

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.11 - phase: 0 /pseudo
(1453 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
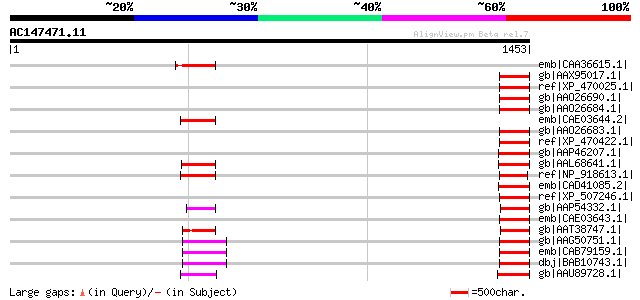
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA36615.1| unnamed protein product [Solanum tuberosum] gi|4... 101 2e-19
gb|AAX95017.1| retrotransposon protein, putative, unclassified [... 97 3e-18
ref|XP_470025.1| putative polyprotein [Oryza sativa (japonica cu... 97 4e-18
gb|AAO26690.1| gag-pol polyprotein [Vitis vinifera] 97 5e-18
gb|AAO26684.1| gag-pol polyprotein [Vitis vinifera] 96 7e-18
emb|CAE03644.2| OSJNBa0060N03.9 [Oryza sativa (japonica cultivar... 96 9e-18
gb|AAO26683.1| gag-pol polyprotein [Vitis vinifera] 96 9e-18
ref|XP_470422.1| putative polyprotein [Oryza sativa (japonica cu... 95 2e-17
gb|AAP46207.1| putative retrotransposon protein [Oryza sativa (j... 95 2e-17
gb|AAL68641.1| polyprotein [Oryza sativa (japonica cultivar-group)] 95 2e-17
ref|NP_918613.1| polyprotein [Oryza sativa (japonica cultivar-gr... 95 2e-17
emb|CAD41085.2| OSJNBb0011N17.2 [Oryza sativa (japonica cultivar... 95 2e-17
ref|XP_507246.1| PREDICTED P0528B09.40 gene product [Oryza sativ... 94 3e-17
gb|AAP54332.1| putative copia-like polyprotein [Oryza sativa (ja... 94 4e-17
emb|CAE03643.1| OSJNBa0060N03.8 [Oryza sativa (japonica cultivar... 94 4e-17
gb|AAT38747.1| putative polyprotein [Solanum demissum] 89 8e-16
gb|AAG50751.1| polyprotein, putative [Arabidopsis thaliana] gi|2... 88 2e-15
emb|CAB79159.1| LTR retrotransposon like protein [Arabidopsis th... 88 2e-15
dbj|BAB10743.1| retroelement pol polyprotein-like [Arabidopsis t... 88 2e-15
gb|AAU89728.1| putative retroelement pol polyprotein-like [Solan... 87 5e-15
>emb|CAA36615.1| unnamed protein product [Solanum tuberosum] gi|421954|pir||S25786
hypothetical protein 3 - potato transposon Tst1
Length = 675
Score = 101 bits (252), Expect = 2e-19
Identities = 49/114 (42%), Positives = 70/114 (60%), Gaps = 8/114 (7%)
Query: 464 YLATTFFLESIKTNREKVLLYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHK 523
+L +TF L KV+L+HY +G PSF ++ L P F+ + FQCE CE+AKH
Sbjct: 91 FLNSTFVLN-------KVMLWHYGLGHPSFYYLRHLLPQLFRNKNPSLFQCEFCEMAKHH 143
Query: 524 RVT-FSVSNKMSTFPFYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
T F ++ PF ++H++VWGPS + + G RWFVTFIDD T ++WV+ L
Sbjct: 144 VDTSFPSQRYQASKPFTMIHSDVWGPSRISTMFGKRWFVTFIDDHTRLSWVFLL 197
>gb|AAX95017.1| retrotransposon protein, putative, unclassified [Oryza sativa
(japonica cultivar-group)]
Length = 277
Score = 97.4 bits (241), Expect = 3e-18
Identities = 45/82 (54%), Positives = 62/82 (74%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
PM+L+CDNK AI+IA+NPVQHDRTKH+E+++ FIKEKLD G++ +V S + +AD LTK
Sbjct: 195 PMKLWCDNKSAISIANNPVQHDRTKHVEIDRFFIKEKLDEGILKLGFVMSGEQVADCLTK 254
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
L+ +K+GMIDI+ P
Sbjct: 255 ALSVGECTSSCNKMGMIDIYRP 276
