

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.13 + phase: 0
(127 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
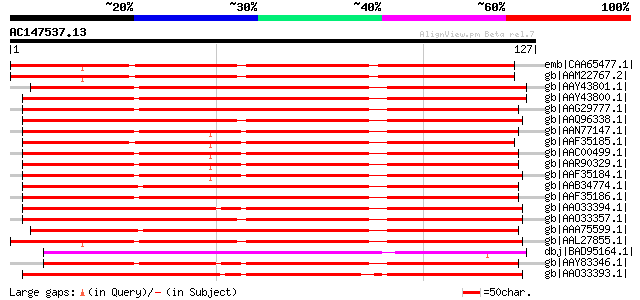
emb|CAA65477.1| lipid transfer protein [Prunus dulcis] gi|249775... 135 2e-31
gb|AAM22767.2| lipid transfer protein 2 precursor [Prunus persica] 133 1e-30
gb|AAY43801.1| FSLTP3 [Gossypium hirsutum] 129 2e-29
gb|AAY43800.1| FSLTP2 [Gossypium hirsutum] 128 3e-29
gb|AAG29777.1| lipid transfer protein 3 precursor [Gossypium hir... 128 4e-29
gb|AAQ96338.1| lipid transfer protein [Vitis aestivalis] 125 2e-28
gb|AAN77147.1| fiber lipid transfer protein [Gossypium barbadense] 125 2e-28
gb|AAF35185.1| lipid transfer protein precursor [Gossypium hirsu... 124 7e-28
gb|AAC00499.1| lipid transfer protein precursor [Gossypium hirsu... 124 7e-28
gb|AAR90329.1| lipid transfer protein precursor [Gossypium barba... 124 7e-28
gb|AAF35184.1| lipid transfer protein precursor [Gossypium hirsu... 123 1e-27
gb|AAB34774.1| LTP [Gossypium hirsutum] gi|66271043|gb|AAY43799.... 122 2e-27
gb|AAF35186.1| lipid transfer protein precursor [Gossypium hirsu... 122 3e-27
gb|AAO33394.1| lipid transfer protein isoform 4 [Vitis vinifera] 121 3e-27
gb|AAO33357.1| nonspecific lipid transfer protein 1 [Vitis berla... 120 6e-27
gb|AAA75599.1| nonspecific lipid transfer protein precursor gi|2... 120 7e-27
gb|AAL27855.1| lipid transfer protein precursor [Davidia involuc... 119 1e-26
dbj|BAD95164.1| putative lipid transfer protein [Arabidopsis tha... 119 2e-26
gb|AAY83346.1| non-specific lipid transfer protein precursor [Fr... 119 2e-26
gb|AAO33393.1| lipid transfer protein isoform 1 [Vitis vinifera] 117 5e-26
>emb|CAA65477.1| lipid transfer protein [Prunus dulcis]
gi|2497753|sp|Q43019|NLT3_PRUDU Nonspecific
lipid-transfer protein 3 precursor (LTP 3)
Length = 123
Score = 135 bits (340), Expect = 2e-31
Identities = 69/124 (55%), Positives = 87/124 (69%), Gaps = 7/124 (5%)
Query: 1 MGYSTMATRLVCLAIV--CLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQC 58
M S +LVCL V C+ GPKA AAV SCG VV +L PC++Y+ NGG P+ C
Sbjct: 1 MASSGQLLKLVCLVAVMCCMAVGGPKAMAAV-SCGQVVNNLTPCINYVANGGALNPS--C 57
Query: 59 CNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISP 118
C G+R+L ++AQTT DR+++C C+K AV+ G YTN N LAAGLP KCGVNIPY+ISP
Sbjct: 58 CTGVRSLYSLAQTTADRQSICNCLKQAVN--GIPYTNANAGLAAGLPGKCGVNIPYKISP 115
Query: 119 NTDC 122
+TDC
Sbjct: 116 STDC 119
>gb|AAM22767.2| lipid transfer protein 2 precursor [Prunus persica]
Length = 123
Score = 133 bits (334), Expect = 1e-30
Identities = 68/124 (54%), Positives = 86/124 (68%), Gaps = 7/124 (5%)
Query: 1 MGYSTMATRLVCLAIV--CLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQC 58
M S +LVCL V C+ GPKA AAV SCG VV +L PC++Y+ NGG P+ C
Sbjct: 1 MASSGQLLKLVCLVAVMCCMAVGGPKAMAAV-SCGQVVNNLTPCINYVANGGALNPS--C 57
Query: 59 CNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISP 118
C G+R+L ++AQTT DR+++C C+K AV+ G Y N N LAAGLP KCGVNIPY+ISP
Sbjct: 58 CTGVRSLYSLAQTTADRQSICNCLKQAVN--GIPYPNANAGLAAGLPGKCGVNIPYKISP 115
