96725 hypothetical protein F20P5.3 [imported] - Arabidopsi... 378 e-103
gb|AAF98210.1| Unknown protein [Arabidopsis thaliana] gi|2540465... 376 e-103
ref|NP_176870.1| protein kinase family protein / glycerophosphor... 376 e-103
ref|NP_176863.2| protein kinase, putative [Arabidopsis thaliana] 375 e-102
gb|AAG50590.1| receptor serine/threonine kinase PR5K, putative [... 375 e-102
dbj|BAB10826.1| receptor protein kinase-like protein [Arabidopsi... 374 e-102
dbj|BAB11292.1| receptor serine/threonine kinase [Arabidopsis th... 374 e-102
dbj|BAD82485.1| receptor serine/threonine kinase PR5K-like [Oryz... 372 e-102
ref|NP_176864.1| serine/threonine protein kinase, putative [Arab... 368 e-100
dbj|BAD82483.1| receptor serine/threonine kinase PR5K-like [Oryz... 367 e-100
ref|NP_176871.1| protein kinase family protein [Arabidopsis thal... 366 e-100
ref|XP_550060.1| putative receptor serine/threonine kinase PR5K ... 361 2e-98
ref|XP_550064.1| putative receptor serine/threonine kinase PR5K ... 359 8e-98
>dbj|BAD95895.1| Ser/Thr protein kinase [Lotus corniculatus var. japonicus]
Length = 691
Score = 516 bits (1329), Expect = e-145
Identities = 253/363 (69%), Positives = 310/363 (84%), Gaps = 2/363 (0%)
Query: 11 SLIPQTIL-RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVY 69
SL QT++ R+R+ V+++VE FM+S+ PRRYSY+EVK ITN F KLGQGGYGVVY
Sbjct: 314 SLSGQTMISRKRSTFVEHDVEAFMQSYGSLAPRRYSYSEVKRITNSFVHKLGQGGYGVVY 373
Query: 70 KASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFM 129
KA+L + R VAVKVISE+ G+GE+FINEV+SISRTSH+NIVSLLG+CY++N+R LIYEFM
Sbjct: 374 KATLPDGRLVAVKVISESGGSGEDFINEVSSISRTSHVNIVSLLGFCYDKNRRVLIYEFM 433
Query: 130 PKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILL 189
P GSLD FI PNAIC FDWNTL+++AIGIA+GLEYLHQGC++RILHLDIKPQNILL
Sbjct: 434 PNGSLDNFINGMGSPNAICCFDWNTLYKVAIGIARGLEYLHQGCNTRILHLDIKPQNILL 493
Query: 190 DEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGML 249
DED CPKI+DFGLAKIC+RK+SIVS+LG RGT GY+APE+FSRAFGGVS++SDVYS+GML
Sbjct: 494 DEDLCPKIADFGLAKICKRKESIVSMLGTRGTPGYIAPEVFSRAFGGVSHKSDVYSFGML 553
Query: 250 ILEMIGGRKNYDT-GGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSL 308
ILEM+GGRKNYD+ GGS +SEM+FPDWIYKDLEQG+ N I+EEE++M RK+ LVSL
Sbjct: 554 ILEMVGGRKNYDSGGGSQSSEMFFPDWIYKDLEQGDVHTNFLVITEEEHEMARKMILVSL 613
Query: 309 WCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLYETNSVT 368
WCIQT+ S+RP MNKV+EML+G L SV YPPKP+L+SPE+ LQ SD++S + ET+S+T
Sbjct: 614 WCIQTRSSERPSMNKVVEMLEGTLESVPYPPKPILHSPEKLLLQNSDLTSGNTLETDSMT 673
Query: 369 VSK 371
K
Sbjct: 674 TEK 676
>dbj|BAD95981.1| Ser/Thr protein kinase [Lotus corniculatus var. japonicus]
Length = 472
Score = 503 bits (1296), Expect = e-141
Identities = 250/373 (67%), Positives = 307/373 (82%), Gaps = 3/373 (0%)
Query: 1 MIILRQQQQSSLIPQTILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKL 60
+II + + S + + R+R + VD+NVE FM+S+ PR+YSY+E+K ITN F +KL
Sbjct: 86 LIICHRMRSMSFRQKMLFRKRRKYVDHNVEAFMKSYGSLAPRQYSYSEIKRITNSFLDKL 145
Query: 61 GQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEEN 120
GQGGYGVVYKA+L + R VAVKVISE+ G+GE+FINEVASIS TSH+NIVSLLG+CY N
Sbjct: 146 GQGGYGVVYKATLPDGRLVAVKVISESNGSGEDFINEVASISNTSHVNIVSLLGFCYM-N 204
Query: 121 KRALIYEFMPKGSLDKFIY-KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILH 179
KRALIYEFMP GSLD FI + PNA+ FDWNTL ++AIGIA+GLEYLHQGC++RILH
Sbjct: 205 KRALIYEFMPNGSLDNFINGRGGSPNAVSYFDWNTLSKVAIGIARGLEYLHQGCNTRILH 264
Query: 180 LDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSY 239
LDIKPQNILLDE+ CP I+DFGLAKIC+RK+SIVS+LG RGT GY+APEIFSRAFGG S+
Sbjct: 265 LDIKPQNILLDENLCPNIADFGLAKICKRKESIVSMLGTRGTPGYIAPEIFSRAFGGASH 324
Query: 240 RSDVYSYGMLILEMIGGRKNYDT-GGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEEND 298
+SDVYSYGMLILEM+GGR+NYD+ GGS +SEM FPDWIYKDLEQG+ NC I+ EE++
Sbjct: 325 KSDVYSYGMLILEMVGGRENYDSGGGSQSSEMSFPDWIYKDLEQGDVHTNCLVITGEEHE 384
Query: 299 MIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSS 358
M RK+ LVSLWCIQT PS+RP M+KV+EML+G L SV YPPKP+L+SPE+ LQ SD++S
