

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
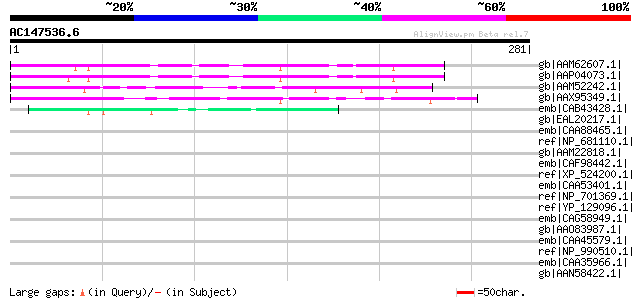
Query= AC147536.6 + phase: 0
(281 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM62607.1| unknown [Arabidopsis thaliana] 152 1e-35
gb|AAP04073.1| unknown protein [Arabidopsis thaliana] gi|8885541... 152 1e-35
gb|AAM52242.1| At2g39560/F12L6.22 [Arabidopsis thaliana] gi|3355... 135 1e-30
gb|AAX95349.1| conserved hypothetical protein [Oryza sativa (jap... 124 2e-27
emb|CAB43428.1| putative protein [Arabidopsis thaliana] gi|21537... 47 8e-04
gb|EAL20217.1| hypothetical protein CNBF0290 [Cryptococcus neofo... 38 0.37
emb|CAA88465.1| Hypothetical protein F27E5.3 [Caenorhabditis ele... 36 1.1
ref|NP_681110.1| hypothetical protein tlr0320 [Thermosynechococc... 36 1.1
gb|AAM22818.1| hv80H14.14a [Hordeum vulgare] gi|20513856|gb|AAM2... 35 1.8
emb|CAF98442.1| unnamed protein product [Tetraodon nigroviridis] 35 1.8
ref|XP_524200.1| PREDICTED: similar to RPB5-mediating protein is... 35 2.4
emb|CAA53401.1| A881; Hsp70 homologue [Saccharomyces cerevisiae]... 34 4.1
ref|NP_701369.1| erythrocyte membrane protein 1 (PfEMP1) [Plasmo... 34 4.1
ref|YP_129096.1| putative DNA methylase HsdM [Photobacterium pro... 34 4.1
emb|CAG58949.1| unnamed protein product [Candida glabrata CBS138... 34 4.1
gb|AAO83987.1| structural glycoprotein [Yellow head virus] 33 6.9
emb|CAA45579.1| BEN glycoprotein precursor [Gallus gallus] 33 9.1
ref|NP_990510.1| SC1 protein [Gallus gallus] gi|238001|gb|AAB201... 33 9.1
emb|CAA35966.1| ARS-binding protein 1 [Saccharomyces cerevisiae]... 33 9.1
gb|AAN58422.1| hypothetical protein SMU.689 [Streptococcus mutan... 33 9.1
>gb|AAM62607.1| unknown [Arabidopsis thaliana]
Length = 287
Score = 152 bits (384), Expect = 1e-35
Identities = 106/249 (42%), Positives = 146/249 (58%), Gaps = 33/249 (13%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLLWWKK---KIIKREI----ENDYGNGNPLKE 53
MK++S +G+GLS++FG LLLALV EVYYLL KK ++I +E E + KE
Sbjct: 1 MKTISGLGIGLSLMFGFLLLALVGEVYYLLRCKKHKNRVISQESEEEKEEEQQQNGYAKE 60
Query: 54 LFYMFCWKRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADY 113
L +FC+K+P + + G E + +N++ D ++ K G+ +A+
Sbjct: 61 LIQLFCFKKPQSLQQNNGGREGE----VSMNEDGNPDLELGLMKHL---NGGDLGFEAEL 113
Query: 114 MMQHHHDGLLGHPRFLFTIVEESKEDLESEDG---KCGKDSRGRSLGDL-LDVETPYLTP 169
M H+ RFLFTI+EE+K DLESEDG + G SR RSL D+ D TP TP
Sbjct: 114 MKLHNQ-------RFLFTIMEETKADLESEDGGKSRLGSRSRRRSLSDVPNDCNTPGFTP 166
Query: 170 IASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFN---RLKSSPPPKFKFLQEAEEKL 226
+ASP + SP S Y HGFNPLFES + EFN R SSPPPKFKF+++AEEKL
Sbjct: 167 LASP----KKSSSPLES-YPHHGFNPLFESDGELEFNKFFRSSSSPPPKFKFMRDAEEKL 221
Query: 227 RRKMQDDNK 235
R+++ ++ K
Sbjct: 222 RKRLIEEAK 230
>gb|AAP04073.1| unknown protein [Arabidopsis thaliana] gi|8885541|dbj|BAA97471.1|
unnamed protein product [Arabidopsis thaliana]