Query: 119 NTDC 122
+TDC
Sbjct: 116 STDC 119
>gb|AAY43801.1| FSLTP3 [Gossypium hirsutum]
Length = 116
Score = 129 bits (324), Expect = 2e-29
Identities = 60/120 (50%), Positives = 82/120 (68%), Gaps = 5/120 (4%)
Query: 6 MATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNL 65
M+ +L C+ ++C+V P A+ AVT CG V SL PC++Y+ G CC+GI++L
Sbjct: 1 MSLKLACVVVLCMVVGAPLAQGAVT-CGQVTNSLAPCINYLRGSGAGAVPPGCCSGIKSL 59
Query: 66 NTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCNRY 125
N+ AQTT DR+A C CIK+A + T +N LA+GLP KCGVNIPY+ISP+TDCNR+
Sbjct: 60 NSAAQTTPDRQAACRCIKSAAA----GITGINFGLASGLPGKCGVNIPYKISPSTDCNRF 115
>gb|AAY43800.1| FSLTP2 [Gossypium hirsutum]
Length = 120
Score = 128 bits (322), Expect = 3e-29
Identities = 61/122 (50%), Positives = 83/122 (68%), Gaps = 5/122 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M+ +L C+ ++ +V P A+ AVT CG V SL PC++Y+ G CC+GI+
Sbjct: 3 SSMSLKLACVVVLGMVVGAPLAQGAVT-CGQVTNSLAPCINYLRGSGAGAVPPGCCSGIK 61
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ AQTT DR+A C CIK+A + T +N LA+GLP KCGVNIPY+ISPNTDCN
Sbjct: 62 SLNSAAQTTPDRQAACRCIKSAAA----GITGINFGLASGLPGKCGVNIPYKISPNTDCN 117
Query: 124 RY 125
R+
Sbjct: 118 RF 119
>gb|AAG29777.1| lipid transfer protein 3 precursor [Gossypium hirsutum]
Length = 120
Score = 128 bits (321), Expect = 4e-29
Identities = 60/120 (50%), Positives = 82/120 (68%), Gaps = 5/120 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M+ +L C+ ++C+V P A+ AVT CG V SL PC++Y+ G CC+GI+
Sbjct: 3 SSMSLKLACVVVLCMVVGAPLAQGAVT-CGQVTNSLAPCINYLRGSGAGAVPPGCCSGIK 61
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ AQTT DR+A C CIK+A + T +N LA+GLP KCGVNIPY+ISP+TDCN
Sbjct: 62 SLNSAAQTTPDRQAACRCIKSAAA----GITGINFGLASGLPGKCGVNIPYKISPSTDCN 117
>gb|AAQ96338.1| lipid transfer protein [Vitis aestivalis]
Length = 119
Score = 125 bits (315), Expect = 2e-28
Identities = 57/121 (47%), Positives = 85/121 (70%), Gaps = 6/121 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+ A +L C+ ++C+V P A A +CG V ++L PC+SY+ GG PA CC+GI+
Sbjct: 3 SSGAVKLACVMVICMVMAAPAAVEAAITCGQVASALSPCISYLQKGGAVPPA--CCSGIK 60
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ A+TT DR+A C C+KN S + + +NL+LA+GLP KCGV++PY+ISP+TDC
Sbjct: 61 SLNSSAKTTADRQAACKCLKNFSS----TVSGINLSLASGLPGKCGVSVPYKISPSTDCT 116
Query: 124 R 124
+
Sbjct: 117 K 117
>gb|AAN77147.1| fiber lipid transfer protein [Gossypium barbadense]
Length = 120
Score = 125 bits (315), Expect = 2e-28
Identities = 62/121 (51%), Positives = 84/121 (69%), Gaps = 7/121 (5%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIM-NGGNTVPAAQCCNGI 62
S+M+ +L C+A++C+V P A AVT CG V +SL PC+ Y+ NG VP CC GI
Sbjct: 3 SSMSLKLACVAVLCMVVGAPLAHGAVT-CGQVTSSLAPCIGYLTGNGAGGVPPG-CCGGI 60
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
++LN+ AQTT DR+A C CIK+A + + +N +A+GLP KCGVNIPY+ISP+TDC
Sbjct: 61 KSLNSAAQTTPDRQAACKCIKSAAA----GISGINYGIASGLPGKCGVNIPYKISPSTDC 116
Query: 123 N 123
N
Sbjct: 117 N 117
>gb|AAF35185.1| lipid transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 124 bits (310), Expect = 7e-28