Sbjct: 385 MARKMILVSLWCIQTHPSERPSMSKVVEMLEGTLQSVPYPPKPILHSPEKLLLQNSDLTS 444
Query: 359 SDLYETNSVTVSK 371
+ ET+S+T K
Sbjct: 445 GNTLETDSMTTEK 457
>dbj|BAB11294.1| receptor serine/threonine kinase [Arabidopsis thaliana]
gi|15240873|ref|NP_198644.1| serine/threonine protein
kinase (PR5K) [Arabidopsis thaliana]
Length = 665
Score = 385 bits (988), Expect = e-105
Identities = 191/340 (56%), Positives = 252/340 (73%), Gaps = 14/340 (4%)
Query: 12 LIPQTILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKA 71
+I +T +E D NVE ++M +RYSY VK +TN F LG+GG+G VYK
Sbjct: 295 IIVRTKNMRNSEWNDQNVEA------VAMLKRYSYTRVKKMTNSFAHVLGKGGFGTVYKG 348
Query: 72 SLYNS-RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMP 130
L +S R VAVK++ ++GNGEEFINEVAS+SRTSH+NIVSLLG+CYE+NKRA+IYEFMP
Sbjct: 349 KLADSGRDVAVKILKVSEGNGEEFINEVASMSRTSHVNIVSLLGFCYEKNKRAIIYEFMP 408
Query: 131 KGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLD 190
GSLDK+I N +W L+ +A+GI++GLEYLH C +RI+H DIKPQNIL+D
Sbjct: 409 NGSLDKYISA----NMSTKMEWERLYDVAVGISRGLEYLHNRCVTRIVHFDIKPQNILMD 464
Query: 191 EDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLI 250
E+ CPKISDFGLAK+C+ K+SI+S+L RGT GY+APE+FS+ FG VS++SDVYSYGM++
Sbjct: 465 ENLCPKISDFGLAKLCKNKESIISMLHMRGTFGYIAPEMFSKNFGAVSHKSDVYSYGMVV 524
Query: 251 LEMIGGR--KNYDTGGSCTSEMYFPDWIYKDLEQGN-TLLNCSTISEEENDMIRKITLVS 307
LEMIG + + + GS MYFP+W+YKD E+G T + +I++EE + +K+ LV+
Sbjct: 525 LEMIGAKNIEKVEYSGSNNGSMYFPEWVYKDFEKGEITRIFGDSITDEEEKIAKKLVLVA 584
Query: 308 LWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPE 347
LWCIQ PSDRPPM KVIEML+G L ++ PP P+L+SPE
Sbjct: 585 LWCIQMNPSDRPPMIKVIEMLEGNLEALQVPPNPLLFSPE 624
>emb|CAB78827.1| receptor serine/threonine kinase-like protein [Arabidopsis
thaliana] gi|4375833|emb|CAA16797.1| receptor
serine/threonine kinase-like protein [Arabidopsis
thaliana] gi|7488208|pir||T04927 probable
serine/threonine-specific protein kinase (EC 2.7.1.-)
T9A21.100 - Arabidopsis thaliana
Length = 687
Score = 384 bits (985), Expect = e-105
Identities = 192/360 (53%), Positives = 262/360 (72%), Gaps = 25/360 (6%)
Query: 19 RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNS-- 76
+ ++EL D N+E + M +RYS+ +VK +TN F +G+GG+G VYK L ++
Sbjct: 324 KRKSELNDENIEAVV------MLKRYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASG 377
Query: 77 RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDK 136
R +A+K++ E+KGNGEEFINE+ S+SR SH+NIVSL G+CYE ++RA+IYEFMP GSLDK
Sbjct: 378 RDIALKILKESKGNGEEFINELVSMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDK 437
Query: 137 FIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPK 196
FI + N +W TL+ IA+G+A+GLEYLH C S+I+H DIKPQNIL+DED CPK
Sbjct: 438 FISE----NMSTKIEWKTLYNIAVGVARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPK 493
Query: 197 ISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGG 256
ISDFGLAK+C++K+SI+S+L ARGT+GY+APE+FS+ +GGVS++SDVYSYGM++LEMIG
Sbjct: 494 ISDFGLAKLCKKKESIISMLDARGTVGYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGA 553
Query: 257 --RKNYDTGGSCTSEMYFPDWIYKDLEQGNT--LLNCSTISEEEND-MIRKITLVSLWCI 311
R+ +T + S MYFPDW+Y+DLE+ T LL I EEE + +++++TLV LWCI
Sbjct: 554 TKREEVETSATDKSSMYFPDWVYEDLERKETMRLLEDHIIEEEEEEKIVKRMTLVGLWCI 613
Query: 312 QTKPSDRPPMNKVIEMLQGP-LSSVSYPPKPVL-------YSPERPELQVSDMSSSDLYE 363
QT PSDRPPM KV+EML+G L ++ PPKP+L + Q S +S+ L E
Sbjct: 614 QTNPSDRPPMRKVVEMLEGSRLEALQVPPKPLLNLHVVTDWETSEDSQQTSRLSTQSLLE 673
>ref|NP_193559.2| receptor serine/threonine kinase, putative [Arabidopsis thaliana]
Length = 853
Score = 384 bits (985), Expect = e-105
Identities = 192/360 (53%), Positives = 262/360 (72%), Gaps = 25/360 (6%)
Query: 19 RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNS-- 76
+ ++EL D N+E + M +RYS+ +VK +TN F +G+GG+G VYK L ++
Sbjct: 490 KRKSELNDENIEAVV------MLKRYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASG 543
Query: 77 RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDK 136
R +A+K++ E+KGNGEEFINE+ S+SR SH+NIVSL G+CYE ++RA+IYEFMP GSLDK
Sbjct: 544 RDIALKILKESKGNGEEFINELVSMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDK 603