gi|28973705|gb|AAO64169.1| unknown protein [Arabidopsis
thaliana] gi|15238384|ref|NP_200743.1| expressed protein
[Arabidopsis thaliana]
Length = 287
Score = 152 bits (384), Expect = 1e-35
Identities = 106/249 (42%), Positives = 146/249 (58%), Gaps = 33/249 (13%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLL---WWKKKIIKREI----ENDYGNGNPLKE 53
MK++S +G+GLS++FG LLLALV EVYYLL KK++I +E E + KE
Sbjct: 1 MKTISGLGIGLSLMFGFLLLALVGEVYYLLRCKKHKKRVISQESEEEKEEEQQQNGYAKE 60
Query: 54 LFYMFCWKRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADY 113
L +FC+K+P + + G E + +N++ D ++ K G+ +A+
Sbjct: 61 LIQLFCFKKPQSLQQNNGGREGE----VSMNEDGNPDLELGLMKHL---NGGDLGFEAEL 113
Query: 114 MMQHHHDGLLGHPRFLFTIVEESKEDLESEDG---KCGKDSRGRSLGDL-LDVETPYLTP 169
M H+ RFLFTI+EE+K DLESEDG + G SR RSL D+ D TP TP
Sbjct: 114 MKLHNQ-------RFLFTIMEETKADLESEDGGKSRLGSRSRRRSLSDVPNDCNTPGFTP 166
Query: 170 IASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFN---RLKSSPPPKFKFLQEAEEKL 226
+ASP + SP S Y HGFNPLFES + EFN R SSPPPKFKF+++AEEKL
Sbjct: 167 LASP----KKSSSPLES-YPHHGFNPLFESDGELEFNKFFRSSSSPPPKFKFMRDAEEKL 221
Query: 227 RRKMQDDNK 235
R+++ ++ K
Sbjct: 222 RKRLIEEAK 230
>gb|AAM52242.1| At2g39560/F12L6.22 [Arabidopsis thaliana] gi|3355485|gb|AAC27847.1|
hypothetical protein [Arabidopsis thaliana]
gi|18700123|gb|AAL77673.1| At2g39560/F12L6.22
[Arabidopsis thaliana] gi|15225496|ref|NP_181488.1|
expressed protein [Arabidopsis thaliana]
gi|7485412|pir||T00566 hypothetical protein At2g39560
[imported] - Arabidopsis thaliana
Length = 233
Score = 135 bits (341), Expect = 1e-30
Identities = 106/251 (42%), Positives = 139/251 (55%), Gaps = 48/251 (19%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLLWWKKKIIKR--EIENDYGNGNPLKELFYMF 58
M+SLSSVGL LSIVFGCLLLAL+AE+YYLLW KK+ R + NDY +EL ++F
Sbjct: 1 MRSLSSVGLALSIVFGCLLLALLAELYYLLWCKKRSTTRRPDFRNDYSTPG-TRELLFIF 59
Query: 59 CWKRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMMQHH 118
C S+S + +P SS ++ D Q Q F+ G
Sbjct: 60 CC---SSSTNPSSSSP----SSSSFSNPKPIDTQQQCPLNNGFENVG------------- 99
Query: 119 HDGLLGHPRFLFTIVEESKEDLESEDGKCGKDSRGRSLGDL-LDVET-----PYLTPIAS 172
GL+ PRFLFTI+EE+ E++ESED ++G+SL DL L++E+ PYLTP AS
Sbjct: 100 GPGLV--PRFLFTIMEETVEEMESED---VVSTKGKSLNDLFLNMESGVITPPYLTPRAS 154
Query: 173 PHFFTPMNCSPYCSPYN--QHGFNPLFESTTDAEFNRL------------KSSPPPKFKF 218
P FTP N + + LFES++DAEFNRL SSP +FKF
Sbjct: 155 PSLFTPPLTPLLMESCNGRKEEISSLFESSSDAEFNRLVRSSPLSSSHSPSSSPLSRFKF 214
Query: 219 LQEAEEKLRRK 229
L++AEEKL +K
Sbjct: 215 LRDAEEKLYKK 225
>gb|AAX95349.1| conserved hypothetical protein [Oryza sativa (japonica
cultivar-group)]
Length = 247
Score = 124 bits (312), Expect = 2e-27
Identities = 99/268 (36%), Positives = 137/268 (50%), Gaps = 55/268 (20%)
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLLWWKKKIIKREIENDYGNGNPLKELFYMFCW 60
M++LS VG+GL++V LLLAL AE+YYL +K++ + + +EL +FC+
