

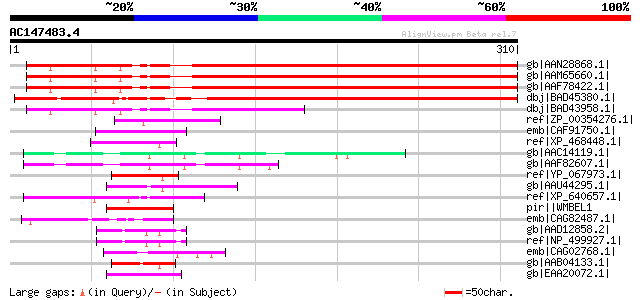
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147483.4 + phase: 0
(310 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN28868.1| At1g53640/F22G10.8 [Arabidopsis thaliana] gi|1840... 291 2e-77
gb|AAM65660.1| Contains similarity to RNA-binding protein from A... 291 2e-77
gb|AAF78422.1| Contains similarity to RNA-binding protein from A... 291 2e-77
dbj|BAD45380.1| hydroxyproline-rich glycoprotein-like [Oryza sat... 278 2e-73
dbj|BAD43958.1| unknown protein [Arabidopsis thaliana] gi|519705... 108 2e-22
ref|ZP_00354276.1| hypothetical protein Krad07002061 [Kineococcu... 52 2e-05
emb|CAF91750.1| unnamed protein product [Tetraodon nigroviridis] 52 3e-05
ref|XP_468448.1| putative fibrillarin [Oryza sativa (japonica cu... 51 4e-05
gb|AAC14119.1| AUT1 [Schistosoma mansoni] 51 5e-05
gb|AAF82607.1| AUT1 [Schistosoma mansoni] 50 6e-05
ref|YP_067973.1| EBNA-1 [Cercopithecine herpesvirus 15] gi|18025... 50 8e-05
gb|AAU44295.1| putative fibrillarin [Oryza sativa (japonica cult... 50 8e-05
ref|XP_640657.1| hypothetical protein DDB0205563 [Dictyostelium ... 50 8e-05
pir||WMBEL1 latency-related protein 1 - human herpesvirus 1 (str... 50 8e-05
emb|CAG82487.1| unnamed protein product [Yarrowia lipolytica CLI... 50 1e-04
gb|AAD12858.2| Hypothetical protein Y66H1A.4 [Caenorhabditis ele... 50 1e-04
ref|NP_499927.1| nucleolar protein family A member 1 (4B179) [Ca... 50 1e-04
emb|CAG02768.1| unnamed protein product [Tetraodon nigroviridis] 49 1e-04
gb|AAB04133.1| cutinase negative acting protein 49 1e-04
gb|EAA20072.1| multidomain scavenger receptor protein PbSR precu... 49 1e-04
>gb|AAN28868.1| At1g53640/F22G10.8 [Arabidopsis thaliana]
gi|18404554|ref|NP_564639.1| hydroxyproline-rich
glycoprotein family protein [Arabidopsis thaliana]
gi|16323139|gb|AAL15304.1| At1g53640/F22G10.8
[Arabidopsis thaliana] gi|12324041|gb|AAG51990.1|
unknown protein; 43598-45751 [Arabidopsis thaliana]
gi|25405656|pir||E96576 unknown protein, 43598-45751
[imported] - Arabidopsis thaliana
Length = 523
Score = 291 bits (745), Expect = 2e-77
Identities = 162/326 (49%), Positives = 205/326 (62%), Gaps = 44/326 (13%)
Query: 11 GRGRGKPLEEAAQ---------EAPQAPVVNRHIRVRQTPADAESDNVPR---------R 52
G GRGKPL E+A P P + ++ +Q A D P+ R
Sbjct: 216 GAGRGKPLVESAPIRQEDNRQIRRPPPPPQQQRVQPQQKRAPTVKDGTPKPQLSAEEAGR 275
Query: 53 QPMNRFVRDDGDGS------GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
+ + R + +GS GRGRGRGRG ARGRG RGRGG G R D +
Sbjct: 276 RARSELSRGEAEGSSVGGRGGRGRGRGRG----ARGRG-RGRGGDGWRDDKK-------- 322