Identities = 58/120 (48%), Positives = 86/120 (71%), Gaps = 7/120 (5%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIM-NGGNTVPAAQCCNGI 62
S+M+ +L C+ + C+V P A+ A+ SCG + ++L PC++Y+ NG + P A CCNGI
Sbjct: 3 SSMSLKLTCVVVFCMVVGAPLAQGAI-SCGQITSALAPCIAYLKGNGAGSAPPA-CCNGI 60
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
R+LN+ A+TT DR+A C+CIK+A + + +N + AAGLP KCG+NIPY+ISP+TDC
Sbjct: 61 RSLNSAAKTTPDRQAACSCIKSAAT----GISGINYSTAAGLPGKCGINIPYKISPSTDC 116
>gb|AAC00499.1| lipid transfer protein precursor [Gossypium hirsutum]
gi|7438303|pir||T09790 lipid transfer protein precursor
- upland cotton
Length = 120
Score = 124 bits (310), Expect = 7e-28
Identities = 61/121 (50%), Positives = 84/121 (69%), Gaps = 7/121 (5%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIM-NGGNTVPAAQCCNGI 62
S+M+ + C+A++C+V P A+ AVT CG V +SL PC+ Y+ NG VP CC GI
Sbjct: 3 SSMSLKPACVAVLCMVVGAPLAQGAVT-CGQVTSSLAPCIGYLTGNGAGGVPPG-CCGGI 60
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
++LN+ AQTT DR+A C CIK+A + + +N +A+GLP KCGVNIPY+ISP+TDC
Sbjct: 61 KSLNSAAQTTPDRQAACKCIKSAAA----GISGINYGIASGLPGKCGVNIPYKISPSTDC 116
Query: 123 N 123
N
Sbjct: 117 N 117
>gb|AAR90329.1| lipid transfer protein precursor [Gossypium barbadense]
Length = 120
Score = 124 bits (310), Expect = 7e-28
Identities = 61/121 (50%), Positives = 84/121 (69%), Gaps = 7/121 (5%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIM-NGGNTVPAAQCCNGI 62
S+M+ +L C+A++C+V P A+ AVT CG V +SL PC+ Y+ NG VP CC GI
Sbjct: 3 SSMSLKLACVAVLCMVVGAPLAQGAVT-CGQVTSSLAPCIGYLTGNGAGGVPPG-CCGGI 60
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
++LN+ AQTT DR+A C CIK+A + + +N +A+G P KCGVNIPY+ISP+TDC
Sbjct: 61 KSLNSAAQTTPDRQAACKCIKSAAA----GISGINYGIASGPPGKCGVNIPYKISPSTDC 116
Query: 123 N 123
N
Sbjct: 117 N 117
>gb|AAF35184.1| lipid transfer protein precursor [Gossypium hirsutum]
gi|11273807|pir||T51144 lipid transfer protein precursor
[imported] - upland cotton
Length = 120
Score = 123 bits (308), Expect = 1e-27
Identities = 60/122 (49%), Positives = 84/122 (68%), Gaps = 7/122 (5%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIM-NGGNTVPAAQCCNGI 62
S+M+ +L C+ ++C+V P A+ VT CG V SL PC++Y+ NG VP CC+GI
Sbjct: 3 SSMSLKLACVVVLCMVVGAPLAQGTVT-CGQVTGSLAPCINYLRGNGAGAVPQG-CCSGI 60
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
++LN+ AQTT DR+A C CIK+A + +N +A+GLP KCGVNIPY+ISP+TDC
Sbjct: 61 KSLNSAAQTTPDRQAACKCIKSAAA----GIPGINYGIASGLPGKCGVNIPYKISPSTDC 116
Query: 123 NR 124
+R
Sbjct: 117 SR 118
>gb|AAB34774.1| LTP [Gossypium hirsutum] gi|66271043|gb|AAY43799.1| NSLTP1
[Gossypium hirsutum] gi|7438304|pir||T10812 lipid
transfer protein - upland cotton
gi|2497743|sp|Q43129|NLT2_GOSHI NONSPECIFIC
LIPID-TRANSFER PROTEIN PRECURSOR (LTP) (GH3)
Length = 120
Score = 122 bits (306), Expect = 2e-27
Identities = 59/120 (49%), Positives = 80/120 (66%), Gaps = 5/120 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M+ +L C+ ++C+V P A+ AVTS G V SL PC++Y+ G CC GI+
Sbjct: 3 SSMSLKLACVVVLCMVVGAPLAQGAVTS-GQVTNSLAPCINYLRGSGAGAVPPGCCTGIK 61