Query: 137 FIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPK 196
FI + N +W TL+ IA+G+A+GLEYLH C S+I+H DIKPQNIL+DED CPK
Sbjct: 604 FISE----NMSTKIEWKTLYNIAVGVARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPK 659
Query: 197 ISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGG 256
ISDFGLAK+C++K+SI+S+L ARGT+GY+APE+FS+ +GGVS++SDVYSYGM++LEMIG
Sbjct: 660 ISDFGLAKLCKKKESIISMLDARGTVGYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGA 719
Query: 257 --RKNYDTGGSCTSEMYFPDWIYKDLEQGNT--LLNCSTISEEEND-MIRKITLVSLWCI 311
R+ +T + S MYFPDW+Y+DLE+ T LL I EEE + +++++TLV LWCI
Sbjct: 720 TKREEVETSATDKSSMYFPDWVYEDLERKETMRLLEDHIIEEEEEEKIVKRMTLVGLWCI 779
Query: 312 QTKPSDRPPMNKVIEMLQGP-LSSVSYPPKPVL-------YSPERPELQVSDMSSSDLYE 363
QT PSDRPPM KV+EML+G L ++ PPKP+L + Q S +S+ L E
Sbjct: 780 QTNPSDRPPMRKVVEMLEGSRLEALQVPPKPLLNLHVVTDWETSEDSQQTSRLSTQSLLE 839
>gb|AAC49208.1| receptor serine/threonine kinase PR5K gi|1589714|prf||2211427A
receptor protein kinase
Length = 665
Score = 381 bits (978), Expect = e-104
Identities = 190/340 (55%), Positives = 250/340 (72%), Gaps = 14/340 (4%)
Query: 12 LIPQTILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKA 71
+I +T +E D NVE ++M +RYSY VK +TN F LG+GG+G VYK
Sbjct: 295 IIVRTKNMRNSEWNDQNVEA------VAMLKRYSYTRVKKMTNSFAHVLGKGGFGTVYKG 348
Query: 72 SLYNS-RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMP 130
L +S R VAVK++ ++GNGEEFINEVAS+SRTSH+NIVSLLG+CYE+NKRA+IYEFMP
Sbjct: 349 KLADSGRDVAVKILKVSEGNGEEFINEVASMSRTSHVNIVSLLGFCYEKNKRAIIYEFMP 408
Query: 131 KGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLD 190
GSLDK+I N +W L+ +A+GI++GLEYLH C +RI+H DIKPQNIL+D
Sbjct: 409 NGSLDKYISA----NMSTKMEWERLYDVAVGISRGLEYLHNRCVTRIVHFDIKPQNILMD 464
Query: 191 EDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLI 250
E+ CPKISDFGLAK+C+ K+SI+S+L RGT GY+APE+FS+ FG VS++SDVYSYGM++
Sbjct: 465 ENLCPKISDFGLAKLCKNKESIISMLHMRGTFGYIAPEMFSKNFGAVSHKSDVYSYGMVV 524
Query: 251 LEMIGGR--KNYDTGGSCTSEMYFPDWIYKDLEQGN-TLLNCSTISEEENDMIRKITLVS 307
LEMIG + + + S MYFP+W+YKD E+G T + ++I+EEE +K+ LV+
Sbjct: 525 LEMIGAKNIEKVEYSESNNGSMYFPEWVYKDFEKGEITRIFGNSITEEEEKFAKKLVLVA 584
Query: 308 LWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPE 347
LWCIQ PSDRPPM KV EML+G L ++ PP P+L+SPE
Sbjct: 585 LWCIQMNPSDRPPMIKVTEMLEGNLEALQVPPNPLLFSPE 624
>ref|NP_177182.2| receptor serine/threonine kinase, putative [Arabidopsis thaliana]
Length = 799
Score = 378 bits (970), Expect = e-103
Identities = 190/351 (54%), Positives = 249/351 (70%), Gaps = 18/351 (5%)
Query: 21 RTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYN-SRQV 79
+++L + N+E + M +R+SY +VK +T F LG+GG+G VYK L + SR V
Sbjct: 432 KSDLNEKNMEAVV------MLKRFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDV 485
Query: 80 AVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY 139
AVK++ E+ +GE+FINE+AS+SRTSH NIVSLLG+CYE K+A+IYE MP GSLDKFI
Sbjct: 486 AVKILKESNEDGEDFINEIASMSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFIS 545
Query: 140 KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISD 199
K N +W TL+ IA+G++ GLEYLH C SRI+H DIKPQNIL+D D CPKISD
Sbjct: 546 K----NMSAKMEWKTLYNIAVGVSHGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISD 601
Query: 200 FGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK- 258
FGLAK+C+ +SI+S+L ARGTIGY+APE+FS+ FGGVS++SDVYSYGM++LEMIG R
Sbjct: 602 FGLAKLCKNNESIISMLHARGTIGYIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNI 661
Query: 259 -NYDTGGSCTSEMYFPDWIYKDLEQGN--TLLNCSTISEEENDMIRKITLVSLWCIQTKP 315
GS + MYFPDWIYKDLE+G + L EE+ +++K+ LV LWCIQT P
Sbjct: 662 GRAQNAGSSNTSMYFPDWIYKDLEKGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNP 721
Query: 316 SDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLYETNS 366
DRPPM+KV+EML+G L ++ PPKP+L P + D+ ET+S
Sbjct: 722 YDRPPMSKVVEMLEGSLEALQIPPKPLLC---LPAITAPITVDEDIQETSS 769
>pir||E96725 hypothetical protein F20P5.3 [imported] - Arabidopsis thaliana
gi|2194117|gb|AAB61092.1| Strong similarity to
Arabidopsis receptor protein kinase PR5K (gb|ATU48698).
