

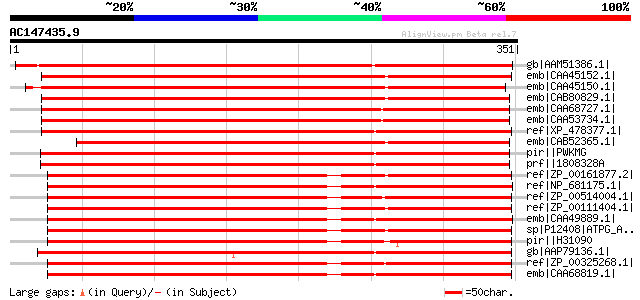
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.9 + phase: 0
(351 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM51386.1| putative ATP synthase gamma-subunit [Arabidopsis ... 456 e-127
emb|CAA45152.1| ATP synthase (gamma subunit) [Nicotiana tabacum]... 454 e-126
emb|CAA45150.1| ATP synthase (gamma subunit) [Pisum sativum] gi|... 440 e-122
emb|CAB80829.1| AT4g04640 [Arabidopsis thaliana] gi|21594056|gb|... 439 e-122
emb|CAA68727.1| ATP synthase [Spinacia oleracea] 436 e-121
emb|CAA53734.1| gamma subunit of the chloroplast ATP synthase [S... 436 e-121
ref|XP_478377.1| putative ATP synthase gamma chain 1, chloroplas... 428 e-118
emb|CAB52365.1| ATP synthase gamma chain, chloroplast precursor ... 405 e-111
pir||PWKMG H+-transporting two-sector ATPase (EC 3.6.3.14) gamma... 356 5e-97
prf||1808328A CF1 ATP synthase:SUBUNIT=gamma 353 4e-96
ref|ZP_00161877.2| COG0224: F0F1-type ATP synthase, gamma subuni... 306 5e-82
ref|NP_681175.1| H+-transporting ATP synthase gamma chain [Therm... 306 6e-82
ref|ZP_00514004.1| H+-transporting two-sector ATPase, gamma subu... 305 1e-81
ref|ZP_00111404.1| COG0224: F0F1-type ATP synthase, gamma subuni... 305 1e-81
emb|CAA49889.1| ATP synthase (gamma); H(+)-transporting ATP synt... 304 3e-81
sp|P12408|ATPG_ANASP ATP synthase gamma chain gi|17134982|dbj|BA... 304 3e-81
pir||H31090 H+-transporting two-sector ATPase (EC 3.6.3.14) gamm... 302 1e-80
gb|AAP79136.1| ATP synthase gamma subunit [Bigelowiella natans] 299 7e-80
ref|ZP_00325268.1| COG0224: F0F1-type ATP synthase, gamma subuni... 299 1e-79
emb|CAA68819.1| unnamed protein product [Synechocystis sp. PCC 6... 296 6e-79
>gb|AAM51386.1| putative ATP synthase gamma-subunit [Arabidopsis thaliana]
gi|17529042|gb|AAL38731.1| putative ATP synthase
gamma-subunit [Arabidopsis thaliana]
gi|15218281|ref|NP_173022.1| ATP synthase gamma chain 2,
chloroplast (ATPC2) [Arabidopsis thaliana]
gi|81636|pir||A39732 H+-transporting two-sector ATPase
(EC 3.6.3.14) gamma-2 chain precursor, chloroplast -
Arabidopsis thaliana gi|461551|sp|Q01909|ATPG2_ARATH ATP
synthase gamma chain 2, chloroplast precursor
gi|166788|gb|AAA32833.1| ATP synthase gamma-subunit
gi|8927649|gb|AAF82140.1| Identical to ATP sythase gamma
subunit (atpC2) from Arabidopsis thaliana gb|M61742 and
contains an ATP synthase PF|00231 domain
Length = 386
Score = 456 bits (1172), Expect = e-127
Identities = 243/345 (70%), Positives = 284/345 (81%), Gaps = 3/345 (0%)
Query: 5 TITNGRIPSKPHFPQHFQIRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVIN 64
++T RI S+ P QIR GIRE+R+RI+SVK TQKITEAM+LVAAARVRRAQ+AVI
Sbjct: 42 SLTPNRISSRSPLPS-IQIRAGIRELRERIDSVKNTQKITEAMRLVAAARVRRAQDAVIK 100
Query: 65 SRPFSEAFAETLHSINQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKA 124
RPF+E E L+SINQS Q +D+ PL+ VRPVK VAL+V+TGD+GLCGGFNN+V KKA
Sbjct: 101 GRPFTETLVEILYSINQSAQLEDIDFPLSIVRPVKRVALVVVTGDKGLCGGFNNAVTKKA 160
Query: 125 EARVMELKNLGINCVVISVGKKGSSYFNRSGFVEVDRFIDNVG-FPTTKDAQIIADDVFS 183
RV ELK GI+CVVISVGKKG++YF+R +VD+ I+ G FPTTK+AQ+IADDVFS
Sbjct: 161 TLRVQELKQRGIDCVVISVGKKGNAYFSRRDEFDVDKCIEGGGVFPTTKEAQVIADDVFS 220
Query: 184 LFVTEEVDKVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSK 243
LFV+EEVDKVELVYTKFVSLV+ +PVI TLLPLS KGE DV G VD +EDE FRLTSK
Sbjct: 221 LFVSEEVDKVELVYTKFVSLVKSDPVIHTLLPLSMKGESCDVKGECVDAIEDEMFRLTSK 280
Query: 244 DGKLALKRDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAAR 303
DGKLA++R K + K P+M+FEQDP QILDAMMPLYLNSQ+L+ALQESLASELA+R
Sbjct: 281 DGKLAVER-TKLEVEKPEISPLMQFEQDPVQILDAMMPLYLNSQILRALQESLASELASR 339
Query: 304 MGAMSNATDNAVELTKELSVAYNRERQAKITGEILEIVAGAEALR 348
M AMSNATDNAVEL K L++AYNR RQAKITGE+LEIVAGAEALR
Sbjct: 340 MNAMSNATDNAVELKKNLTMAYNRARQAKITGELLEIVAGAEALR 384
>emb|CAA45152.1| ATP synthase (gamma subunit) [Nicotiana tabacum]
gi|67880|pir||PWNTG H+-transporting two-sector ATPase