Query: 107 DRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYV 166
++ A ++ GD+ADGEK A+K+GPE+M + E +EEI E+ LPS D +
Sbjct: 323 -----EEEGEQEAMRIFAGDSADGEKFAEKMGPELMKTLAEGFEEICEKALPSTTHDAII 377
Query: 167 EAMDINCAIEFEPEYAV-EF-DNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEV 224
+A D N IE EPEY + +F NPDIDEK P++LR+ LEK+KPF++ YEGI+ QEEWEE
Sbjct: 378 DAYDTNLMIECEPEYIMPDFGSNPDIDEKPPMSLRECLEKVKPFIVAYEGIKDQEEWEEA 437
Query: 225 IEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQ 284
I E M + PL+K+IVDHYSGPDRVTAKKQ EEL+R+A TLP SAP SVK F +RA ++L+
Sbjct: 438 INEAMTQAPLMKEIVDHYSGPDRVTAKKQNEELDRIATTLPASAPDSVKRFADRAALTLK 497
Query: 285 SNPGWGFDKKCQFMDKLVFEVSQHHK 310
SNPGWGFDKK QFMDKLV EVSQ +K
Sbjct: 498 SNPGWGFDKKYQFMDKLVLEVSQSYK 523
>gb|AAM65660.1| Contains similarity to RNA-binding protein from Arabidopsis
thaliana gi|2129727 and contains RNA recognition
PF|00076 domain
Length = 523
Score = 291 bits (745), Expect = 2e-77
Identities = 162/326 (49%), Positives = 205/326 (62%), Gaps = 44/326 (13%)
Query: 11 GRGRGKPLEEAAQ---------EAPQAPVVNRHIRVRQTPADAESDNVPR---------R 52
G GRGKPL E+A P P + ++ +Q A D P+ R
Sbjct: 216 GAGRGKPLVESAPIRQEDNRQIRRPPPPPQQQRVQPQQKRAPTVKDGTPKPQLSAEEAGR 275
Query: 53 QPMNRFVRDDGDGS------GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
+ + R + +GS GRGRGRGRG ARGRG RGRGG G R D +
Sbjct: 276 RARSELSRGEAEGSSVGGRGGRGRGRGRG----ARGRG-RGRGGDGWRDDKK-------- 322
Query: 107 DRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYV 166
++ A ++ GD+ADGEK A+K+GPE+M + E +EEI E+ LPS D +
Sbjct: 323 -----EEEGEQEAMRIFAGDSADGEKFAEKMGPELMKTLAEGFEEICEKALPSTTHDAII 377
Query: 167 EAMDINCAIEFEPEYAV-EF-DNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEV 224
+A D N IE EPEY + +F NPDIDEK P++LR+ LEK+KPF++ YEGI+ QEEWEE
Sbjct: 378 DAYDTNLMIECEPEYIMPDFGSNPDIDEKPPMSLRECLEKVKPFIVAYEGIKDQEEWEEA 437
Query: 225 IEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQ 284
I E M + PL+K+IVDHYSGPDRVTAKKQ EEL+R+A TLP SAP SVK F +RA ++L+
Sbjct: 438 INEAMTQAPLMKEIVDHYSGPDRVTAKKQNEELDRIATTLPASAPDSVKRFADRAALTLK 497
Query: 285 SNPGWGFDKKCQFMDKLVFEVSQHHK 310
SNPGWGFDKK QFMDKLV EVSQ +K
Sbjct: 498 SNPGWGFDKKYQFMDKLVLEVSQSYK 523
>gb|AAF78422.1| Contains similarity to RNA-binding protein from Arabidopsis
thaliana gi|2129727 and contains RNA recognition
PF|00076 domain. ESTs gb|H37317, gb|F14415, gb|AA651290
come from this gene
Length = 829
Score = 291 bits (745), Expect = 2e-77
Identities = 162/326 (49%), Positives = 205/326 (62%), Gaps = 44/326 (13%)
Query: 11 GRGRGKPLEEAAQ---------EAPQAPVVNRHIRVRQTPADAESDNVPR---------R 52