>ref|XP_470025.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|30103001|gb|AAP21414.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1393
Score = 97.1 bits (240), Expect = 4e-18
Identities = 44/83 (53%), Positives = 62/83 (74%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ + L+CDNK AI+IA+NPVQHDRTKH+E+++ FIKEK+DSG++ Y+ S + LAD LT
Sbjct: 1310 DTVMLHCDNKSAISIANNPVQHDRTKHVEIDRFFIKEKIDSGVLRLEYIKSCEQLADCLT 1369
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L + + +K+GMIDI P
Sbjct: 1370 KGLGPSEIQSICNKMGMIDIFCP 1392
>gb|AAO26690.1| gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 96.7 bits (239), Expect = 5e-18
Identities = 42/82 (51%), Positives = 61/82 (74%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P +L+CDN+ A++IA NPV H+RTKH+EV+ HFI+EK++ L+ T YV + + L D+ TK
Sbjct: 244 PAKLWCDNQAALHIAANPVYHERTKHIEVDCHFIREKIEENLVSTDYVKTGEQLGDIFTK 303
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
LN E F +KLGMI+I++P
Sbjct: 304 ALNGTRVEYFCNKLGMINIYAP 325
>gb|AAO26684.1| gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 96.3 bits (238), Expect = 7e-18
Identities = 42/82 (51%), Positives = 61/82 (74%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P +L+CDN+ A++IA NPV H+RTKH+EV+ HFI+EK++ L+ T YV + + L D+ TK
Sbjct: 244 PAKLWCDNQAALHIAANPVYHERTKHIEVDCHFIREKIEENLVSTGYVKTGEQLGDIFTK 303
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
LN E F +KLGMI+I++P
Sbjct: 304 ALNGTRVEYFCNKLGMINIYAP 325
>emb|CAE03644.2| OSJNBa0060N03.9 [Oryza sativa (japonica cultivar-group)]
gi|50928597|ref|XP_473826.1| OSJNBa0060N03.9 [Oryza
sativa (japonica cultivar-group)]
Length = 1045
Score = 95.9 bits (237), Expect = 9e-18
Identities = 41/98 (41%), Positives = 62/98 (62%)
Query: 479 EKVLLYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPF 538
+++ L H ++G PSF ++ +L+P F +D C+ CEL KH R T+ + PF
Sbjct: 407 KEISLLHCQLGHPSFEILSKLYPDLFSRVDKHRLVCDACELGKHTRSTYVGIGLRNCEPF 466
Query: 539 YLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
L+H++VWGP V ++SG +WFVTFID T +TW+Y L
Sbjct: 467 ILIHSDVWGPCPVTSVSGFKWFVTFIDCHTRMTWIYML 504
>gb|AAO26683.1| gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 95.9 bits (237), Expect = 9e-18
Identities = 42/82 (51%), Positives = 61/82 (74%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P +L+CDN+ A++IA NPV H+RTKH+EV+ HFI+EK++ L+ T YV + + L D+ TK
Sbjct: 244 PAKLWCDNQAALHIAANPVYHERTKHIEVDCHFIREKIEENLVSTGYVKTGEQLGDIFTK 303
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
LN E F +KLGMI+I++P
Sbjct: 304 ALNGTRVECFCNKLGMINIYAP 325
>ref|XP_470422.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|27573360|gb|AAO20078.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1299
Score = 95.1 bits (235), Expect = 2e-17
Identities = 46/82 (56%), Positives = 59/82 (71%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
PM+L CDNK AINIA+NPVQHDRTKH+E+++ FIKEKLD G++ +V+S +AD LTK