(EC 3.6.3.14) gamma chain precursor, chloroplast -
common tobacco gi|231610|sp|P29790|ATPG_TOBAC ATP
synthase gamma chain, chloroplast precursor
Length = 377
Score = 454 bits (1169), Expect = e-126
Identities = 228/325 (70%), Positives = 273/325 (83%), Gaps = 1/325 (0%)
Query: 23 IRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQS 82
+ CG+R++RDRI SVK TQKITEAMKLVAAA+VRRAQEAV+ +RPFSE E L++IN+
Sbjct: 53 VHCGLRDLRDRIESVKNTQKITEAMKLVAAAKVRRAQEAVVGARPFSETLVEVLYNINEQ 112
Query: 83 LQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVIS 142
LQ DD+ VPLT VRPVK VAL+V+TGDRGLCGGFNN + KKAEAR+ +LK LGI+ +IS
Sbjct: 113 LQTDDIDVPLTKVRPVKKVALVVVTGDRGLCGGFNNYLIKKAEARIRDLKALGIDYTIIS 172
Query: 143 VGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVS 202
VGKKG+SYF R ++ VD+F++ PT KDAQ IADDVFSLFV+EEVDKVEL+YTKFVS
Sbjct: 173 VGKKGNSYFIRRPYIPVDKFLEGSNLPTAKDAQAIADDVFSLFVSEEVDKVELLYTKFVS 232
Query: 203 LVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGF 262
LV+ PVI TLLPLS KGE+ D+NGN VD DEFFRLT+K+GKL ++RD+ + K D F
Sbjct: 233 LVKSEPVIHTLLPLSPKGEICDINGNCVDAANDEFFRLTTKEGKLTVERDIIRTKTTD-F 291
Query: 263 VPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELS 322
P+++FEQDP QILDA++PLYLNSQ+L+ALQESLASELAARM AMS+ATDNA EL K LS
Sbjct: 292 SPILQFEQDPVQILDALLPLYLNSQILRALQESLASELAARMSAMSSATDNATELKKNLS 351
Query: 323 VAYNRERQAKITGEILEIVAGAEAL 347
YNR+RQAKITGEILEIVAGA+AL
Sbjct: 352 RVYNRQRQAKITGEILEIVAGADAL 376
>emb|CAA45150.1| ATP synthase (gamma subunit) [Pisum sativum] gi|282923|pir||S27976
H+-transporting two-sector ATPase (EC 3.6.3.14) gamma
chain precursor, chloroplast - garden pea
gi|114640|sp|P28552|ATPG_PEA ATP synthase gamma chain,
chloroplast precursor
Length = 376
Score = 440 bits (1131), Expect = e-122
Identities = 222/332 (66%), Positives = 271/332 (80%), Gaps = 6/332 (1%)
Query: 12 PSKPHFPQHFQIRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEA 71
PS+P I+CG++++++RI+SVK TQKITEAMKLVAAA+VRRAQEAV+N RPFSE
Sbjct: 44 PSRPT-----SIQCGLKDLKNRIDSVKNTQKITEAMKLVAAAKVRRAQEAVVNGRPFSET 98
Query: 72 FAETLHSINQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMEL 131
E L+SIN+ LQ DD+ PLT +RPVK VAL+V TGDRGLCGGFNN++ KKAEAR+ EL
Sbjct: 99 LVEVLYSINEQLQTDDIESPLTKLRPVKKVALVVCTGDRGLCGGFNNAILKKAEARIAEL 158
Query: 132 KNLGINCVVISVGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVD 191
K LG+ V+SVG+KG+SYFNR ++ VDRF++ PT K+AQ IADDVFSLFV+EEVD
Sbjct: 159 KELGLEYTVVSVGRKGNSYFNRRPYIPVDRFLEGGSLPTAKEAQTIADDVFSLFVSEEVD 218
Query: 192 KVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKR 251
KVEL+YTKFVSLV+ NP+I TLLPLS KGE+ D+NGN VD EDE FRLT+K+GKL ++R
Sbjct: 219 KVELLYTKFVSLVKSNPIIHTLLPLSPKGEICDINGNCVDAAEDELFRLTTKEGKLTVER 278
Query: 252 DVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNAT 311
DV + K D F P+++FEQDP QILDA++PLYLNSQ+L+ LQESLASELAARM AMS+A
Sbjct: 279 DVIRSKTVD-FSPILQFEQDPVQILDALLPLYLNSQILRPLQESLASELAARMSAMSSAF 337
Query: 312 DNAVELTKELSVAYNRERQAKITGEILEIVAG 343
DNA EL +L+ YNR QAKITGEILEIVAG
Sbjct: 338 DNASELKTDLTRVYNRATQAKITGEILEIVAG 369
>emb|CAB80829.1| AT4g04640 [Arabidopsis thaliana] gi|21594056|gb|AAM65974.1| ATP
synthase gamma-subunit, putative [Arabidopsis thaliana]
gi|20148353|gb|AAM10067.1| unknown protein [Arabidopsis
thaliana] gi|17473929|gb|AAL38375.1| unknown protein
[Arabidopsis thaliana] gi|16226524|gb|AAL16191.1|
AT4g04640/T19J18_4 [Arabidopsis thaliana]
gi|5732056|gb|AAD48955.1| Arabidopsis thaliana APC1-ATP
synthase gamma chain 1 (GB:M61741); contains similarity
to Pfam PF00231 -ATP synthase; score=658.6, E=3.1e-194n
n+1 gi|18412632|ref|NP_567265.1| ATP synthase gamma
chain 1, chloroplast (ATPC1) [Arabidopsis thaliana]
gi|81635|pir||B39732 H+-transporting two-sector ATPase
(EC 3.6.3.14) gamma-1 chain precursor, chloroplast -
Arabidopsis thaliana gi|461550|sp|Q01908|ATPG1_ARATH ATP
synthase gamma chain 1, chloroplast precursor