Sbjct: 1 MRALSRVGVGLAVVSALLLLALAAELYYLFVYKRRRSAAISDAASSPSSSSRELLQLFCF 60
Query: 61 KRPSTSLRQTGFTPPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMMQHHHD 120
K+ PP L S+ + +P + + +D ++A M
Sbjct: 61 KK-----------PPALAST------YAQEPHAGEAVVAVAVDDDDDTVEAQLM---RLG 100
Query: 121 GLLGHPRFLFTIVEESKEDLESEDG--KCGKDSRGRSLGDLL-DVETPYLTPIASPHFFT 177
L+G R LFTI EE+KEDLESEDG +CG R RSL +LL ETP++TP +SP
Sbjct: 101 SLVGPTRLLFTIKEETKEDLESEDGRSRCG---RSRSLAELLHSSETPFMTPASSP---L 154
Query: 178 PMNCSPYCSPYNQHGFNPLFESTTDAEFNRLKSSPPPKFKFLQEAEEKL----------- 226
PM+ S FNPLFE T A + SPPPKF+FL++AEEK+
Sbjct: 155 PMDKS----------FNPLFEPTVAA---AVTVSPPPKFQFLKDAEEKMYRRALAEEAMR 201
Query: 227 -RRKMQDDNKGINGNEVDNSLITIIVDK 253
RR Q + G E D ITI+V K
Sbjct: 202 ARRSPQTRSPAAAGEE-DGGYITIMVGK 228
>emb|CAB43428.1| putative protein [Arabidopsis thaliana] gi|21537012|gb|AAM61353.1|
unknown [Arabidopsis thaliana]
gi|15231230|ref|NP_190816.1| expressed protein
[Arabidopsis thaliana] gi|7460044|pir||T08451
hypothetical protein F22O6.140 - Arabidopsis thaliana
Length = 209
Score = 46.6 bits (109), Expect = 8e-04
Identities = 54/193 (27%), Positives = 78/193 (39%), Gaps = 38/193 (19%)
Query: 11 LSIVFGCLLLALVAEVYYLLWWKKKIIKREI---ENDYGNGN---------PLKELFYMF 58
L VF L+A+ A+ Y+LWWK++ +R I E D + P KEL Y F
Sbjct: 10 LVAVFAFCLVAVTAQFAYVLWWKRRFRRRSIAGSERDAFSSRGGDLTATPPPSKELLYFF 69
Query: 59 CWKRPSTSLRQTGFTPP----------ELCSSMRIN-DNFVHDPQVQTSKEFLFKPYGED 107
+ + R T P ++ S IN +N + P E LF
Sbjct: 70 LFCLENKQFRIGSATAPPLPAAAPPVNDVASKWSINGENLLCGP-----SETLF------ 118
Query: 108 VIDADYMMQ-HHHDGLLGHPRFLFTIVEESKEDLESEDGKCGKDSRGRSLG-DLLDVETP 165
I DY + H G + +FT E+ +D E E+ D + + TP
Sbjct: 119 TIAEDYTSESDHRTGEIDPRGSIFT--EDHVKDDEVEEEVVATDISDDEVDFSHYNQTTP 176
Query: 166 YLTPIASPHFFTP 178
+ TP ASP F+TP
Sbjct: 177 FSTPCASPPFYTP 189
>gb|EAL20217.1| hypothetical protein CNBF0290 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 363
Score = 37.7 bits (86), Expect = 0.37
Identities = 34/106 (32%), Positives = 43/106 (40%), Gaps = 7/106 (6%)
Query: 113 YMMQHHHDGLLGHPRFLFTIVEESKEDL-ESEDGKCGKDSRGRSLGDLLDVETPYLTPIA 171
YM+ H LG V + DL SED D+ G G +LDV P P
Sbjct: 251 YMLTESHLHTLGSLPEGAVFVNIGRGDLIRSEDILAALDAEGGLFGAVLDVTDPEPLPSG 310
Query: 172 SPHFFTPMNCSPYCSPYNQHGFNPLFESTTD---AEFNRLKSSPPP 214
P F P S +P+ F F+S D A+ +RLK PP
Sbjct: 311 HPLFTHP---SVIVTPHTSGSFEGYFDSGADVLLAQGDRLKQDLPP 353
>emb|CAA88465.1| Hypothetical protein F27E5.3 [Caenorhabditis elegans]
gi|17533501|ref|NP_496188.1| predicted CDS, putative
nuclear protein, with a coiled coil-4 domain (2K428)
[Caenorhabditis elegans] gi|7500016|pir||T21457
hypothetical protein F27E5.3 - Caenorhabditis elegans
Length = 490
Score = 36.2 bits (82), Expect = 1.1
Identities = 33/98 (33%), Positives = 47/98 (47%), Gaps = 16/98 (16%)
Query: 136 SKEDLESEDGKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNP 195