[Arabidopsis thaliana]
Length = 676
Score = 378 bits (970), Expect = e-103
Identities = 190/351 (54%), Positives = 249/351 (70%), Gaps = 18/351 (5%)
Query: 21 RTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYN-SRQV 79
+++L + N+E + M +R+SY +VK +T F LG+GG+G VYK L + SR V
Sbjct: 309 KSDLNEKNMEAVV------MLKRFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDV 362
Query: 80 AVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY 139
AVK++ E+ +GE+FINE+AS+SRTSH NIVSLLG+CYE K+A+IYE MP GSLDKFI
Sbjct: 363 AVKILKESNEDGEDFINEIASMSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFIS 422
Query: 140 KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISD 199
K N +W TL+ IA+G++ GLEYLH C SRI+H DIKPQNIL+D D CPKISD
Sbjct: 423 K----NMSAKMEWKTLYNIAVGVSHGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISD 478
Query: 200 FGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK- 258
FGLAK+C+ +SI+S+L ARGTIGY+APE+FS+ FGGVS++SDVYSYGM++LEMIG R
Sbjct: 479 FGLAKLCKNNESIISMLHARGTIGYIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNI 538
Query: 259 -NYDTGGSCTSEMYFPDWIYKDLEQGN--TLLNCSTISEEENDMIRKITLVSLWCIQTKP 315
GS + MYFPDWIYKDLE+G + L EE+ +++K+ LV LWCIQT P
Sbjct: 539 GRAQNAGSSNTSMYFPDWIYKDLEKGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNP 598
Query: 316 SDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLYETNS 366
DRPPM+KV+EML+G L ++ PPKP+L P + D+ ET+S
Sbjct: 599 YDRPPMSKVVEMLEGSLEALQIPPKPLLC---LPAITAPITVDEDIQETSS 646
>gb|AAF98210.1| Unknown protein [Arabidopsis thaliana] gi|25404657|pir||G96693
hypothetical protein F1O19.6 [imported] - Arabidopsis
thaliana
Length = 1111
Score = 376 bits (966), Expect = e-103
Identities = 180/322 (55%), Positives = 240/322 (73%), Gaps = 8/322 (2%)
Query: 44 YSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISR 103
Y+YA+VK IT F E +G+GG+G+VYK +L + R VAVKV+ +TKGNGE+FINEVA++SR
Sbjct: 788 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDTKGNGEDFINEVATMSR 847
Query: 104 TSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIA 163
TSH+NIVSLLG+C E +KRA+IYEF+ GSLDKFI N DW L+RIA+G+A
Sbjct: 848 TSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGKTSVN----MDWTALYRIALGVA 903
Query: 164 KGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIG 223
GLEYLH C +RI+H DIKPQN+LLD+ FCPK+SDFGLAK+C++K+SI+S+L RGTIG
Sbjct: 904 HGLEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIG 963
Query: 224 YMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSC---TSEMYFPDWIYKDL 280
Y+APE+ SR +G VS++SDVYSYGML+LE+IG R +C TS MYFP+W+Y+DL
Sbjct: 964 YIAPEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDL 1023
Query: 281 EQGNTLLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPP 339
E + + I+ EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++ PP
Sbjct: 1024 ESCKSGRHIEDGINSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLEALEVPP 1083
Query: 340 KPVLYSPERPELQVSDMSSSDL 361
+PVL L S + S D+
Sbjct: 1084 RPVLQQIPISNLHESSILSEDV 1105
>ref|NP_176870.1| protein kinase family protein / glycerophosphoryl diester
phosphodiesterase family protein [Arabidopsis thaliana]
Length = 1109
Score = 376 bits (966), Expect = e-103
Identities = 180/322 (55%), Positives = 240/322 (73%), Gaps = 8/322 (2%)
Query: 44 YSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISR 103
Y+YA+VK IT F E +G+GG+G+VYK +L + R VAVKV+ +TKGNGE+FINEVA++SR
Sbjct: 786 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDTKGNGEDFINEVATMSR 845
Query: 104 TSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIA 163
TSH+NIVSLLG+C E +KRA+IYEF+ GSLDKFI N DW L+RIA+G+A
Sbjct: 846 TSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGKTSVN----MDWTALYRIALGVA 901
Query: 164 KGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIG 223
GLEYLH C +RI+H DIKPQN+LLD+ FCPK+SDFGLAK+C++K+SI+S+L RGTIG
Sbjct: 902 HGLEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIG 961