gi|166632|gb|AAA32753.1| ATP synthase gamma-subunit
Length = 373
Score = 439 bits (1129), Expect = e-122
Identities = 221/324 (68%), Positives = 269/324 (82%), Gaps = 1/324 (0%)
Query: 23 IRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQS 82
++ +RE+RDRI+SVK TQKITEAMKLVAAA+VRRAQEAV+N RPFSE E L++IN+
Sbjct: 49 LQASLRELRDRIDSVKNTQKITEAMKLVAAAKVRRAQEAVVNGRPFSETLVEVLYNINEQ 108
Query: 83 LQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVIS 142
LQ DDV VPLT VRPVK VAL+V+TGDRGLCGGFNN + KKAEAR+ ELK LG+ VIS
Sbjct: 109 LQTDDVDVPLTKVRPVKKVALVVVTGDRGLCGGFNNFIIKKAEARIKELKGLGLEYTVIS 168
Query: 143 VGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVS 202
VGKKG+SYF R ++ VD++++ PT K+AQ +ADDVFSLF++EEVDKVEL+YTKFVS
Sbjct: 169 VGKKGNSYFLRRPYIPVDKYLEAGTLPTAKEAQAVADDVFSLFISEEVDKVELLYTKFVS 228
Query: 203 LVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGF 262
LV+ PVI TLLPLS KGE+ D+NG VD EDEFFRLT+K+GKL ++R+ + D F
Sbjct: 229 LVKSEPVIHTLLPLSPKGEICDINGTCVDAAEDEFFRLTTKEGKLTVERETFRTPTAD-F 287
Query: 263 VPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELS 322
P+++FEQDP QILDA++PLYLNSQ+L+ALQESLASELAARM AMS+A+DNA +L K LS
Sbjct: 288 SPILQFEQDPVQILDALLPLYLNSQILRALQESLASELAARMSAMSSASDNASDLKKSLS 347
Query: 323 VAYNRERQAKITGEILEIVAGAEA 346
+ YNR+RQAKITGEILEIVAGA A
Sbjct: 348 MVYNRKRQAKITGEILEIVAGANA 371
>emb|CAA68727.1| ATP synthase [Spinacia oleracea]
Length = 329
Score = 436 bits (1120), Expect = e-121
Identities = 223/325 (68%), Positives = 268/325 (81%), Gaps = 2/325 (0%)
Query: 23 IRCG-IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQ 81
I+C +RE+RDRI SVK TQKITEAMKLVAAA+VRRAQEAV+N RPFSE E L+++N+
Sbjct: 4 IQCANLRELRDRIGSVKNTQKITEAMKLVAAAKVRRAQEAVVNGRPFSETLVEVLYNMNE 63
Query: 82 SLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVI 141
LQ +DV VPLT +R VK VAL+V+TGDRGLCGGFNN + KKAE+R+ ELK LG++ +I
Sbjct: 64 QLQTEDVDVPLTKIRTVKKVALMVVTGDRGLCGGFNNMLLKKAESRIAELKKLGVDYTII 123
Query: 142 SVGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFV 201
S+GKKG++YF R + VDR+ D PT K+AQ IADDVFSLFV+EEVDKVE++YTKFV
Sbjct: 124 SIGKKGNTYFIRRPEIPVDRYFDGTNLPTAKEAQAIADDVFSLFVSEEVDKVEMLYTKFV 183
Query: 202 SLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDG 261
SLV+ +PVI TLLPLS KGE+ D+NG VD EDE FRLT+K+GKL ++RD+ K +
Sbjct: 184 SLVKSDPVIHTLLPLSPKGEICDINGKCVDAAEDELFRLTTKEGKLTVERDMIKTE-TPA 242
Query: 262 FVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKEL 321
F P++EFEQDPAQILDA++PLYLNSQ+L+ALQESLASELAARM AMSNATDNA EL K L
Sbjct: 243 FSPILEFEQDPAQILDALLPLYLNSQILRALQESLASELAARMTAMSNATDNANELKKTL 302
Query: 322 SVAYNRERQAKITGEILEIVAGAEA 346
S+ YNR RQAKITGEILEIVAGA A
Sbjct: 303 SINYNRARQAKITGEILEIVAGANA 327
>emb|CAA53734.1| gamma subunit of the chloroplast ATP synthase [Spinacia oleracea]
gi|21238|emb|CAA35158.1| gamma-subunit of chloroplast
ATP synthase [Spinacia oleracea] gi|67879|pir||PWSPG
H+-transporting two-sector ATPase (EC 3.6.3.14) gamma
chain precursor, chloroplast - spinach
gi|114643|sp|P05435|ATPG_SPIOL ATP synthase gamma chain,
chloroplast precursor
Length = 364
Score = 436 bits (1120), Expect = e-121
Identities = 223/325 (68%), Positives = 268/325 (81%), Gaps = 2/325 (0%)
Query: 23 IRCG-IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQ 81
I+C +RE+RDRI SVK TQKITEAMKLVAAA+VRRAQEAV+N RPFSE E L+++N+
Sbjct: 39 IQCANLRELRDRIGSVKNTQKITEAMKLVAAAKVRRAQEAVVNGRPFSETLVEVLYNMNE 98
Query: 82 SLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVI 141
LQ +DV VPLT +R VK VAL+V+TGDRGLCGGFNN + KKAE+R+ ELK LG++ +I
Sbjct: 99 QLQTEDVDVPLTKIRTVKKVALMVVTGDRGLCGGFNNMLLKKAESRIAELKKLGVDYTII 158
Query: 142 SVGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFV 201
S+GKKG++YF R + VDR+ D PT K+AQ IADDVFSLFV+EEVDKVE++YTKFV
Sbjct: 159 SIGKKGNTYFIRRPEIPVDRYFDGTNLPTAKEAQAIADDVFSLFVSEEVDKVEMLYTKFV 218
Query: 202 SLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDG 261