S+ DL+ D + + + + D ET LTPI P PY P +Q G P
Sbjct: 193 SQSDLKKHDSRFAYLPKNQRPRQVSDDETQKLTPIFLP--------QPY-KPSSQQGPPP 243
Query: 196 LFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDD 233
+ T F+ K+SPP K L+E+ EK +KM+DD
Sbjct: 244 ML---TRQPFHAQKTSPPK--KPLEESPEK--KKMKDD 274
>ref|NP_681110.1| hypothetical protein tlr0320 [Thermosynechococcus elongatus BP-1]
gi|22294041|dbj|BAC07872.1| tlr0320 [Thermosynechococcus
elongatus BP-1]
Length = 895
Score = 36.2 bits (82), Expect = 1.1
Identities = 23/71 (32%), Positives = 37/71 (51%), Gaps = 1/71 (1%)
Query: 208 LKSSPPPKFKFLQEAEEKLRRKMQDDNK-GINGNEVDNSLITIIVDKKYEREVNHHQCHL 266
LK PPP K++QE EEKLR ++ N N +V +TI + + + ++ H Q +
Sbjct: 601 LKQLPPPDQKYVQELEEKLREFIEFINAIDCNCADVAEQGMTIRQEPQKDTKIIHFQSII 660
Query: 267 QQYHSSTSQVL 277
Q S+V+
Sbjct: 661 QDIQKRLSEVV 671
>gb|AAM22818.1| hv80H14.14a [Hordeum vulgare] gi|20513856|gb|AAM22817.1|
HV80H14.14B [Hordeum vulgare]
Length = 254
Score = 35.4 bits (80), Expect = 1.8
Identities = 18/65 (27%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 74 PPELCSSMRINDNFVHDPQVQTSKEFLFKPYGED--VIDADYMMQHHHDGLLGHPRFLFT 131
P E C+ M + ++ HDP + LF E +++ + H GLL ++LF
Sbjct: 107 PMESCADMPLTPSYAHDPSFAAASATLFLTLEERKVILEEKKLAMEEHTGLLKWEKYLFF 166
Query: 132 IVEES 136
I++ S
Sbjct: 167 IMDTS 171
>emb|CAF98442.1| unnamed protein product [Tetraodon nigroviridis]
Length = 603
Score = 35.4 bits (80), Expect = 1.8
Identities = 17/74 (22%), Positives = 42/74 (55%), Gaps = 6/74 (8%)
Query: 194 NPLFESTTDAEFNR-LKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLITIIVD 252
N +F + DA+F ++S+P PK ++ +++ R + D K +E+ + ++ +++
Sbjct: 133 NCVFAAGEDAQFTCVIQSAPSPKIRWFKDS-----RLLTDQEKYQTYSELRSGVLVLVIK 187
Query: 253 KKYEREVNHHQCHL 266
ER++ H++C +
Sbjct: 188 NLTERDLGHYECEV 201
>ref|XP_524200.1| PREDICTED: similar to RPB5-mediating protein isoform a; RNA
polymerase II, subunit 5-mediating protein;
unconventional prefoldin RPB5 interactor [Pan
troglodytes]
Length = 393
Score = 35.0 bits (79), Expect = 2.4
Identities = 30/146 (20%), Positives = 57/146 (38%), Gaps = 13/146 (8%)
Query: 102 KPYGEDVIDADYMMQHHHDGLLGHPRFL-FTIVEESKEDLESEDGKCGKDSRGRSLGDLL 160
KP D+ +AD LL T++ ++ SE+ K +++ ++ +
Sbjct: 102 KPKTSDIFEADIANDVKSKDLLADKELKPDTVIANGEDTTSSEEEKEDRNTNVNAMHQVT 161
Query: 161 DVETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNPL----FESTTDAEFNRLKSSPPPKF 216
D TP +AS P+ N+ G N + F T + + R+ +
Sbjct: 162 DSHTPCHKDVAS--------SEPFSGQVNRVGDNSIPTIYFSHTVEPKRVRINTGKNTTL 213
Query: 217 KFLQEAEEKLRRKMQDDNKGINGNEV 242
KF ++ EE R++ G + E+
Sbjct: 214 KFSEKKEEAKRKRKNSTGSGHSAQEL 239
>emb|CAA53401.1| A881; Hsp70 homologue [Saccharomyces cerevisiae]
gi|6322777|ref|NP_012850.1| Molecular chaperone of the
endoplasmic reticulum lumen, involved in polypeptide
translocation and folding; member of the Hsp70 family;
localizes to the lumen of the ER; regulated by the