G GRGKPL E+A P P + ++ +Q A D P+ R
Sbjct: 522 GAGRGKPLVESAPIRQEDNRQIRRPPPPPQQQRVQPQQKRAPTVKDGTPKPQLSAEEAGR 581
Query: 53 QPMNRFVRDDGDGS------GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
+ + R + +GS GRGRGRGRG ARGRG RGRGG G R D +
Sbjct: 582 RARSELSRGEAEGSSVGGRGGRGRGRGRG----ARGRG-RGRGGDGWRDDKK-------- 628
Query: 107 DRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYV 166
++ A ++ GD+ADGEK A+K+GPE+M + E +EEI E+ LPS D +
Sbjct: 629 -----EEEGEQEAMRIFAGDSADGEKFAEKMGPELMKTLAEGFEEICEKALPSTTHDAII 683
Query: 167 EAMDINCAIEFEPEYAV-EF-DNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEV 224
+A D N IE EPEY + +F NPDIDEK P++LR+ LEK+KPF++ YEGI+ QEEWEE
Sbjct: 684 DAYDTNLMIECEPEYIMPDFGSNPDIDEKPPMSLRECLEKVKPFIVAYEGIKDQEEWEEA 743
Query: 225 IEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQ 284
I E M + PL+K+IVDHYSGPDRVTAKKQ EEL+R+A TLP SAP SVK F +RA ++L+
Sbjct: 744 INEAMTQAPLMKEIVDHYSGPDRVTAKKQNEELDRIATTLPASAPDSVKRFADRAALTLK 803
Query: 285 SNPGWGFDKKCQFMDKLVFEVSQHHK 310
SNPGWGFDKK QFMDKLV EVSQ +K
Sbjct: 804 SNPGWGFDKKYQFMDKLVLEVSQSYK 829
>dbj|BAD45380.1| hydroxyproline-rich glycoprotein-like [Oryza sativa (japonica
cultivar-group)]
Length = 436
Score = 278 bits (710), Expect = 2e-73
Identities = 154/320 (48%), Positives = 202/320 (63%), Gaps = 33/320 (10%)
Query: 4 PKVLPGGGRGRGKP-LEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDD 62
P+ LP G GRG P +++ E PQ NR IR R+ A S P + +D
Sbjct: 137 PRTLPSAGAGRGVPRMQQPPVEMPQEE--NRFIRRREEKKKAASAARPAPSGQPKLSPED 194
Query: 63 ------------GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTS 110
GD G GRG GRGR R RG RGRG GGRG +R D
Sbjct: 195 AVKRAMELLGGGGDDDG-GRG-GRGRGARGRERG-RGRGRDGGRG------RRSADMEEK 245
Query: 111 HQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMD 170
H G+Y+GDNADG++L K+LG + M EA++E + LP P QD Y+EA
Sbjct: 246 H---------GIYLGDNADGDRLQKRLGEDKMKIFNEAFDEAADNALPDPKQDAYLEACH 296
Query: 171 INCAIEFEPEYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEELMQ 230
N IEFEPEY V F+NPDI+EK P++L D L+K+KPF++ YEGI++QEEWEE ++++M
Sbjct: 297 TNNMIEFEPEYHVNFNNPDIEEKPPMSLEDMLQKVKPFIVAYEGIQNQEEWEEAVKDVMA 356
Query: 231 RVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQSNPGWG 290
R P +K+++D YSGPD VTAK+Q+EEL+RVA TLP + PSSVK FT++ ++SL++NPGWG
Sbjct: 357 RAPHMKELIDMYSGPDVVTAKQQEEELQRVANTLPGNIPSSVKRFTDKTLLSLKNNPGWG 416
Query: 291 FDKKCQFMDKLVFEVSQHHK 310
FDKKCQFMDK EVS+ +K
Sbjct: 417 FDKKCQFMDKFAREVSELYK 436
>dbj|BAD43958.1| unknown protein [Arabidopsis thaliana] gi|51970502|dbj|BAD43943.1|
unknown protein [Arabidopsis thaliana]
Length = 417