Sbjct: 1217 PMKLLCDNKSAINIANNPVQHDRTKHVEIDRFFIKEKLDEGVLELGFVTSGGQVADCLTK 1276
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
L K+GMIDI+ P
Sbjct: 1277 GLGVKECNCSCDKMGMIDIYHP 1298
>gb|AAP46207.1| putative retrotransposon protein [Oryza sativa (japonica
cultivar-group)] gi|50920663|ref|XP_470692.1| putative
retrotransposon protein [Oryza sativa (japonica
cultivar-group)]
Length = 1176
Score = 94.7 bits (234), Expect = 2e-17
Identities = 45/84 (53%), Positives = 59/84 (69%)
Query: 1370 DEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLL 1429
D PM+L+CDNK AI+IA+NPVQHDRTKH+E+++ FIKEKLD G++ +V S +AD
Sbjct: 1092 DTPMKLWCDNKSAISIANNPVQHDRTKHVELDRFFIKEKLDEGVLELEFVMSGGQVADCF 1151
Query: 1430 TKRLNNNNFEKFVSKLGMIDIHSP 1453
TK L K+GMIDI+ P
Sbjct: 1152 TKGLGVKECNSSCDKMGMIDIYHP 1175
>gb|AAL68641.1| polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1472
Score = 94.7 bits (234), Expect = 2e-17
Identities = 45/84 (53%), Positives = 59/84 (69%)
Query: 1370 DEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLL 1429
D PM+L+CDNK AI+IA+NPVQHDRTKH+E+++ FIKEKLD G++ +V S +AD
Sbjct: 1388 DTPMKLWCDNKSAISIANNPVQHDRTKHVELDRFFIKEKLDEGVLELEFVMSGGQVADCF 1447
Query: 1430 TKRLNNNNFEKFVSKLGMIDIHSP 1453
TK L K+GMIDI+ P
Sbjct: 1448 TKGLGVKECNSSCDKMGMIDIYHP 1471
Score = 81.6 bits (200), Expect = 2e-13
Identities = 38/97 (39%), Positives = 60/97 (61%), Gaps = 1/97 (1%)
Query: 480 KVLLYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFY 539
+VLL H R+G SF ++ ++FP+ F +D C+ CE KH R ++ S PF
Sbjct: 500 EVLLLHCRLGHISFEIMSKMFPVEFSKVDKHMLICDACEYGKHTRTSYVSRGLRSILPFM 559
Query: 540 LVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
L+H++VW S V ++SG ++FVTFID + +TW+Y +
Sbjct: 560 LIHSDVW-TSPVVSMSGMKYFVTFIDCYSRMTWLYLM 595
>ref|NP_918613.1| polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1554
Score = 94.7 bits (234), Expect = 2e-17
Identities = 43/80 (53%), Positives = 61/80 (75%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ + L+CDNK AI+IA+NPVQHDRTKH+E+++ FIKEK+DSG++ Y+ S + LAD LT
Sbjct: 1471 DTVMLHCDNKSAISIANNPVQHDRTKHVEIDRFFIKEKIDSGVLRLEYIKSCEQLADCLT 1530
Query: 1431 KRLNNNNFEKFVSKLGMIDI 1450
K L + + +K+GMIDI
Sbjct: 1531 KGLGPSEIQSVCNKMGMIDI 1550
Score = 92.0 bits (227), Expect = 1e-16
Identities = 40/98 (40%), Positives = 61/98 (61%)
Query: 479 EKVLLYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPF 538
+++ L H ++G PSF ++ +L+P F +D C+ CEL KH R T+ + F
Sbjct: 530 KEISLLHCQLGHPSFEILSKLYPDLFSRVDKHRLVCDACELGKHTRSTYVGIGLRNCELF 589
Query: 539 YLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
L+H++VWGP V ++SG +WFVTFID T +TW+Y L
Sbjct: 590 ILIHSDVWGPCPVTSVSGFKWFVTFIDCHTRMTWIYML 627
>emb|CAD41085.2| OSJNBb0011N17.2 [Oryza sativa (japonica cultivar-group)]
gi|50925209|ref|XP_472906.1| OSJNBb0011N17.2 [Oryza
sativa (japonica cultivar-group)]
Length = 1262
Score = 94.7 bits (234), Expect = 2e-17
Identities = 45/84 (53%), Positives = 59/84 (69%)