unfolded protein response pathway; Lhs1p [Saccharomyces
cerevisiae] gi|486103|emb|CAA81910.1| LHS1
[Saccharomyces cerevisiae]
gi|549693|sp|P36016|LHS1_YEAST Heat shock protein 70
homolog LHS1 precursor gi|1587570|prf||2206496B ORF
Length = 881
Score = 34.3 bits (77), Expect = 4.1
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query: 196 LFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLITIIVDK 253
L +S ++ N +K F+ + EEKL+RK++ + K N NE ++++I DK
Sbjct: 801 LIKSGDESRLNEIKKLHLRNFRLQKRKEEKLKRKLEQE-KSRNNNETESTVINSADDK 857
>ref|NP_701369.1| erythrocyte membrane protein 1 (PfEMP1) [Plasmodium falciparum 3D7]
gi|23496535|gb|AAN36093.1| erythrocyte membrane protein 1
(PfEMP1) [Plasmodium falciparum 3D7]
Length = 2860
Score = 34.3 bits (77), Expect = 4.1
Identities = 30/118 (25%), Positives = 50/118 (41%), Gaps = 6/118 (5%)
Query: 133 VEESKEDLESEDGKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNC---SPYCSPYN 189
+++ +E +S K K S + D L TPY T H MNC + +C
Sbjct: 1136 LQKLQEANKSSASKRSKRSTDGTTTDTLTPTTPYSTAEGYVHQEATMNCDTQTQFCEK-K 1194
Query: 190 QHGFNPLFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNKGINGNEVDNSLI 247
G P + TDA + PPP++K E + K + + + + + + NSL+
Sbjct: 1195 HGGTTPTGTNDTDAPYT--FKQPPPEYKDACECDGKSPQAPKKEEEKKDACTIVNSLL 1250
>ref|YP_129096.1| putative DNA methylase HsdM [Photobacterium profundum SS9]
gi|46912502|emb|CAG19294.1| putative DNA methylase HsdM
[Photobacterium profundum SS9]
Length = 793
Score = 34.3 bits (77), Expect = 4.1
Identities = 41/185 (22%), Positives = 80/185 (43%), Gaps = 40/185 (21%)
Query: 79 SSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMMQHHHDGLLGHPRFLFTIVEESKE 138
SS RI + D ++ E+++ YGED A+ + + + + EE +
Sbjct: 519 SSERI-ETLRFDKALREPMEYIYNTYGEDAYKAEILAK--------ESKAILAWCEEKEI 569
Query: 139 DLESED-GKCGKDSRGRSLGDLLDVETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNPLF 197
L +++ K + LGDL+D+ + I + + + YNQ F
Sbjct: 570 SLNTKNRNKLLDVATWTRLGDLIDIANTLMKAIGTDIY----------NDYNQ------F 613
Query: 198 ESTTDAEF--NRLKSSPPPKFKFL-------QEAEEKLRRKMQDDNKGINGNEVDNSLIT 248
++T DAE ++K S P K L + AE+ +++K++ + G+++D L +
Sbjct: 614 KATVDAELKSRKIKLSAPEKNAILNAVSWYHENAEKVIKKKLK-----LTGSKLDELLTS 668
Query: 249 IIVDK 253
D+
Sbjct: 669 CDCDE 673
>emb|CAG58949.1| unnamed protein product [Candida glabrata CBS138]
gi|50287191|ref|XP_446025.1| unnamed protein product
[Candida glabrata]
Length = 818
Score = 34.3 bits (77), Expect = 4.1
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 9/69 (13%)
Query: 200 TTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNK---------GINGNEVDNSLITII 250
+T+ R+ + PP + + +++ LR + D+N G NG+ DNS++ +
Sbjct: 576 STNGSSARISNKPPAEVVPSRHSQDFLRHTVPDNNSTIDNYTLRNGSNGDNSDNSVLYMA 635
Query: 251 VDKKYEREV 259
VDK+Y V
Sbjct: 636 VDKRYNGAV 644
>gb|AAO83987.1| structural glycoprotein [Yellow head virus]
Length = 1666
Score = 33.5 bits (75), Expect = 6.9