Query: 224 YMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSC---TSEMYFPDWIYKDL 280
Y+APE+ SR +G VS++SDVYSYGML+LE+IG R +C TS MYFP+W+Y+DL
Sbjct: 962 YIAPEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDL 1021
Query: 281 EQGNTLLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPP 339
E + + I+ EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++ PP
Sbjct: 1022 ESCKSGRHIEDGINSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLEALEVPP 1081
Query: 340 KPVLYSPERPELQVSDMSSSDL 361
+PVL L S + S D+
Sbjct: 1082 RPVLQQIPISNLHESSILSEDV 1103
>ref|NP_176863.2| protein kinase, putative [Arabidopsis thaliana]
Length = 666
Score = 375 bits (962), Expect = e-102
Identities = 181/323 (56%), Positives = 242/323 (74%), Gaps = 7/323 (2%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
+ YSYA+V IT F E +G+GG+G VY+ +LY+ R VAVKV+ E++GNGE+FINEVAS+
Sbjct: 336 KHYSYAQVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLKESQGNGEDFINEVASM 395
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI + DW L+ IA+G
Sbjct: 396 SQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSST----MDWRELYGIALG 451
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
+A+GLEYLH GC +RI+H DIKPQN+LLD++ PK+SDFGLAK+C+RK+SI+S++ RGT
Sbjct: 452 VARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGT 511
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGG--SCTSEMYFPDWIYKD 279
IGY+APE+FSR +G VS++SDVYSYGML+L++IG R T S TS MYFP+WIY+D
Sbjct: 512 IGYIAPEVFSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRD 571
Query: 280 LEQGNTLLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYP 338
LE+ + + T IS EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++ P
Sbjct: 572 LEKAHNGKSIETAISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVP 631
Query: 339 PKPVLYSPERPELQVSDMSSSDL 361
P+PVL LQ S S D+
Sbjct: 632 PRPVLQQIPTATLQESSTFSEDI 654
>gb|AAG50590.1| receptor serine/threonine kinase PR5K, putative [Arabidopsis
thaliana] gi|25404649|pir||H96692 probable receptor
serine/threonine kinase PR5K T4O24.8 [imported] -
Arabidopsis thaliana
Length = 655
Score = 375 bits (962), Expect = e-102
Identities = 181/323 (56%), Positives = 242/323 (74%), Gaps = 7/323 (2%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
+ YSYA+V IT F E +G+GG+G VY+ +LY+ R VAVKV+ E++GNGE+FINEVAS+
Sbjct: 325 KHYSYAQVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLKESQGNGEDFINEVASM 384
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI + DW L+ IA+G
Sbjct: 385 SQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSST----MDWRELYGIALG 440
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
+A+GLEYLH GC +RI+H DIKPQN+LLD++ PK+SDFGLAK+C+RK+SI+S++ RGT
Sbjct: 441 VARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGT 500
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGG--SCTSEMYFPDWIYKD 279
IGY+APE+FSR +G VS++SDVYSYGML+L++IG R T S TS MYFP+WIY+D
Sbjct: 501 IGYIAPEVFSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRD 560
Query: 280 LEQGNTLLNCST-ISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYP 338
LE+ + + T IS EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++ P
Sbjct: 561 LEKAHNGKSIETAISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVP 620
Query: 339 PKPVLYSPERPELQVSDMSSSDL 361
P+PVL LQ S S D+
Sbjct: 621 PRPVLQQIPTATLQESSTFSEDI 643
>dbj|BAB10826.1| receptor protein kinase-like protein [Arabidopsis thaliana]
gi|15241610|ref|NP_198718.1| protein kinase family
protein [Arabidopsis thaliana]
Length = 813
Score = 374 bits (960), Expect = e-102
Identities = 182/331 (54%), Positives = 247/331 (73%), Gaps = 9/331 (2%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
++Y YAE+K IT F +G+GG+G VY+ +L N R VAVKV+ + KGNG++FINEV S+
Sbjct: 484 KQYIYAELKKITKSFSHTVGKGGFGTVYRGNLSNGRTVAVKVLKDLKGNGDDFINEVTSM 543
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY--KSEFPNAICDFDWNTLFRIA 159
S+TSH+NIVSLLG+CYE +KRA+I EF+ GSLD+FI KS PN TL+ IA