Query: 1370 DEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLL 1429
D PM+L+CDNK AI+IA+NPVQHDRTKH+E+++ FIKEKLD G++ +V S +AD
Sbjct: 1178 DTPMKLWCDNKSAISIANNPVQHDRTKHVELDRFFIKEKLDEGVLELEFVMSGGQVADCF 1237
Query: 1430 TKRLNNNNFEKFVSKLGMIDIHSP 1453
TK L K+GMIDI+ P
Sbjct: 1238 TKGLGVKECNSSCDKMGMIDIYHP 1261
>ref|XP_507246.1| PREDICTED P0528B09.40 gene product [Oryza sativa (japonica
cultivar-group)]
Length = 147
Score = 94.0 bits (232), Expect = 3e-17
Identities = 43/83 (51%), Positives = 60/83 (71%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ + L+CDNK AINI +N VQHDRTKH+E+++ FIKEK+DSG++ Y+ S + LAD LT
Sbjct: 64 DTVMLHCDNKSAINIGNNLVQHDRTKHVEIDRFFIKEKIDSGVLMLEYIKSCEQLADCLT 123
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L + + +K+GMIDI P
Sbjct: 124 KGLGPSEIQSICNKMGMIDIFYP 146
>gb|AAP54332.1| putative copia-like polyprotein [Oryza sativa (japonica
cultivar-group)] gi|37535486|ref|NP_922045.1| putative
copia-like polyprotein [Oryza sativa (japonica
cultivar-group)] gi|22094347|gb|AAM91874.1| putative
copia-like polyprotein [Oryza sativa (japonica
cultivar-group)]
Length = 894
Score = 93.6 bits (231), Expect = 4e-17
Identities = 43/81 (53%), Positives = 59/81 (72%)
Query: 1373 MRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKR 1432
M+L+CDNK AINIA+NPVQHDRTKH+E+++ FIKE++D G + +V+S + + D LTK
Sbjct: 813 MKLWCDNKSAINIANNPVQHDRTKHVEIDRFFIKERMDEGTLNLGFVNSGEQVVDSLTKA 872
Query: 1433 LNNNNFEKFVSKLGMIDIHSP 1453
L SK+GMIDI+ P
Sbjct: 873 LGARECTSSCSKMGMIDIYRP 893
Score = 68.9 bits (167), Expect = 1e-09
Identities = 31/81 (38%), Positives = 48/81 (58%), Gaps = 1/81 (1%)
Query: 496 IKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLVHTNVWGPSNVPNIS 555
+ ++FP +D C+ CE KH R T+ S PF LVH++VW S + ++S
Sbjct: 1 MSKVFPDEMSKVDKSKLMCDACEYGKHTRTTYISRGLQSILPFVLVHSDVW-TSPIASVS 59
Query: 556 GARWFVTFIDDCTWVTWVYFL 576
GA++FVTFID + +TW+Y +
Sbjct: 60 GAKYFVTFIDCHSRMTWIYLM 80
>emb|CAE03643.1| OSJNBa0060N03.8 [Oryza sativa (japonica cultivar-group)]
gi|50928595|ref|XP_473825.1| OSJNBa0060N03.8 [Oryza
sativa (japonica cultivar-group)]
Length = 343
Score = 93.6 bits (231), Expect = 4e-17
Identities = 42/83 (50%), Positives = 62/83 (74%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ + L+CDN+ AI+IA+NPVQHDRTKH+E+++ FIKEK++SG++ Y+ S + LAD LT
Sbjct: 260 DTVMLHCDNRSAISIANNPVQHDRTKHVEIDRFFIKEKINSGVLRLEYIKSCEQLADYLT 319
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L + + +K+GMIDI P
Sbjct: 320 KGLGPSEIQSICNKMGMIDIFFP 342
>gb|AAT38747.1| putative polyprotein [Solanum demissum]
Length = 1336
Score = 89.4 bits (220), Expect = 8e-16
Identities = 41/80 (51%), Positives = 57/80 (71%)
Query: 1374 RLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRL 1433
+L+CDN+ A++IA NPV H+RTKH+EV+ HFI+EK+ LI T YV + + LADL TK L
Sbjct: 1256 KLWCDNQAALHIASNPVYHERTKHIEVDCHFIREKIQENLISTSYVKTGEQLADLFTKAL 1315
Query: 1434 NNNNFEKFVSKLGMIDIHSP 1453