Score = 108 bits (270), Expect = 2e-22
Identities = 72/194 (37%), Positives = 96/194 (49%), Gaps = 42/194 (21%)
Query: 11 GRGRGKPLEEAAQ---------EAPQAPVVNRHIRVRQTPADAESDNVPR---------R 52
G GRGKPL E+A P P + ++ +Q A D P+ R
Sbjct: 216 GAGRGKPLVESAPIRQEDNRQIRRPPPPPQQQRVQPQQKRAPTVKDGTPKPQLSAEEAGR 275
Query: 53 QPMNRFVRDDGDGS------GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
+ + R + +GS GRGRGRGRG ARGRG RGRGG G R D +
Sbjct: 276 RARSELSRGEAEGSSVGGRGGRGRGRGRG----ARGRG-RGRGGDGWRDDKK-------- 322
Query: 107 DRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYV 166
++ A ++ GD+ADGEK A+K+GPE+M + E +EEI E+ LPS D +
Sbjct: 323 -----EEEGEQEAMRIFAGDSADGEKFAEKMGPELMKTLAEGFEEICEKALPSTTHDAII 377
Query: 167 EAMDINCAIEFEPE 180
+A D N IE EPE
Sbjct: 378 DAYDTNLMIECEPE 391
>ref|ZP_00354276.1| hypothetical protein Krad07002061 [Kineococcus radiotolerans
SRS30216]
Length = 784
Score = 52.0 bits (123), Expect = 2e-05
Identities = 33/69 (47%), Positives = 36/69 (51%), Gaps = 4/69 (5%)
Query: 65 GSGRGRGRGRGRDVYA---RGRGDRGRGGRGGRGDGRG-GFKRYGDDRTSHQDIARSNAD 120
G GR RG GR RD RGRGDRGRG R R GRG G +R DDR D+ R
Sbjct: 114 GGGRQRGHGRDRDHGGRGHRGRGDRGRGDRSRRDGGRGDGSRRDRDDRGRRGDLDRGGRR 173
Query: 121 GLYVGDNAD 129
G + D D
Sbjct: 174 GDGLDDGLD 182
Score = 34.3 bits (77), Expect = 4.8
Identities = 22/69 (31%), Positives = 28/69 (39%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNAD 120
D+G+ RG GRG G GRG RG G G G+G D + +
Sbjct: 192 DEGELRLRGAGRGDGGRDGGGGRGGRGAGAERDEGGGQGDGGDDPDGGVGRAAAGQRHGH 251
Query: 121 GLYVGDNAD 129
G VG+ D
Sbjct: 252 GPAVGEQPD 260
Score = 33.9 bits (76), Expect = 6.2
Identities = 26/55 (47%), Positives = 27/55 (48%), Gaps = 9/55 (16%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGR---GGRGGRGDGR-----GGFKRYGDD 107
DDG G G G R + GRGD GR GGRGGRG G GG GDD
Sbjct: 182 DDGLRPGGGPDEGELR-LRGAGRGDGGRDGGGGRGGRGAGAERDEGGGQGDGGDD 235
>emb|CAF91750.1| unnamed protein product [Tetraodon nigroviridis]
Length = 512
Score = 51.6 bits (122), Expect = 3e-05
Identities = 29/57 (50%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDG-RGGFKRYGDDR 108
+P RF RD G+ S G RGRGR Y G +RGR G G G G RGG+K YG R
Sbjct: 108 RPGPRFGRDQGERSDGGGYRGRGRGGYDSGGYERGRRGPPGMGGGDRGGYKNYGGSR 164
>ref|XP_468448.1| putative fibrillarin [Oryza sativa (japonica cultivar-group)]
gi|48716271|dbj|BAD22886.1| putative fibrillarin [Oryza
sativa (japonica cultivar-group)]
gi|48716513|dbj|BAD23118.1| putative fibrillarin [Oryza
sativa (japonica cultivar-group)]
Length = 306
Score = 51.2 bits (121), Expect = 4e-05
Identities = 30/60 (50%), Positives = 32/60 (53%), Gaps = 7/60 (11%)