SLV+ +PVI TLLPLS KGE+ D+NG VD EDE FRLT+K+GKL ++RD+ K +
Sbjct: 219 SLVKSDPVIHTLLPLSPKGEICDINGKCVDAAEDELFRLTTKEGKLTVERDMIKTE-TPA 277
Query: 262 FVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKEL 321
F P++EFEQDPAQILDA++PLYLNSQ+L+ALQESLASELAARM AMSNATDNA EL K L
Sbjct: 278 FSPILEFEQDPAQILDALLPLYLNSQILRALQESLASELAARMTAMSNATDNANELKKTL 337
Query: 322 SVAYNRERQAKITGEILEIVAGAEA 346
S+ YNR RQAKITGEILEIVAGA A
Sbjct: 338 SINYNRARQAKITGEILEIVAGANA 362
>ref|XP_478377.1| putative ATP synthase gamma chain 1, chloroplast (H(+)-transporting
two-sector ATPase/F(1)-ATPase/ATPC1) [Oryza sativa
(japonica cultivar-group)] gi|50509502|dbj|BAD31183.1|
putative ATP synthase gamma chain 1, chloroplast
(H(+)-transporting two-sector ATPase/F(1)-ATPase/ATPC1)
[Oryza sativa (japonica cultivar-group)]
gi|27818005|dbj|BAC55768.1| putative ATP synthase gamma
chain 1, chloroplast (H(+)-transporting two-sector
ATPase/F(1)-ATPase/ATPC1) [Oryza sativa (japonica
cultivar-group)]
Length = 358
Score = 428 bits (1100), Expect = e-118
Identities = 215/325 (66%), Positives = 270/325 (82%), Gaps = 1/325 (0%)
Query: 23 IRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQS 82
+RC +RE+R RI+SV+ TQKITEAMKLVAAA+VRRAQEAV++SRPFSEA E L+++NQ
Sbjct: 34 VRCSLRELRSRIDSVRNTQKITEAMKLVAAAKVRRAQEAVVSSRPFSEALVEVLYNMNQE 93
Query: 83 LQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVIS 142
+Q +D+ +PLT +RPVK VAL+V+TG+RGLCG FNN+V KKAE R+ ELK LG+ V+S
Sbjct: 94 IQTEDIDLPLTRIRPVKKVALVVLTGERGLCGSFNNNVLKKAETRIEELKQLGLEYTVVS 153
Query: 143 VGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVS 202
VGKKG++YF R F+ +R ++ G PT KD+Q I D V+SLFV+E VDKVEL+Y+KFVS
Sbjct: 154 VGKKGNAYFIRRPFIPTERTLEVNGIPTVKDSQSICDLVYSLFVSEAVDKVELLYSKFVS 213
Query: 203 LVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGF 262
LVR +P+IQTLLP+S KGE+ D+NG VD EDE FRLT+K+GKL ++R+ K K F
Sbjct: 214 LVRSDPIIQTLLPMSPKGEICDINGVCVDATEDELFRLTTKEGKLTVERE-KVKIETQPF 272
Query: 263 VPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELS 322
PV++FEQDP QILDA++PLYLNSQ+L+ALQESLASELAARM AMS+ATDNA+EL K LS
Sbjct: 273 SPVVQFEQDPVQILDALLPLYLNSQILRALQESLASELAARMSAMSSATDNAIELRKNLS 332
Query: 323 VAYNRERQAKITGEILEIVAGAEAL 347
+ YNR+RQAKITGEILEIVAGA+AL
Sbjct: 333 MVYNRQRQAKITGEILEIVAGADAL 357
>emb|CAB52365.1| ATP synthase gamma chain, chloroplast precursor [Arabidopsis
thaliana]
Length = 302
Score = 405 bits (1040), Expect = e-111
Identities = 205/300 (68%), Positives = 247/300 (82%), Gaps = 1/300 (0%)
Query: 47 MKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQNDDVVVPLTAVRPVKTVALIVI 106
MKLVAAA+VRRAQEAV+N RPFSE E L++IN+ LQ DDV VPLT VRPVK VAL+V+
Sbjct: 2 MKLVAAAKVRRAQEAVVNGRPFSETLVEVLYNINEQLQTDDVDVPLTKVRPVKKVALVVV 61
Query: 107 TGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKKGSSYFNRSGFVEVDRFIDNV 166
TGDRGLCGGFNN + KKAEAR+ ELK LG+ VISVGKKG+SYF R ++ VD++++
Sbjct: 62 TGDRGLCGGFNNFIIKKAEARIKELKGLGLEYTVISVGKKGNSYFLRRPYIPVDKYLEAG 121
Query: 167 GFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVN 226
PT K+AQ +ADDVFSLF++EEVDKVEL+YTKFVSLV+ PVI TLLPLS KGE+ D+N
Sbjct: 122 TLPTAKEAQAVADDVFSLFISEEVDKVELLYTKFVSLVKSEPVIHTLLPLSPKGEICDIN 181
Query: 227 GNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNS 286
G VD EDEFFRLT+K+GKL ++R+ + D F P++ FEQDP QILDA++PLYLNS
Sbjct: 182 GTCVDAAEDEFFRLTTKEGKLTVERETFRTPTAD-FSPILLFEQDPVQILDALLPLYLNS 240
Query: 287 QVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYNRERQAKITGEILEIVAGAEA 346
Q+L+ALQESLASELAARM AMS+A+DNA +L K LS+ YNR+RQAKITGEILEIVAGA A
Sbjct: 241 QILRALQESLASELAARMSAMSSASDNASDLKKSLSMVYNRKRQAKITGEILEIVAGANA 300
>pir||PWKMG H+-transporting two-sector ATPase (EC 3.6.3.14) gamma chain
precursor, chloroplast - Chlamydomonas reinhardtii
gi|114638|sp|P12113|ATPG_CHLRE ATP synthase gamma chain,
chloroplast precursor gi|167405|gb|AAA33080.1| ATP
synthase precursor gi|167403|gb|AAA33079.1| ATP synthase
gamma-subunit
Length = 358