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ AQTT R+A C CIK+A + T +N LA+GLP KCGVNIPY+ISP+TDCN
Sbjct: 62 SLNSAAQTTPVRQAACRCIKSAAA----GITGINFGLASGLPGKCGVNIPYKISPSTDCN 117
>gb|AAF35186.1| lipid transfer protein precursor [Gossypium hirsutum]
Length = 120
Score = 122 bits (305), Expect = 3e-27
Identities = 58/120 (48%), Positives = 81/120 (67%), Gaps = 5/120 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M +L C+ ++C+V P A+ AVT CG V +SL PC++Y+ G CC+GI+
Sbjct: 3 SSMYLKLACVVVLCMVVGAPLAQGAVT-CGQVTSSLAPCINYLRGTGAGAIPPGCCSGIK 61
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ AQTT DR+A C CIK+A + +N LA+GLP KCGVNIPY+ISP+TDC+
Sbjct: 62 SLNSAAQTTPDRQAACKCIKSAAA----GIPGINFGLASGLPGKCGVNIPYKISPSTDCS 117
>gb|AAO33394.1| lipid transfer protein isoform 4 [Vitis vinifera]
Length = 119
Score = 121 bits (304), Expect = 3e-27
Identities = 56/121 (46%), Positives = 83/121 (68%), Gaps = 6/121 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+ A +L C+ ++C+V P A +CG V ++L PC+SY+ GG VPA CC+GI+
Sbjct: 3 SSGAVKLACVMVICMVVAAPAVVEATVTCGQVASALSPCISYLQKGG-AVPAG-CCSGIK 60
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ A+TT DR+A C C+K S S + +N LA+GLP KCGV++PY+ISP+TDC+
Sbjct: 61 SLNSAAKTTGDRQAACKCLKTFSS----SVSGINYGLASGLPGKCGVSVPYKISPSTDCS 116
Query: 124 R 124
+
Sbjct: 117 K 117
>gb|AAO33357.1| nonspecific lipid transfer protein 1 [Vitis berlandieri x Vitis
vinifera]
Length = 119
Score = 120 bits (302), Expect = 6e-27
Identities = 56/121 (46%), Positives = 81/121 (66%), Gaps = 6/121 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+ A +L C+ ++C+V P A A +CG V ++L C+ Y+ NGG P CC+GI+
Sbjct: 3 SSGAVKLACVMVICMVVAAPAAVEAAITCGQVSSALSSCLGYLKNGGAVPPG--CCSGIK 60
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
NLN+ AQTT DR+A C C+K + S +NL LA+GLP KCGV++PY+ISP+TDC+
Sbjct: 61 NLNSAAQTTADRQAACKCLKTFSN----SIPGINLGLASGLPGKCGVSVPYKISPSTDCS 116
Query: 124 R 124
+
Sbjct: 117 K 117
>gb|AAA75599.1| nonspecific lipid transfer protein precursor
gi|2497742|sp|Q42762|NLT1_GOSHI NONSPECIFIC
LIPID-TRANSFER PROTEIN PRECURSOR (LTP)
Length = 116
Score = 120 bits (301), Expect = 7e-27
Identities = 58/118 (49%), Positives = 78/118 (65%), Gaps = 5/118 (4%)
Query: 6 MATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNL 65
M+ +L C+ ++C+V P A+ AVTS G V SL PC++Y+ G CC GI++L
Sbjct: 1 MSLKLACVVVLCMVVGAPLAQGAVTS-GQVTNSLAPCINYLRGSGAGAVPPGCCTGIKSL 59
Query: 66 NTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
N+ AQTT R+A C CIK+A + T +N LA+GLP KCGVNIPY+ISP+TDCN
Sbjct: 60 NSAAQTTPVRQAACRCIKSAAA----GITGINFGLASGLPGKCGVNIPYKISPSTDCN 113
>gb|AAL27855.1| lipid transfer protein precursor [Davidia involucrata]
Length = 120
Score = 119 bits (299), Expect = 1e-26
Identities = 60/125 (48%), Positives = 85/125 (68%), Gaps = 8/125 (6%)
Query: 1 MGYSTMATRLVCLAIV-CLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCC 59
MG S + ++ +A++ C+V P AEAA+T CG V SL PC++Y+ GG PA CC
Sbjct: 1 MGRSGVVIKVGIVAVLMCMVVSAPHAEAAIT-CGTVTVSLAPCLNYLKKGGPVPPA--CC 57
Query: 60 NGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPN 119