Sbjct: 544 SQTSHVNIVSLLGFCYEGSKRAIISEFLEHGSLDQFISRNKSLTPNV------TTLYGIA 597
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
+GIA+GLEYLH GC +RI+H DIKPQNILLD++FCPK++DFGLAK+C++++SI+S++ R
Sbjct: 598 LGIARGLEYLHYGCKTRIVHFDIKPQNILLDDNFCPKVADFGLAKLCEKRESILSLIDTR 657
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKD 279
GTIGY+APE+ SR +GG+S++SDVYSYGML+L+MIG R +T S YFPDWIYKD
Sbjct: 658 GTIGYIAPEVVSRMYGGISHKSDVYSYGMLVLDMIGARNKVETTTCNGSTAYFPDWIYKD 717
Query: 280 LEQGN-TLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYP 338
LE G+ T + I+EE+N +++K+ LVSLWCI+ PSDRPPMNKV+EM++G L ++ P
Sbjct: 718 LENGDQTWIIGDEINEEDNKIVKKMILVSLWCIRPCPSDRPPMNKVVEMIEGSLDALELP 777
Query: 339 PKPVLYSPERPELQVSDMSSSDLYETNSVTV 369
PKP + L+ S +S E + T+
Sbjct: 778 PKPSRHISTELVLESSSLSDGQEAEKQTQTL 808
>dbj|BAB11292.1| receptor serine/threonine kinase [Arabidopsis thaliana]
gi|15240865|ref|NP_198642.1| serine/threonine protein
kinase, putative [Arabidopsis thaliana]
Length = 638
Score = 374 bits (959), Expect = e-102
Identities = 181/338 (53%), Positives = 251/338 (73%), Gaps = 13/338 (3%)
Query: 26 DNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVIS 85
DNN++ L ++YSYAEV+ IT F LG+GG+G VY +L + R+VAVK++
Sbjct: 299 DNNLK------GLVQLKQYSYAEVRKITKLFSHTLGKGGFGTVYGGNLCDGRKVAVKILK 352
Query: 86 ETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPN 145
+ K NGE+FINEVAS+S+TSH+NIVSLLG+CYE +KRA++YEF+ GSLD+F+ + + N
Sbjct: 353 DFKSNGEDFINEVASMSQTSHVNIVSLLGFCYEGSKRAIVYEFLENGSLDQFLSEKKSLN 412
Query: 146 AICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKI 205
D +TL+RIA+G+A+GL+YLH GC +RI+H DIKPQNILLD+ FCPK+SDFGLAK+
Sbjct: 413 ----LDVSTLYRIALGVARGLDYLHHGCKTRIVHFDIKPQNILLDDTFCPKVSDFGLAKL 468
Query: 206 CQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN--YDTG 263
C++++SI+S+L ARGTIGY+APE+FS +G VS++SDVYSYGML+LEMIG + +T
Sbjct: 469 CEKRESILSLLDARGTIGYIAPEVFSGMYGRVSHKSDVYSYGMLVLEMIGAKNKEIEETA 528
Query: 264 GSCTSEMYFPDWIYKDLEQG-NTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMN 322
S +S YFPDWIYK+LE G +T IS E+ ++ +K+TLV LWCIQ P +RPPMN
Sbjct: 529 ASNSSSAYFPDWIYKNLENGEDTWKFGDEISREDKEVAKKMTLVGLWCIQPSPLNRPPMN 588
Query: 323 KVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSD 360
+++EM++G L + PPKP ++ P Q+S S +
Sbjct: 589 RIVEMMEGSLDVLEVPPKPSIHYSAEPLPQLSSFSEEN 626
>dbj|BAD82485.1| receptor serine/threonine kinase PR5K-like [Oryza sativa (japonica
cultivar-group)]
Length = 658
Score = 372 bits (956), Expect = e-102
Identities = 172/315 (54%), Positives = 240/315 (75%), Gaps = 3/315 (0%)
Query: 29 VEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETK 88
+E F++ + P+RY+Y EVK +T F EKLG GG+G VY+ +L + RQVAVK++ ++K
Sbjct: 345 IESFLQRNGTLHPKRYTYTEVKRMTKSFAEKLGHGGFGAVYRGNLSDGRQVAVKMLKDSK 404
Query: 89 GNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAIC 148
G+GEEFINEVASISRTSH+N+V+LLG+C +KRALIYE+MP GSL+++ +++ +
Sbjct: 405 GDGEEFINEVASISRTSHVNVVTLLGFCLHRSKRALIYEYMPNGSLERYAFRNNSKGEL- 463
Query: 149 DFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
W LF +A+GIA+GLEYLH+GCS+RI+H DIKP NILLD++FCPKISDFG+AK+C
Sbjct: 464 SLTWEKLFDVAVGIARGLEYLHRGCSTRIVHFDIKPHNILLDQEFCPKISDFGMAKLCAN 523
Query: 209 KDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTS 268
K+SIVSI GARGTIGY+APE++S+ FG +S +SDVYSYGM+ILEM+G R+ S +S
Sbjct: 524 KESIVSIAGARGTIGYIAPEVYSKQFGAISSKSDVYSYGMMILEMVGARERNIEANSESS 583
Query: 269 EMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEML 328
YFP WIY+ L++ ++ S I E +++RK+ +V+LWCIQ P++RP M +V+EML
Sbjct: 584 SHYFPQWIYEHLDE--YCISSSEIDGETTELVRKMVVVALWCIQVVPTNRPTMTRVVEML 641
Query: 329 QGPLSSVSYPPKPVL 343
+G S + PPK +L
Sbjct: 642 EGSTSGLELPPKVLL 656
>ref|NP_176864.1| serine/threonine protein kinase, putative [Arabidopsis thaliana]
gi|12320924|gb|AAG50589.1| receptor serine/threonine
kinase PR5K, putative [Arabidopsis thaliana]
gi|25404651|pir||A96693 probable receptor