Score = 356 bits (914), Expect = 5e-97
Identities = 181/325 (55%), Positives = 245/325 (74%), Gaps = 1/325 (0%)
Query: 22 QIRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQ 81
Q+ G++E+RDRI SVK TQKIT+AMKLVAAA+VRRAQEAV+N RPFSE + L+ +NQ
Sbjct: 32 QVVAGLKEVRDRIASVKNTQKITDAMKLVAAAKVRRAQEAVVNGRPFSENLVKVLYGVNQ 91
Query: 82 SLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVI 141
++ +DV PL AVRPVK+V L+V+TGDRGLCGG+NN + KK EAR EL +G+ ++
Sbjct: 92 RVRQEDVDSPLCAVRPVKSVLLVVLTGDRGLCGGYNNFIIKKTEARYRELTAMGVKVNLV 151
Query: 142 SVGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFV 201
VG+KG+ YF R + + P+TK+AQ IAD++F+ F+ +E DKVELV+TKF+
Sbjct: 152 CVGRKGAQYFARRKQYNIVKSFSLGAAPSTKEAQGIADEIFASFIAQESDKVELVFTKFI 211
Query: 202 SLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDG 261
SL+ NP IQTLLP++ GE+ DV+G VD +DE F+LT+K G+ A++R+ K +
Sbjct: 212 SLINSNPTIQTLLPMTPMGELCDVDGKCVDAADDEIFKLTTKGGEFAVERE-KTTIETEA 270
Query: 262 FVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKEL 321
P + FEQ+PAQILDA++PLY++S +L++LQE+LASELAARM AM+NA+DNA EL K L
Sbjct: 271 LDPSLIFEQEPAQILDALLPLYMSSCLLRSLQEALASELAARMNAMNNASDNAKELKKGL 330
Query: 322 SVAYNRERQAKITGEILEIVAGAEA 346
+V YN++RQAKIT E+ EIV GA A
Sbjct: 331 TVQYNKQRQAKITQELAEIVGGAAA 355
>prf||1808328A CF1 ATP synthase:SUBUNIT=gamma
Length = 358
Score = 353 bits (906), Expect = 4e-96
Identities = 180/325 (55%), Positives = 244/325 (74%), Gaps = 1/325 (0%)
Query: 22 QIRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQ 81
Q+ G++E+RDRI SVK TQKIT+AMKLVAAA+VRRAQEAV+N R FSE + L+ +NQ
Sbjct: 32 QVVAGLKEVRDRIASVKNTQKITDAMKLVAAAKVRRAQEAVVNGRTFSENLVKVLYGVNQ 91
Query: 82 SLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVI 141
++ +DV PL AVRPVK+V L+V+TGDRGLCGG+NN + KK EAR EL +G+ ++
Sbjct: 92 RVRQEDVDSPLCAVRPVKSVLLVVLTGDRGLCGGYNNFIIKKTEARYRELTAMGVKVNLV 151
Query: 142 SVGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFV 201
VG+KG+ YF R + + P+TK+AQ IAD++F+ F+ +E DKVELV+TKF+
Sbjct: 152 CVGRKGAQYFARRKQYNIVKSFSLGAAPSTKEAQGIADEIFASFIAQESDKVELVFTKFI 211
Query: 202 SLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDG 261
SL+ NP IQTLLP++ GE+ DV+G VD +DE F+LT+K G+ A++R+ K +
Sbjct: 212 SLINSNPTIQTLLPMTPMGELCDVDGKCVDAADDEIFKLTTKGGEFAVERE-KTTIETEA 270
Query: 262 FVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKEL 321
P + FEQ+PAQILDA++PLY++S +L++LQE+LASELAARM AM+NA+DNA EL K L
Sbjct: 271 LDPSLIFEQEPAQILDALLPLYMSSCLLRSLQEALASELAARMNAMNNASDNAKELKKGL 330
Query: 322 SVAYNRERQAKITGEILEIVAGAEA 346
+V YN++RQAKIT E+ EIV GA A
Sbjct: 331 TVQYNKQRQAKITQELAEIVGGAAA 355
>ref|ZP_00161877.2| COG0224: F0F1-type ATP synthase, gamma subunit [Anabaena variabilis
ATCC 29413]
Length = 315
Score = 306 bits (785), Expect = 5e-82
Identities = 167/321 (52%), Positives = 226/321 (70%), Gaps = 10/321 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI SVK T+KITEAM+LVAAARVRRAQE VI +RPF++ A+ L+ + L+ +
Sbjct: 4 LKSIRDRIQSVKNTKKITEAMRLVAAARVRRAQEQVIATRPFADRLAQVLYGLQTRLRFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
DV +PL R VK+V L+VI+GDRGLCGG+N +V ++AE R ELK G++ + VG+K
Sbjct: 64 DVDLPLLKKREVKSVGLLVISGDRGLCGGYNTNVIRRAENRAKELKKEGLDYTFVIVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
YF R + PT ++A IAD++ SLF++E+VD++ELVYT+FVSLV
Sbjct: 124 AEQYFRRREQPIDASYTGLEQIPTAEEANKIADELLSLFLSEKVDRIELVYTRFVSLVSS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PVIQTLLPL +G ++ +DE FRLT++ G+ ++R + + M
Sbjct: 184 RPVIQTLLPLDTQG---------LEAADDEIFRLTTRGGQFEVERQTVTTQARP-LPRDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILD+++PLYL++Q+L+ALQES ASELAARM AMSNA+DNA EL K LS++YN
Sbjct: 234 IFEQDPVQILDSLLPLYLSNQLLRALQESAASELAARMTAMSNASDNAGELIKSLSLSYN 293
Query: 327 RERQAKITGEILEIVAGAEAL 347
+ RQA IT E+LE+V GAEAL
Sbjct: 294 KARQAAITQELLEVVGGAEAL 314
>ref|NP_681175.1| H+-transporting ATP synthase gamma chain [Thermosynechococcus
elongatus BP-1] gi|22294106|dbj|BAC07937.1|