Query: 50 PRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGG-------RGGRGDGRGGFK 102
PR + R R DG G G GRG GR + GRG RGRGG GGRG GRGG K
Sbjct: 4 PRGRGFGRGGRGDGGGRSGGGGRGFGRGGDSGGRGGRGRGGGRTPRGRGGGRGGGRGGMK 63
Score = 36.2 bits (82), Expect = 1.3
Identities = 20/35 (57%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDG 97
GD GRG GRGRG RGRG GGRGG G
Sbjct: 32 GDSGGRG-GRGRGGGRTPRGRGGGRGGGRGGMKGG 65
Score = 34.3 bits (77), Expect = 4.8
Identities = 23/40 (57%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 68 RGRGRGRGRDVYARGR-GDRGRG-GRGGRGDGRGGFKRYG 105
RGRG GRG GR G GRG GRGG GRGG R G
Sbjct: 5 RGRGFGRGGRGDGGGRSGGGGRGFGRGGDSGGRGGRGRGG 44
>gb|AAC14119.1| AUT1 [Schistosoma mansoni]
Length = 335
Score = 50.8 bits (120), Expect = 5e-05
Identities = 66/248 (26%), Positives = 97/248 (38%), Gaps = 47/248 (18%)
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESD-NVPRRQPMNRFVRDDGDGSG 67
G GRG + + + P+ I + P D+ SD N PR
Sbjct: 121 GSGRGTPRGMRVGRGQGPR-------IAPTEAPQDSVSDLNAPRGSSFEP---------- 163
Query: 68 RGRGRGRGRDVYARGRG---DRGRGGRGGRGDGRGGFKRYG--DDRTSHQDIARSNADGL 122
RGRGRGRGR ++ RGRG + R G R G ++YG D + QD+ DGL
Sbjct: 164 RGRGRGRGRGMFGRGRGMPFNSNRDFENQDGPDRQGPRQYGRRDGNWNSQDV-----DGL 218
Query: 123 YVGDNADGEKLAKKLGP--EIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEPE 180
+ ++ D E++ + E+ DQ A E E V+ + +E EP+
Sbjct: 219 IMPESGDSEQVVRFADDRNEVEDQPEHATAENEEGVV-----------VGTETPVEEEPK 267
Query: 181 -YAVEFDNPDIDEKEPIAL--RDALEKM---KPFLMTYEGIRSQEEWEEVIEELMQRVPL 234
Y +E +P L L K K R +E E + E+ +R
Sbjct: 268 SYTLEGYKAMRQSSKPAVLLNNKGLRKANDGKDVFANMVAHRKLQEVSEDVYEVEERKTS 327
Query: 235 LKKIVDHY 242
L+ VD Y
Sbjct: 328 LRASVDRY 335
>gb|AAF82607.1| AUT1 [Schistosoma mansoni]
Length = 349
Score = 50.4 bits (119), Expect = 6e-05
Identities = 49/167 (29%), Positives = 74/167 (43%), Gaps = 33/167 (19%)
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESD-NVPRRQPMNRFVRDDGDGSG 67
G GRG + + + P+ I + P D+ SD N PR
Sbjct: 121 GSGRGTPRGMRVGRGQGPR-------IAPTEAPQDSVSDLNAPRGSSFEP---------- 163
Query: 68 RGRGRGRGRDVYARGRG---DRGRGGRGGRGDGRGGFKRYG--DDRTSHQDIARSNADGL 122
RGRGRGRGR ++ RGRG + R G R G ++YG D + QD+ DGL
Sbjct: 164 RGRGRGRGRGMFGRGRGMPFNSNRDFENQDGPDRQGPRQYGRRDGNWNSQDV-----DGL 218
Query: 123 YVGDNADGEKLAKKLGP--EIMDQITEAYEEIIERVL---PSPLQDE 164
+ ++ D E++ + E+ DQ A E E V+ +P+++E
Sbjct: 219 IMPESGDSEQVVRFADDRNEVEDQPEHATAENEEGVVVGTETPVEEE 265
>ref|YP_067973.1| EBNA-1 [Cercopithecine herpesvirus 15] gi|18025496|gb|AAK95440.1|
EBNA-1 [cercopithicine herpesvirus 15]
Length = 511
Score = 50.1 bits (118), Expect = 8e-05