NGI++LN A+TT DR+A C C+K A + S +NL+ A+ LP KCGVN+PY+ISP+
Sbjct: 58 NGIKSLNAAAKTTADRQAACNCLKTAST----SIAGINLSYASSLPGKCGVNVPYKISPS 113
Query: 120 TDCNR 124
TDC++
Sbjct: 114 TDCSK 118
>dbj|BAD95164.1| putative lipid transfer protein [Arabidopsis thaliana]
gi|4115360|gb|AAD03362.1| putative lipid transfer
protein [Arabidopsis thaliana]
gi|13272391|gb|AAK17134.1| putative lipid transfer
protein [Arabidopsis thaliana]
gi|15226046|ref|NP_179109.1| lipid transfer protein,
putative [Arabidopsis thaliana] gi|25294078|pir||D84524
probable lipid transfer protein [imported] - Arabidopsis
thaliana
Length = 123
Score = 119 bits (298), Expect = 2e-26
Identities = 60/119 (50%), Positives = 71/119 (59%), Gaps = 5/119 (4%)
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
+L CL V ++ GP A SCG V ++L PC Y+ NGG T P QCCNG+R LN M
Sbjct: 6 KLGCLVFVFVIAAGPITAKAALSCGEVNSNLKPCTGYLTNGGITSPGPQCCNGVRKLNGM 65
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPY--QISPNTDCNRY 125
TT DRR C CIKNA G LN + AAG+PR+CG+ IPY QI NT CN Y
Sbjct: 66 VLTTLDRRQACRCIKNAARNVG---PGLNADRAAGIPRRCGIKIPYSTQIRFNTKCNTY 121
>gb|AAY83346.1| non-specific lipid transfer protein precursor [Fragaria x ananassa]
Length = 117
Score = 119 bits (297), Expect = 2e-26
Identities = 59/115 (51%), Positives = 81/115 (70%), Gaps = 7/115 (6%)
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
+L +A++C+V P A+AA+T CG V ++ PC +Y+ +GG VPAA CC G+ NLN+M
Sbjct: 7 KLALVALICIVVAVPVAQAAIT-CGQVTHNVAPCFNYVKSGG-AVPAA-CCKGVSNLNSM 63
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
A+TT DR+ C C+K+A S LN NLAAGLP KCGVN+PY+IS +T+CN
Sbjct: 64 AKTTADRQQTCNCLKSAAG----SIKGLNANLAAGLPGKCGVNVPYKISTSTNCN 114
>gb|AAO33393.1| lipid transfer protein isoform 1 [Vitis vinifera]
Length = 119
Score = 117 bits (294), Expect = 5e-26
Identities = 57/121 (47%), Positives = 84/121 (69%), Gaps = 6/121 (4%)
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+ A +L C+ ++C+V P A A +CG V +++ PC SY+ GG+ VPA CC+GI+
Sbjct: 3 SSGAVKLACVMVICMVVAAPAAVEATITCGQVASAVGPCASYLQKGGS-VPAG-CCSGIK 60
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN+ A+TT DR+A C C+K SG S +N LA+GLP KCGV++PY+ISP+TDC+
Sbjct: 61 SLNSAAKTTVDRQAACKCLKTF---SG-SIPGINFGLASGLPGKCGVSVPYKISPSTDCS 116
Query: 124 R 124
+
Sbjct: 117 K 117
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 209,117,513
Number of Sequences: 2540612
Number of extensions: 7064389
Number of successful extensions: 15037
Number of sequences better than 10.0: 320
Number of HSP's better than 10.0 without gapping: 240
Number of HSP's successfully gapped in prelim test: 80
Number of HSP's that attempted gapping in prelim test: 14332
Number of HSP's gapped (non-prelim): 341
length of query: 127
length of database: 863,360,394
effective HSP length: 103
effective length of query: 24
effective length of database: 601,677,358
effective search space: 14440256592
effective search space used: 14440256592
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Medicago: description of AC147537.13