H+-transporting ATP synthase gamma chain
[Thermosynechococcus elongatus BP-1]
Length = 315
Score = 306 bits (784), Expect = 6e-82
Identities = 165/322 (51%), Positives = 228/322 (70%), Gaps = 10/322 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI ++K T+KITEAM+LVAAA+VRRAQE V+ SRPF++ A+ L+S+ L+ +
Sbjct: 4 LKAIRDRIKTIKDTRKITEAMRLVAAAKVRRAQEQVMASRPFADRLAQVLYSLQTRLRFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
DV +PL A RPVKTVAL+V+TGDRGLCGG+N +V ++A+ R+ EL+ G+ ++ VG+K
Sbjct: 64 DVDLPLLAKRPVKTVALLVVTGDRGLCGGYNTNVIRRAKERLQELEAEGLKYTLVIVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
+ YF R + + P+ +A IA ++ SLF++E VD+VEL+YTKFVSL+
Sbjct: 124 AAQYFQRRDYPIDAVYSGLEQIPSASEAGQIASELLSLFLSETVDRVELIYTKFVSLISS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PV+QTLLPL +G ++ +DE FRLT++ L + R+ K M
Sbjct: 184 KPVVQTLLPLDPQG---------LETADDEIFRLTTRGSHLEVNRE-KVTSTLPALPSDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILDA++PLYLN+Q+L+ALQE+ ASELAARM AM+NA+DNA L L+++YN
Sbjct: 234 IFEQDPLQILDALLPLYLNNQLLRALQEAAASELAARMTAMNNASDNAQALIGTLTLSYN 293
Query: 327 RERQAKITGEILEIVAGAEALR 348
+ RQA IT EILE+VAGAEALR
Sbjct: 294 KARQAAITQEILEVVAGAEALR 315
>ref|ZP_00514004.1| H+-transporting two-sector ATPase, gamma subunit [Crocosphaera
watsonii WH 8501] gi|67857968|gb|EAM53207.1|
H+-transporting two-sector ATPase, gamma subunit
[Crocosphaera watsonii WH 8501]
Length = 314
Score = 305 bits (782), Expect = 1e-81
Identities = 163/321 (50%), Positives = 230/321 (70%), Gaps = 10/321 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI SVK T+KITEAM+LVAAA+VRRAQE VI++RPF++A A L ++ L++
Sbjct: 4 LKAIRDRIQSVKNTKKITEAMRLVAAAKVRRAQEQVISTRPFADALANVLFNLLNRLKSG 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
DV +PL R VKTVA++V++GDRGLCGG+N+ V ++ E R+ EL+ GI +I+VG+K
Sbjct: 64 DVSLPLLEQREVKTVAIVVVSGDRGLCGGYNSYVIRRTEQRIKELQAQGIGYRLITVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
+ YF+R + + PT +A IAD++ SLF++E VD+VEL+YT+FVSL+
Sbjct: 124 ATQYFSRRSAPIENTYTGLNQIPTADEAASIADELLSLFLSETVDRVELIYTRFVSLISS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PV+QTL PL+ +G ++ +DE FRL S +GKL ++R + M F M
Sbjct: 184 RPVVQTLAPLTIQG---------LETEDDEIFRLISSEGKLGVERSKVTQNM-SSFPQDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILDA++PLY+N+Q+L++LQES ASELAARM AMSNA+DNA +L L+++YN
Sbjct: 234 IFEQDPVQILDALLPLYVNNQLLRSLQESAASELAARMTAMSNASDNAGQLIGTLTLSYN 293
Query: 327 RERQAKITGEILEIVAGAEAL 347
+ RQA IT +++E+VAGA AL
Sbjct: 294 KARQAAITQQLMEVVAGANAL 314
>ref|ZP_00111404.1| COG0224: F0F1-type ATP synthase, gamma subunit [Nostoc punctiforme
PCC 73102]
Length = 315
Score = 305 bits (781), Expect = 1e-81
Identities = 162/321 (50%), Positives = 227/321 (70%), Gaps = 10/321 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI SVK T+KITEAM+LVAAARVRRAQE V+ +RPF++ A+ L+ + L+ +
Sbjct: 4 LKAIRDRIQSVKNTKKITEAMRLVAAARVRRAQEQVLATRPFADRLAQVLYGLQSRLRFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
+ +PL R VK+V L+VI+GDRGLCGG+NN+V ++AE R E+K G+N + VG+K
Sbjct: 64 EANLPLLRKREVKSVGLLVISGDRGLCGGYNNNVIRRAENRAKEIKAEGLNYQFVLVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
+ YF R + PT +A IAD + SLF++EEVD++EL+YT+F+SLV
Sbjct: 124 STQYFQRRDQPIDATYSGLEQIPTAAEANQIADQLLSLFLSEEVDRIELIYTRFLSLVSS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PV+QTLLPL +G ++ +DE FRLT++ GK ++R+ +++ P M
Sbjct: 184 RPVVQTLLPLDPQG---------LEAADDEIFRLTTRGGKFEVEREKVASQVRP-LAPDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILD+++PLYL++Q+L+ALQES ASELAARM AMSNA++NA EL L+++YN
Sbjct: 234 IFEQDPVQILDSLLPLYLSNQLLRALQESAASELAARMTAMSNASENAGELINTLTLSYN 293
Query: 327 RERQAKITGEILEIVAGAEAL 347
+ RQA IT E+LE+V GAEAL
Sbjct: 294 KARQAAITQELLEVVGGAEAL 314
>emb|CAA49889.1| ATP synthase (gamma); H(+)-transporting ATP synthase [Synechococcus
sp.] gi|480506|pir||S36979 H+-transporting two-sector
ATPase (EC 3.6.3.14) gamma chain - Synechococcus sp.