Identities = 27/44 (61%), Positives = 28/44 (63%), Gaps = 3/44 (6%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRG---GRGDGRGGFKR 103
G G+G RGRGRGR A GRG RG GG G GRG GRGG R
Sbjct: 158 GSGAGGSRGRGRGRGGSAGGRGGRGGGGGGGSRGRGRGRGGGSR 201
Score = 38.5 bits (88), Expect = 0.25
Identities = 28/67 (41%), Positives = 33/67 (48%), Gaps = 9/67 (13%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRG-GRGGRG--DGRGGFKRYGDDR-----TSHQDI 114
G SG G G RGR + RGRG RGRG GRGG G G F +G + H+D
Sbjct: 23 GGASGSGSGGNRGRGAHGRGRG-RGRGRGRGGGGVLGETGEFGGHGSESETRHGNGHRDK 81
Query: 115 ARSNADG 121
R + G
Sbjct: 82 KRRSCVG 88
Score = 34.3 bits (77), Expect = 4.8
Identities = 18/46 (39%), Positives = 20/46 (43%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
G G+G G G G G G G GG G G G GG + G R
Sbjct: 126 GGGAGGSLGGGAGGSSGGSGAGGSGAGGSGAGGSGAGGSRGRGRGR 171
>gb|AAU44295.1| putative fibrillarin [Oryza sativa (japonica cultivar-group)]
gi|50511418|gb|AAT77341.1| putative fibrillarin protein
[Oryza sativa (japonica cultivar-group)]
Length = 351
Score = 50.1 bits (118), Expect = 8e-05
Identities = 34/85 (40%), Positives = 41/85 (48%), Gaps = 7/85 (8%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRG-----GRGDGRGGFKRYGDDRTSHQDI 114
R DG G G G GRG GR + GRG GRGGRG GRG GRGG R G S +
Sbjct: 16 RGDGGGRGGGGGRGFGRVGDSGGRG--GRGGRGGRTPRGRGGGRGGGGRGGMKGGSKVVV 73
Query: 115 ARSNADGLYVGDNADGEKLAKKLGP 139
DG+++ + K + P
Sbjct: 74 VPHKHDGVFIAKAKEDALCTKNMVP 98
Score = 33.9 bits (76), Expect = 6.2
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 12/41 (29%)
Query: 68 RGRGRGRGRDVYARGRGDRG-RGGRGGRG-------DGRGG 100
RGRG GRG GRGD G RGG GGRG GRGG
Sbjct: 5 RGRGFGRG----GGGRGDGGGRGGGGGRGFGRVGDSGGRGG 41
>ref|XP_640657.1| hypothetical protein DDB0205563 [Dictyostelium discoideum]
gi|60468684|gb|EAL66686.1| hypothetical protein
DDB0205563 [Dictyostelium discoideum]
Length = 369
Score = 50.1 bits (118), Expect = 8e-05
Identities = 38/119 (31%), Positives = 52/119 (42%), Gaps = 10/119 (8%)
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVP----RRQPMNRFVRDDGD 64
GG RG A +P + P ++Q ++ D
Sbjct: 175 GGFRGSSSHPSTATDNTAATTTTTSPSTSTTSPTTTTTTTAPVEQQQQQQQQQYQPSDSG 234
Query: 65 GSGRGRG----RGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNA 119
GRGRG RGRGR + RGRG RGRGG GRG GRGGF+ +++Q+ S++
Sbjct: 235 FRGRGRGGFRGRGRGRGGF-RGRG-RGRGGFRGRGRGRGGFRGGSPLNSNYQESESSSS 291
>pir||WMBEL1 latency-related protein 1 - human herpesvirus 1 (strain F)
gi|126465|sp|P17588|LRP1_HHV1F Latency-related protein 1
gi|330134|gb|AAA45799.1| latency-related protein 1