+KLGMI+I++P
Sbjct: 1316 TGARVNYLCNKLGMINIYAP 1335
Score = 88.6 bits (218), Expect = 1e-15
Identities = 44/93 (47%), Positives = 61/93 (65%), Gaps = 4/93 (4%)
Query: 485 HYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSV-SNKMSTFPFYLVHT 543
H R+G PS V+K+L P F +V S CE C AKH R++ S +NK + F F LVH+
Sbjct: 495 HCRLGHPSLPVLKKLCPQFH---NVPSIDCESCHFAKHHRISLSPRNNKRANFAFELVHS 551
Query: 544 NVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
+VWGP V + G R+FVTF+DD + +TW+YF+
Sbjct: 552 DVWGPCPVVSKVGFRYFVTFMDDFSRMTWIYFM 584
>gb|AAG50751.1| polyprotein, putative [Arabidopsis thaliana] gi|25301686|pir||F96610
probable polyprotein T8L23.26 [imported] - Arabidopsis
thaliana
Length = 1468
Score = 88.2 bits (217), Expect = 2e-15
Identities = 39/83 (46%), Positives = 59/83 (70%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ MR++ D+K AI ++ NPVQH+RTKH+EV+ HFI++ + G+I T +V S LAD+LT
Sbjct: 1385 QAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILT 1444
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L F+ KLG++D+H+P
Sbjct: 1445 KALGEKEVRYFLRKLGILDVHAP 1467
Score = 75.9 bits (185), Expect = 1e-11
Identities = 45/126 (35%), Positives = 68/126 (53%), Gaps = 2/126 (1%)
Query: 483 LYHYRVGDPSFRVIKQL-FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G S +++ L L ++ C+ C AK R TF +S+ S F L+
Sbjct: 515 LWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLSDNRSMDSFQLI 574
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD*KSV*CEH*TD*IR*CQRLF*SQ 601
H +VWGP P+ SGAR+F+T +DD + WVY + D KS +H D I +R F ++
Sbjct: 575 HCDVWGPYRAPSYSGARYFLTIVDDYSRGVWVYLMTD-KSETQKHLKDFIALVERQFDTE 633
Query: 602 FKFILS 607
K + S
Sbjct: 634 IKIVRS 639
>emb|CAB79159.1| LTR retrotransposon like protein [Arabidopsis thaliana]
gi|2961349|emb|CAA18107.1| LTR retrotransposon like
protein [Arabidopsis thaliana] gi|11358464|pir||T49111
hypothetical retrovirus-related pol polyprotein AT4g22040
- Arabidopsis thaliana
Length = 1109
Score = 88.2 bits (217), Expect = 2e-15
Identities = 39/83 (46%), Positives = 59/83 (70%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ MR++ D+K AI ++ NPVQH+RTKH+EV+ HFI++ + G+I T +V S LAD+LT
Sbjct: 1026 QAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILT 1085
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L F+ KLG++D+H+P
Sbjct: 1086 KALGEKEVRYFLRKLGILDVHAP 1108
Score = 72.0 bits (175), Expect = 1e-10
Identities = 43/126 (34%), Positives = 67/126 (53%), Gaps = 2/126 (1%)
Query: 483 LYHYRVGDPSFRVIKQL-FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G S +++ L L ++ C+ C AK R TF + + S F L+
Sbjct: 291 LWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLRDNRSMDSFQLI 350
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD*KSV*CEH*TD*IR*CQRLF*SQ 601
H +VWGP P+ SGAR+F+T +DD + WVY + D KS +H D + +R F ++
Sbjct: 351 HCDVWGPYRTPSYSGARYFLTIVDDYSRGVWVYLMTD-KSETQKHLKDFMALVERQFDTE 409
Query: 602 FKFILS 607
K + S