serine/threonine kinase PR5K T4O24.7 [imported] -
Arabidopsis thaliana
Length = 609
Score = 368 bits (944), Expect = e-100
Identities = 184/333 (55%), Positives = 248/333 (74%), Gaps = 19/333 (5%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGN-GEEFINEVAS 100
++YSY +VK ITN F E +G+GG+G+VY+ +L + R VAVKV+ + KGN GE+FINEVAS
Sbjct: 287 KQYSYEQVKRITNSFAEVVGRGGFGIVYRGTLSDGRMVAVKVLKDLKGNNGEDFINEVAS 346
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
+S+TSH+NIV+LLG+C E KRA+IYEFM GSLDKFI + DW L+ IA+
Sbjct: 347 MSQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSST----MDWRELYGIAL 402
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+A+GLEYLH GC +RI+H DIKPQN+LLD++ PK+SDFGLAK+C+RK+SI+S++ RG
Sbjct: 403 GVARGLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRG 462
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGG--SCTSEMYFPDWIYK 278
TIGY+APE+FSR +G VS++SDVYSYGML+L++IG R T S TS MYFP+WIYK
Sbjct: 463 TIGYIAPEVFSRVYGSVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYK 522
Query: 279 DLEQGNT---LLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
DLE+G+ ++N S EE+++ +K+TLV LWCIQ P DRP MN+V+EM++G L ++
Sbjct: 523 DLEKGDNGRLIVNRS----EEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDAL 578
Query: 336 SYPPKPVLYSPERPELQVSDMSSSDLYETNSVT 368
PP+PVL P L SS + E NS++
Sbjct: 579 EVPPRPVLQCSVVPHL-----DSSWISEENSIS 606
>dbj|BAD82483.1| receptor serine/threonine kinase PR5K-like [Oryza sativa (japonica
cultivar-group)]
Length = 913
Score = 367 bits (943), Expect = e-100
Identities = 169/315 (53%), Positives = 237/315 (74%), Gaps = 3/315 (0%)
Query: 29 VEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETK 88
+E F++ + P+RY+Y EVK +T F EKLG GG+G VY+ +L + RQVAVK++ ++K
Sbjct: 600 IESFLQRNGTLHPKRYTYTEVKRMTKSFAEKLGHGGFGAVYRGNLSDGRQVAVKMLKDSK 659
Query: 89 GNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAIC 148
G+GEEFINEVASISRTSH+N+V+LLG+C +KR LIYE+MP GSL+++ +++
Sbjct: 660 GDGEEFINEVASISRTSHVNVVTLLGFCLHGSKRVLIYEYMPNGSLERYAFRNNSEGEH- 718
Query: 149 DFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
W LF + +GIA+GLEYLH+GC++RI+H DIKP NILLD++FCPKISDFG+AK+C
Sbjct: 719 SLTWEKLFDVVVGIARGLEYLHRGCNTRIVHFDIKPHNILLDQEFCPKISDFGMAKLCSN 778
Query: 209 KDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTS 268
K+SI+SI GARGTIGY+APE++S+ FG +S +SDVYSYGM+ILEM+G R+ S +S
Sbjct: 779 KESIISIAGARGTIGYIAPEVYSKQFGAISSKSDVYSYGMMILEMVGARERNIDANSESS 838
Query: 269 EMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEML 328
YFP WIY+ L++ +N S I E +++RK+ +V+LWCIQ P++RP M +V+EML
Sbjct: 839 SHYFPQWIYEHLDE--YCINSSEIDGETTELVRKMVVVALWCIQVVPTNRPTMTRVVEML 896
Query: 329 QGPLSSVSYPPKPVL 343
+G S + PPK +L
Sbjct: 897 EGSTSGLELPPKVLL 911
>ref|NP_176871.1| protein kinase family protein [Arabidopsis thaliana]
Length = 717
Score = 366 bits (940), Expect = e-100
Identities = 186/334 (55%), Positives = 247/334 (73%), Gaps = 24/334 (7%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKG-NGEEFINEVAS 100
+ Y+YAEVK +T F E +G+GG+G+VY +L +S VAVKV+ ++KG +GE+FINEVAS
Sbjct: 369 KHYTYAEVKKMTKSFTEVVGRGGFGIVYSGTLSDSSMVAVKVLKDSKGTDGEDFINEVAS 428
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
+S+TSH+NIVSLLG+C E ++RA+IYEF+ GSLDKFI N D TL+ IA+
Sbjct: 429 MSQTSHVNIVSLLGFCCEGSRRAIIYEFLGNGSLDKFISDKSSVN----LDLKTLYGIAL 484
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+A+GLEYLH GC +RI+H DIKPQN+LLD++ CPK+SDFGLAK+C++K+SI+S+L RG
Sbjct: 485 GVARGLEYLHYGCKTRIVHFDIKPQNVLLDDNLCPKVSDFGLAKLCEKKESILSLLDTRG 544
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK--NYDTGG-SCTSEMYFPDWIY 277
TIGY+APE+ SR +G VS++SDVYSYGML+LEMIG RK +D S S +YFP+WIY
Sbjct: 545 TIGYIAPEMISRLYGSVSHKSDVYSYGMLVLEMIGARKKERFDQNSRSDGSSIYFPEWIY 604
Query: 278 KDLEQGNTL---------LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEML 328
KDLE+ N L + IS EE ++ RK+TLV LWCIQ+ PSDRPPMNKV+EM+
Sbjct: 605 KDLEKANIKDIEKTENGGLIENGISSEEEEIARKMTLVGLWCIQSSPSDRPPMNKVVEMM 664