Length = 340
Score = 50.1 bits (118), Expect = 8e-05
Identities = 25/41 (60%), Positives = 26/41 (62%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
R G G GRG G GRG +RGRG RGRGGRGG GR G
Sbjct: 288 RGGGSGGGRGPGGGRGGPRGSRGRGGRGRGGRGGGRRGRQG 328
>emb|CAG82487.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50549393|ref|XP_502167.1| hypothetical protein
[Yarrowia lipolytica]
Length = 1526
Score = 49.7 bits (117), Expect = 1e-04
Identities = 40/96 (41%), Positives = 48/96 (49%), Gaps = 15/96 (15%)
Query: 8 PGGG--RGRGKPLEEAAQEAPQA-PVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGD 64
PG G +G G ++ Q APQ P N + + A SDN +QP +
Sbjct: 1427 PGQGYPQGPGPQNQQHQQGAPQGTPQGNEDLSAQLMAALQVSDN--SQQPATGLANE--- 1481
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
GRGRG GRGR GRG RGRGGRG G G+GG
Sbjct: 1482 -GGRGRG-GRGR-----GRGGRGRGGRGRGGQGQGG 1510
>gb|AAD12858.2| Hypothetical protein Y66H1A.4 [Caenorhabditis elegans]
gi|51701700|sp|Q9TYK1|NOLA1_CAEEL Putative nucleolar
protein family A member 1 (snoRNP protein GAR1) (H/ACA
ribonucleoprotein GAR1)
Length = 244
Score = 49.7 bits (117), Expect = 1e-04
Identities = 34/68 (50%), Positives = 38/68 (55%), Gaps = 16/68 (23%)
Query: 54 PMNRFVRDDGDGSGRGRGRGRGRDVYARG-----RGDRGRGG--------RGGRGDGRGG 100
P++RF+ G G GRG GRGRG D RG RG GRGG RGG G GRGG
Sbjct: 141 PVDRFLPQAGGGRGRG-GRGRGGDRGGRGSDRGGRGGFGRGGGGGFRGGDRGGFGGGRGG 199
Query: 101 FKRYGDDR 108
F+ G DR
Sbjct: 200 FR--GGDR 205
Score = 37.4 bits (85), Expect = 0.56
Identities = 24/55 (43%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
R G G GRG G G GDRG G RGGRG G GG R G D+ +++
Sbjct: 7 RGGGGGFRGGRGGGGGGGFRGGRGGDRGGGFRGGRG-GFGGGGRGGYDQGPPEEV 60
>ref|NP_499927.1| nucleolar protein family A member 1 (4B179) [Caenorhabditis
elegans] gi|7510373|pir||T33925 hypothetical protein
Y66H1A.4 - Caenorhabditis elegans
Length = 276
Score = 49.7 bits (117), Expect = 1e-04
Identities = 34/68 (50%), Positives = 38/68 (55%), Gaps = 16/68 (23%)
Query: 54 PMNRFVRDDGDGSGRGRGRGRGRDVYARG-----RGDRGRGG--------RGGRGDGRGG 100
P++RF+ G G GRG GRGRG D RG RG GRGG RGG G GRGG
Sbjct: 173 PVDRFLPQAGGGRGRG-GRGRGGDRGGRGSDRGGRGGFGRGGGGGFRGGDRGGFGGGRGG 231
Query: 101 FKRYGDDR 108
F+ G DR
Sbjct: 232 FR--GGDR 237
Score = 37.4 bits (85), Expect = 0.56
Identities = 24/55 (43%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
R G G GRG G G GDRG G RGGRG G GG R G D+ +++
Sbjct: 39 RGGGGGFRGGRGGGGGGGFRGGRGGDRGGGFRGGRG-GFGGGGRGGYDQGPPEEV 92
>emb|CAG02768.1| unnamed protein product [Tetraodon nigroviridis]
Length = 311
Score = 49.3 bits (116), Expect = 1e-04