Sbjct: 410 IKTVRS 415
>dbj|BAB10743.1| retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1109
Score = 88.2 bits (217), Expect = 2e-15
Identities = 39/83 (46%), Positives = 59/83 (70%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ MR++ D+K AI ++ NPVQH+RTKH+EV+ HFI++ + G+I T +V S LAD+LT
Sbjct: 1026 QAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILT 1085
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L F+ KLG++D+H+P
Sbjct: 1086 KALGEKEVRYFLRKLGILDVHAP 1108
Score = 75.1 bits (183), Expect = 2e-11
Identities = 45/126 (35%), Positives = 68/126 (53%), Gaps = 2/126 (1%)
Query: 483 LYHYRVGDPSFRVIKQL-FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G S +++ L L ++ C+ C AK R TF +S+ S F L+
Sbjct: 291 LWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLSDNRSMDSFQLI 350
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD*KSV*CEH*TD*IR*CQRLF*SQ 601
H +VWGP P+ SGAR+F+T +DD + WVY + D KS +H D I +R F ++
Sbjct: 351 HCDVWGPYRTPSYSGARYFLTIVDDYSRGVWVYLMTD-KSETQKHLKDFIALVERQFDTE 409
Query: 602 FKFILS 607
K + S
Sbjct: 410 IKTVRS 415
>gb|AAU89728.1| putative retroelement pol polyprotein-like [Solanum tuberosum]
Length = 1476
Score = 86.7 bits (213), Expect = 5e-15
Identities = 41/88 (46%), Positives = 60/88 (67%)
Query: 1366 DKDFDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNL 1425
D P+ LYCD+K AI IA NPV H+RTKH++++ HFI+EK+ +GL+ Y+ +Q+
Sbjct: 1370 DMPLSLPVSLYCDSKAAIQIAANPVFHERTKHIDIDCHFIREKVQAGLVMIHYLPTQEQP 1429
Query: 1426 ADLLTKRLNNNNFEKFVSKLGMIDIHSP 1453
AD+LTK L++ VSKLG+ +I P
Sbjct: 1430 ADILTKGLSSAQHSYLVSKLGLKNIFIP 1457
Score = 61.2 bits (147), Expect = 2e-07
Identities = 29/99 (29%), Positives = 54/99 (54%), Gaps = 1/99 (1%)
Query: 479 EKVLLYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPF 538
E+ ++H R+G V++++ +F + C++C LA+ R+ F +S S F
Sbjct: 548 EEAEMWHKRLGHIPMSVLRKI-KMFDSPQKLVLPSCDVCPLARQVRLPFPISQSRSENCF 606
Query: 539 YLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLH 577
L+H +VWGP + R+F+T +DD + TW++ +H
Sbjct: 607 DLIHLDVWGPYKAATHNKMRYFLTVVDDHSRWTWIFLMH 645
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.364 0.163 0.626
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,008,227,583
Number of Sequences: 2540612
Number of extensions: 70163963
Number of successful extensions: 343868
Number of sequences better than 10.0: 767
Number of HSP's better than 10.0 without gapping: 708
Number of HSP's successfully gapped in prelim test: 59
Number of HSP's that attempted gapping in prelim test: 342123
Number of HSP's gapped (non-prelim): 1664
length of query: 1453
length of database: 863,360,394
effective HSP length: 141
effective length of query: 1312
effective length of database: 505,134,102
effective search space: 662735941824
effective search space used: 662735941824
T: 11
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 82 (36.2 bits)
Medicago: description of AC147471.11