(PCC 6716) gi|461582|sp|Q05384|ATPG_SYNP1 ATP synthase
gamma chain
Length = 315
Score = 304 bits (778), Expect = 3e-81
Identities = 164/322 (50%), Positives = 227/322 (69%), Gaps = 10/322 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI ++K T+KITEAM+LVAAA+VRRAQE V+ SRPF++ A+ L+ + L+ +
Sbjct: 4 LKAIRDRIKTIKDTRKITEAMRLVAAAKVRRAQEQVMASRPFADRLAQVLYGLQTRLRFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
D +PL A RPVKTVAL+V+TGDRGLCGG+N +V ++A+ R EL+ GI ++ VG+K
Sbjct: 64 DANLPLLAKRPVKTVALLVVTGDRGLCGGYNTNVIRRAKERTEELEAEGIKYTLVIVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
+ YF R + + P+ +A IA+++ SLF++E VD+VEL+YTKFVSL+
Sbjct: 124 AAQYFQRRDYPIDAVYSGLEQIPSASEAGQIANELLSLFLSETVDRVELIYTKFVSLISS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PV+QTLLPL +G ++ +DE FRLT++ L + R+ K P M
Sbjct: 184 KPVVQTLLPLDPQG---------LEAADDEIFRLTTRASHLEVNRE-KVTSNLPALPPDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILDA++PLYL++Q+L+ALQE+ ASELAARM AM+NA+DNA L L+++YN
Sbjct: 234 IFEQDPVQILDALLPLYLSNQLLRALQEAAASELAARMTAMNNASDNAQTLIGTLTLSYN 293
Query: 327 RERQAKITGEILEIVAGAEALR 348
+ RQA IT EILE+VAGAEALR
Sbjct: 294 KARQAAITQEILEVVAGAEALR 315
>sp|P12408|ATPG_ANASP ATP synthase gamma chain gi|17134982|dbj|BAB77528.1| ATP synthase
subunit gamma [Nostoc sp. PCC 7120]
gi|17227500|ref|NP_484048.1| ATP synthase subunit gamma
[Nostoc sp. PCC 7120]
Length = 315
Score = 304 bits (778), Expect = 3e-81
Identities = 166/321 (51%), Positives = 225/321 (69%), Gaps = 10/321 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI SVK T+KITEAM+LVAAARVRRAQE VI +RPF++ A+ L+ + L+ +
Sbjct: 4 LKSIRDRIQSVKNTKKITEAMRLVAAARVRRAQEQVIATRPFADRLAQVLYGLQTRLRFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
DV +PL R VK+V L+VI+GDRGLCGG+N +V ++AE R ELK G++ + VG+K
Sbjct: 64 DVDLPLLKKREVKSVGLLVISGDRGLCGGYNTNVIRRAENRAKELKAEGLDYTFVIVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
YF R + PT +A IAD++ SLF++E+VD++ELVYT+FVSLV
Sbjct: 124 AEQYFRRREQPIDASYTGLEQIPTADEANKIADELLSLFLSEKVDRIELVYTRFVSLVSS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PVIQTLLPL +G ++ +DE FRLT++ G+ ++R + + M
Sbjct: 184 RPVIQTLLPLDTQG---------LEAADDEIFRLTTRGGQFQVERQTVTSQARP-LPRDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILD+++PLYL++Q+L+ALQES ASELAARM AMSNA++NA EL K LS++YN
Sbjct: 234 IFEQDPVQILDSLLPLYLSNQLLRALQESAASELAARMTAMSNASENAGELIKSLSLSYN 293
Query: 327 RERQAKITGEILEIVAGAEAL 347
+ RQA IT E+LE+V GAEAL
Sbjct: 294 KARQAAITQELLEVVGGAEAL 314
>pir||H31090 H+-transporting two-sector ATPase (EC 3.6.3.14) gamma chain -
Anabaena sp. (strain PCC 7120) gi|142004|gb|AAA21992.1|
ATP synthase subunit gamma [Nostoc sp. PCC 7120]
Length = 315
Score = 302 bits (773), Expect = 1e-80
Identities = 166/324 (51%), Positives = 226/324 (69%), Gaps = 16/324 (4%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI SVK T+KITEAM+LVAAARVRRAQE VI +RPF++ A+ L+ + L+ +
Sbjct: 4 LKSIRDRIQSVKNTKKITEAMRLVAAARVRRAQEQVIATRPFADRLAQVLYGLQTRLRFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
DV +PL R VK+V L+VI+GDRGLCGG+N +V ++AE R ELK G++ + VG+K
Sbjct: 64 DVDLPLLKKREVKSVGLLVISGDRGLCGGYNTNVIRRAENRAKELKAEGLDYTFVIVGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
YF R + PT +A IAD++ SLF++E+VD++ELVYT+FVSLV
Sbjct: 124 AEQYFRRREQPIDASYTGLEQIPTADEANKIADELLSLFLSEKVDRIELVYTRFVSLVSS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PVIQTLLPL +G ++ +DE FRLT++ G+ ++R + + P+
Sbjct: 184 RPVIQTLLPLDTQG---------LEAADDEIFRLTTRGGQFQVERQTVTSQAR----PLP 230
Query: 267 E---FEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSV 323
FEQDP QILD+++PLYL++Q+L+ALQES ASELAARM AMSNA++NA EL K LS+
Sbjct: 231 RDSIFEQDPVQILDSLLPLYLSNQLLRALQESAASELAARMTAMSNASENAGELIKSLSL 290
Query: 324 AYNRERQAKITGEILEIVAGAEAL 347
+YN+ RQA IT E+LE+V GAEAL
Sbjct: 291 SYNKARQAAITQELLEVVGGAEAL 314
>gb|AAP79136.1| ATP synthase gamma subunit [Bigelowiella natans]
Length = 390
Score = 299 bits (766), Expect = 7e-80
Identities = 164/337 (48%), Positives = 227/337 (66%), Gaps = 10/337 (2%)
Query: 20 HFQIRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSI 79
+ + ++E+R RI SV T+KIT +MKLVAAA+VR+AQ AV+ RPF+E +TL+ I
Sbjct: 54 NIRANANLKEVRGRIESVSNTKKITSSMKLVAAAKVRKAQAAVLGGRPFAENLVKTLYGI 113
Query: 80 NQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCV 139
NQ ++ + + LT VRPVK V ++ ++GDRGLCG +N V KK R EL+ +GI
Sbjct: 114 NQKVRAEALTSALTEVRPVKNVLILTVSGDRGLCGAYNTFVLKKLLTRTQELQGMGIGVK 173