Identities = 36/85 (42%), Positives = 42/85 (49%), Gaps = 16/85 (18%)
Query: 58 FVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGF---KRYGDDRTSHQD- 113
F DG G G RGRG GR RGRGGRGGRG GRGGF + + H+
Sbjct: 33 FRSPDGGGGGGFRGRGGGRGT------PRGRGGRGGRGGGRGGFGGGNKVLIEPHRHEGV 86
Query: 114 -IARSNADGL-----YVGDNADGEK 132
I+R D L VG++ GEK
Sbjct: 87 FISRGKEDALVTKNMVVGESVYGEK 111
>gb|AAB04133.1| cutinase negative acting protein
Length = 507
Score = 49.3 bits (116), Expect = 1e-04
Identities = 27/42 (64%), Positives = 28/42 (66%), Gaps = 4/42 (9%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGG---RGGRGDGRGGF 101
G G GRG GRG GR + GRG GRGG RGGRG GRGGF
Sbjct: 448 GFGGGRGGGRGGGRGGFG-GRGGGGRGGGRGRGGRGGGRGGF 488
Score = 38.1 bits (87), Expect = 0.33
Identities = 23/43 (53%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G G G G GRG GR GRG G G GGRG GRG R G
Sbjct: 444 GGGGGFGGGRGGGR---GGGRGGFGGRGGGGRGGGRGRGGRGG 483
Score = 35.0 bits (79), Expect = 2.8
Identities = 21/39 (53%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGR-GGRGGRGDG 97
R G G GRG GRG GRG GR GGRGG G G
Sbjct: 453 RGGGRGGGRGGFGGRGGGGRGGGRGRGGRGGGRGGFGTG 491
>gb|EAA20072.1| multidomain scavenger receptor protein PbSR precursor [Plasmodium
yoelii yoelii]
Length = 1615
Score = 49.3 bits (116), Expect = 1e-04
Identities = 28/47 (59%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query: 60 RDDGDGSGRG-RGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
R GDG GRG RG G GR RG G RG GGRGG G G GG R G
Sbjct: 1328 RGGGDGGGRGGRGGGGGRGGGGRGGGGRGGGGRGGGGRGGGGGGRGG 1374
Score = 36.6 bits (83), Expect = 0.96
Identities = 24/47 (51%), Positives = 24/47 (51%), Gaps = 5/47 (10%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGR-GDGRGGFKRYGDDR 108
G GRG G G GR GRG G G GGR G GRGG R G R
Sbjct: 1323 GSNGGRGGGDGGGRG----GRGGGGGRGGGGRGGGGRGGGGRGGGGR 1365
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 600,017,390
Number of Sequences: 2540612
Number of extensions: 31289073
Number of successful extensions: 193670
Number of sequences better than 10.0: 2781
Number of HSP's better than 10.0 without gapping: 969
Number of HSP's successfully gapped in prelim test: 2028
Number of HSP's that attempted gapping in prelim test: 147741
Number of HSP's gapped (non-prelim): 23307
length of query: 310
length of database: 863,360,394
effective HSP length: 127
effective length of query: 183
effective length of database: 540,702,670
effective search space: 98948588610
effective search space used: 98948588610
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC147483.4