Query: 329 QGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLY 362
+G L ++ PP+PVL Q+S S SD +
Sbjct: 665 EGSLDALEVPPRPVLQ-------QISASSVSDSF 691
>ref|XP_550060.1| putative receptor serine/threonine kinase PR5K [Oryza sativa
(japonica cultivar-group)] gi|54290827|dbj|BAD61466.1|
putative receptor serine/threonine kinase PR5K [Oryza
sativa (japonica cultivar-group)]
Length = 598
Score = 361 bits (927), Expect = 2e-98
Identities = 175/315 (55%), Positives = 231/315 (72%), Gaps = 4/315 (1%)
Query: 26 DNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVIS 85
++N++ + S+ P+RY Y+EV IT++ KLG+GGYGVV+K L + R VAVK +
Sbjct: 281 ESNIQKLIVSYGSLAPKRYKYSEVAKITSFLSNKLGEGGYGVVFKGKLQDGRLVAVKFLH 340
Query: 86 ETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPN 145
++KGNGEEF+NEV SI RTSH+NIVSL G+C E +KRALIY++MP SLD +IY SE P
Sbjct: 341 DSKGNGEEFVNEVMSIGRTSHVNIVSLFGFCLEGSKRALIYDYMPNSSLDNYIY-SEKPK 399
Query: 146 AICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKI 205
W L+ IAIGIA+GLEYLH GC++RI+H DIKPQNILLD+DFCPKI+DFGLAK+
Sbjct: 400 ET--LGWEKLYDIAIGIARGLEYLHHGCNTRIVHFDIKPQNILLDQDFCPKIADFGLAKL 457
Query: 206 CQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGS 265
C K+S +S+ GARGTIG++APE+ R+FG VS +SDVYSYGM++LEMIGGRKN +
Sbjct: 458 CCTKESKLSMTGARGTIGFIAPEVLYRSFGVVSIKSDVYSYGMMLLEMIGGRKNVKSMVQ 517
Query: 266 CTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVI 325
+SE YFPDWIY QG+ L C SE E ++ +K+TL+ LWC+Q P RP + +V+
Sbjct: 518 NSSEKYFPDWIYDHFYQGDGLQACEVTSEVE-EIAKKMTLIGLWCVQVLPMHRPTITQVL 576
Query: 326 EMLQGPLSSVSYPPK 340
+M + L + PPK
Sbjct: 577 DMFEKALDELDMPPK 591
>ref|XP_550064.1| putative receptor serine/threonine kinase PR5K [Oryza sativa
(japonica cultivar-group)] gi|54290831|dbj|BAD61470.1|
putative receptor serine/threonine kinase PR5K [Oryza
sativa (japonica cultivar-group)]
Length = 419
Score = 359 bits (921), Expect = 8e-98
Identities = 179/342 (52%), Positives = 238/342 (69%), Gaps = 5/342 (1%)
Query: 26 DNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVIS 85
+ N+E + S+ P RY Y+EV IT++ KLG+GGYGVV+K L + R VAVK +
Sbjct: 52 ERNIEALIISYGSIAPTRYKYSEVTKITSFLNYKLGEGGYGVVFKGRLQDGRLVAVKFLH 111
Query: 86 ETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPN 145
++KGNGEEF+NEV SI RTSH+NIVSL G+C E +KRAL+YE+MP GSLD +IY SE P
Sbjct: 112 DSKGNGEEFVNEVMSIGRTSHINIVSLFGFCLEGSKRALLYEYMPNGSLDDYIY-SENPK 170
Query: 146 AICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKI 205
I W L+ IAIGIA+GLEYLH C++RI+H DIKPQNILLD+DFCPKI+DFGLAK+
Sbjct: 171 EI--LGWEKLYGIAIGIARGLEYLHHSCNTRIIHFDIKPQNILLDQDFCPKIADFGLAKL 228
Query: 206 CQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGS 265
C+ K+S +S+ GARGTIG++APE+ R+FG VS +SDVYSYGM++LEM+GGRKN +
Sbjct: 229 CRTKESKLSMTGARGTIGFIAPEVIYRSFGIVSTKSDVYSYGMMLLEMVGGRKNAKSMVE 288
Query: 266 CTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVI 325
+SE YFPDWIY + L C SE E + +K+TL+ LWC+Q P RP + +V+
Sbjct: 289 NSSEKYFPDWIYDHFALDDGLQACEVTSEVE-QIAKKMTLIGLWCVQVLPMHRPTITQVL 347
Query: 326 EMLQGPLSSVSYPPKPVLYS-PERPELQVSDMSSSDLYETNS 366
+M + L + PPK E P +++ S+S T +
Sbjct: 348 DMFERSLDELEMPPKQNFSELLEHPAQEINTESTSSTINTKA 389
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 629,526,292
Number of Sequences: 2540612
Number of extensions: 27184319
Number of successful extensions: 108750
Number of sequences better than 10.0: 20430
Number of HSP's better than 10.0 without gapping: 8626
Number of HSP's successfully gapped in prelim test: 11804
Number of HSP's that attempted gapping in prelim test: 70110
Number of HSP's gapped (non-prelim): 22800
length of query: 371
length of database: 863,360,394
effective HSP length: 129
effective length of query: 242
effective length of database: 535,621,446
effective search space: 129620389932
effective search space used: 129620389932
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147875.1