Query: 140 VISVGKKGSSYFNR--------SGFVEVDRFIDNVGFPTTKDAQI-IADDVFSLFVTEEV 190
++VG+K ++ R + D F G DA +A++V S F EV
Sbjct: 174 HVAVGRKLQTWLKRRTDEYDVMGAYEMTDIFNQKAGDDENADALTGLAEEVLSSFTDGEV 233
Query: 191 DKVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALK 250
DKVE++YTKF SL+ +PVIQTLLPL+ KGE+ DVNGN +D EDE F LT+K G+LA++
Sbjct: 234 DKVEIIYTKFKSLIGSDPVIQTLLPLTPKGEICDVNGNCIDADEDEVFSLTTKGGELAVE 293
Query: 251 RDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNA 310
+ K++ G + FEQ P +++DA++PLY++S +L+++QESLASELAARM AMS+A
Sbjct: 294 TE-KQQLDTAGDLDKFMFEQQPDEVVDAVLPLYMDSTILRSIQESLASELAARMNAMSSA 352
Query: 311 TDNAVELTKELSVAYNRERQAKITGEILEIVAGAEAL 347
DNA EL +LS YNR RQAKIT EILEIVAGA+A+
Sbjct: 353 CDNADELGTKLSRIYNRGRQAKITNEILEIVAGADAV 389
>ref|ZP_00325268.1| COG0224: F0F1-type ATP synthase, gamma subunit [Trichodesmium
erythraeum IMS101]
Length = 314
Score = 299 bits (765), Expect = 1e-79
Identities = 158/321 (49%), Positives = 221/321 (68%), Gaps = 10/321 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI S+K T+KITEAM+LVAAA+VRRAQE VI +RPF++ A+ L+ + + LQ +
Sbjct: 4 LKAIRDRIKSIKNTKKITEAMRLVAAAKVRRAQEQVIGTRPFADRLAQVLYGLAERLQFE 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
+ +PL R KTV L+VI+G+RGLCG +N +V K+AE R EL GINC + +G+K
Sbjct: 64 SLDLPLLQKREAKTVGLLVISGNRGLCGSYNTNVIKRAEMRARELTQQGINCQYVLIGRK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
YF R ++ + + PT +A ++AD++ + F+ +D+VEL++T+F+SL+
Sbjct: 124 AIQYFQRREVPILNTYDNPEKIPTADNASVVADEILAGFMAGSIDRVELIFTRFISLISS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PVIQTLLPL +G ++V +DE FRLTSK G+ + R+ +K F M
Sbjct: 184 KPVIQTLLPLDPQG---------LEVKDDEIFRLTSKGGEFEVSRETVSAAVK-SFPRDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QILDA++PL+L +Q+L+A QES ASELA+RM AMSNA+DNA EL L++AYN
Sbjct: 234 IFEQDPIQILDALLPLFLTNQLLRAWQESSASELASRMTAMSNASDNASELAGTLTLAYN 293
Query: 327 RERQAKITGEILEIVAGAEAL 347
+ RQA IT EILE+V GA AL
Sbjct: 294 KARQAAITQEILEVVGGANAL 314
>emb|CAA68819.1| unnamed protein product [Synechocystis sp. PCC 6803]
gi|47514|emb|CAA41136.1| ATPase subunit gamma
[Synechocystis sp. PCC 6803]
gi|16329326|ref|NP_440054.1| ATP synthase g subunit
[Synechocystis sp. PCC 6803] gi|1651807|dbj|BAA16734.1|
ATP synthase g subunit [Synechocystis sp. PCC 6803]
gi|114645|sp|P17253|ATPG_SYNY3 ATP synthase gamma chain
gi|67877|pir||PWBYG H+-transporting two-sector ATPase
(EC 3.6.3.14) gamma chain - Synechocystis sp
Length = 314
Score = 296 bits (758), Expect = 6e-79
Identities = 160/321 (49%), Positives = 221/321 (68%), Gaps = 10/321 (3%)
Query: 27 IREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ IRDRI SVK T+KITEAM+LVAAA+VRRAQE V+++RPF++A A+ L+++ L
Sbjct: 4 LKAIRDRIQSVKNTKKITEAMRLVAAAKVRRAQEQVLSTRPFADALAQVLYNLQNRLSFA 63
Query: 87 DVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVISVGKK 146
+ +PL R K VAL+V+TGDRGLCGG+N + K+AE R ELKN GI ++ VG K
Sbjct: 64 ETELPLFEQREPKAVALLVVTGDRGLCGGYNVNAIKRAEQRAKELKNQGIAVKLVLVGSK 123
Query: 147 GSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRF 206
YF R + + + P +A IAD + +LFV+E VD+VEL+YT+FVSL+
Sbjct: 124 AKQYFGRRDYDVAASYANLEQIPNASEAAQIADSLVALFVSETVDRVELIYTRFVSLISS 183
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVM 266
PV+QTL PLS +G ++ +DE FRL ++ GK ++R+ K + + F M
Sbjct: 184 QPVVQTLFPLSPQG---------LEAPDDEIFRLITRGGKFQVERE-KVEAPVESFPQDM 233
Query: 267 EFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYN 326
FEQDP QIL+A++PLY +Q+L+ALQES ASELAARM AMSNA+DNA +L L+++YN
Sbjct: 234 IFEQDPVQILEALLPLYNTNQLLRALQESAASELAARMTAMSNASDNAGQLIGTLTLSYN 293
Query: 327 RERQAKITGEILEIVAGAEAL 347
+ RQA IT E+LE+VAGA +L
Sbjct: 294 KARQAAITQELLEVVAGANSL 314
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.135 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,168,118
Number of Sequences: 2540612
Number of extensions: 19255222
Number of successful extensions: 57879
Number of sequences better than 10.0: 472
Number of HSP's better than 10.0 without gapping: 455
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 56357
Number of HSP's gapped (non-prelim): 803
length of query: 351
length of database: 863,360,394
effective HSP length: 129
effective length of query: 222
effective length of database: 535,621,446
effective search space: 118907961012
effective search space used: 118907961012
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC147435.9