Identities = 17/42 (40%), Positives = 27/42 (63%), Gaps = 2/42 (4%)
Query: 74 PPELCSSMRINDNFVHDPQVQTSKEFLFKPYGEDVIDADYMM 115
PP+LC + RIN + +H + S E+LFKP G+ + DY++
Sbjct: 1211 PPKLCDA-RINWSCLHTGSCKNSSEYLFKPLGQHTSN-DYIV 1250
>emb|CAA45579.1| BEN glycoprotein precursor [Gallus gallus]
Length = 588
Score = 33.1 bits (74), Expect = 9.1
Identities = 20/45 (44%), Positives = 27/45 (59%), Gaps = 1/45 (2%)
Query: 11 LSIVFGCLLLALVAEVYYLLWWKK-KIIKREIENDYGNGNPLKEL 54
+ IV G LL+ALVA V Y L+ KK K + ++ D GN K+L
Sbjct: 535 VGIVVGLLLVALVAGVVYWLYVKKSKTASKHVDKDLGNIEENKKL 579
>ref|NP_990510.1| SC1 protein [Gallus gallus] gi|238001|gb|AAB20170.1| SC1 [Gallus
gallus] gi|1173371|sp|P42292|CD166_CHICK CD166 antigen
precursor (Activated leukocyte-cell adhesion molecule)
(SC1 glycoprotein) (BEN glycoprotein) (DM-GRASP protein)
(JC7 protein)
Length = 588
Score = 33.1 bits (74), Expect = 9.1
Identities = 20/45 (44%), Positives = 27/45 (59%), Gaps = 1/45 (2%)
Query: 11 LSIVFGCLLLALVAEVYYLLWWKK-KIIKREIENDYGNGNPLKEL 54
+ IV G LL+ALVA V Y L+ KK K + ++ D GN K+L
Sbjct: 535 VGIVVGLLLVALVAGVVYWLYVKKSKTASKHVDKDLGNIEENKKL 579
>emb|CAA35966.1| ARS-binding protein 1 [Saccharomyces cerevisiae]
gi|114814|sp|P14164|BAF1_YEAST Transcription factor BAF1
(ARS binding factor 1) (Protein ABF1) (Bidirectionally
acting factor) (SFB-B) (DNA replication enhancer-binding
protein OBF1) gi|170968|gb|AAA66311.1| ARS binding
factor 1 gi|172058|gb|AAA34823.1| OBF1
gi|226720|prf||1604243A ARS binding factor I
Length = 731
Score = 33.1 bits (74), Expect = 9.1
Identities = 13/42 (30%), Positives = 22/42 (51%)
Query: 238 NGNEVDNSLITIIVDKKYEREVNHHQCHLQQYHSSTSQVLPL 279
N ++VD+ ++ + V+ +Y + NHH H Q H V L
Sbjct: 668 NDDDVDDVMVDVDVESQYNKNTNHHNNHHSQPHHDEEDVAGL 709
>gb|AAN58422.1| hypothetical protein SMU.689 [Streptococcus mutans UA159]
gi|24379161|ref|NP_721116.1| hypothetical protein
SMU.689 [Streptococcus mutans UA159]
gi|62954735|dbj|BAD97687.1| autolysin [Streptococcus
mutans]
Length = 979
Score = 33.1 bits (74), Expect = 9.1
Identities = 14/51 (27%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query: 231 QDDNKGINGNEVDNSLITIIVD-KKYEREVNHHQCHLQQYHSSTSQVLPLA 280
QDD K N + D+ +++D K ++ ++ H++ H+ Y + T + LA
Sbjct: 310 QDDIKWYNARKADDGSYKVLIDTKNHKNDLGHYEAHIYGYSTVTQSQIGLA 360
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 528,566,485
Number of Sequences: 2540612
Number of extensions: 24013749
Number of successful extensions: 55623
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 55590
Number of HSP's gapped (non-prelim): 29
length of query: 281
length of database: 863,360,394
effective HSP length: 126
effective length of query: 155
effective length of database: 543,243,282
effective search space: 84202708710
effective search space used: 84202708710
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC147536.6


