

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.7 + phase: 0
(1667 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
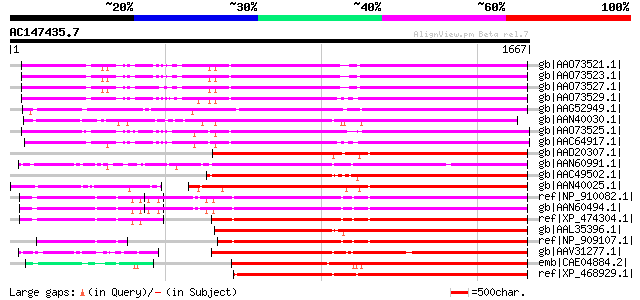
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO73521.1| gag-pol polyprotein [Glycine max] 1065 0.0
gb|AAO73523.1| gag-pol polyprotein [Glycine max] 1064 0.0
gb|AAO73527.1| gag-pol polyprotein [Glycine max] 1062 0.0
gb|AAO73529.1| gag-pol polyprotein [Glycine max] 1053 0.0
gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana] 1049 0.0
gb|AAN40030.1| putative gag-pol polyprotein [Zea mays] 1045 0.0
gb|AAO73525.1| gag-pol polyprotein [Glycine max] 1044 0.0
gb|AAC64917.1| gag-pol polyprotein [Glycine max] 1042 0.0
gb|AAD20307.1| copia-type pol polyprotein [Zea mays] 1018 0.0
gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 1011 0.0
gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein... 990 0.0
gb|AAN40025.1| putative gag-pol polyprotein [Zea mays] 984 0.0
ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (jap... 972 0.0
gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 970 0.0
ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultiva... 969 0.0
gb|AAL35396.1| Opie2a pol [Zea mays] 969 0.0
ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica... 966 0.0
gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cult... 937 0.0
emb|CAE04884.2| OSJNBa0042I15.6 [Oryza sativa (japonica cultivar... 936 0.0
ref|XP_468929.1| putative polyprotein [Oryza sativa (japonica cu... 920 0.0
>gb|AAO73521.1| gag-pol polyprotein [Glycine max]
Length = 1574
Score = 1065 bits (2754), Expect = 0.0
Identities = 640/1673 (38%), Positives = 921/1673 (54%), Gaps = 168/1673 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKISRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D ++
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLNT 243
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K KK D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKKSDVKPSHSKGIQC 302
Query: 315 FNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH I +CP K+ KG S+ S D +SE SD
Sbjct: 303 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 340
Query: 375 ADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 341 SDRDVNALTGIFET------------AEDSSDTDSEITFDELATSYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNK 494
LK+ DL ++++ E+ K E+ GF +++ E+ KS+ + +KGS
Sbjct: 389 AQLKKVIADLEAEKEAHKEEISELKGEV-GF--LNSKLENMTKSI-----KMLNKGSDT- 439
Query: 495 HEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKS 552
LD+ ++ G KN G +G+G++ + + ++ +
Sbjct: 440 ----LDEVLLLG--------------KNAGNQRGLGFNPKSAGRTTM------------T 469
Query: 553 TFVP-EGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTK-SDPKSQKITILKRSETVH 610
FVP + T Q +S G Q K SK + + K K + T
Sbjct: 470 EFVPAKNRTGATMSQHRSRHHGMQQK-KSKRKKWRCHYCGKYGHIKPFCYHLHPHHGTQS 528
Query: 611 QNLIKPESKIPKQKDQKNKAATASEKTIPK-------------GVKPKVLNDQKTLSIHP 657
N K +PK K T+ + + GVK +LN + + +
Sbjct: 529 SNSRKKMMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTSYV 588
Query: 658 --------KVQGRKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLV 709
K+ G + + S+N V LV GL NL+SISQ CD G++V F+K+ C LV
Sbjct: 589 TFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LV 647
Query: 710 NKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISK 769
+ + KG R ++ + CL S D+ +WH+R GH + R + KI
Sbjct: 648 TNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIID 707
Query: 770 LQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLY 829
V+G+PN+ +CG CQ GK VK S + +TSR LELLH+DL GP+ SL
Sbjct: 708 KGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLG 767
Query: 830 GSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENE 889
G +Y V+VDD+SR+TWVKFI+ K EVF ++Q EK+ I ++RSDHG EFEN
Sbjct: 768 GKRYAYVVVDDFSRFTWVKFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENS 827
Query: 890 PFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSC 949
FC GI HEFS+ TPQQNG+VERKNRTLQE AR M+H L + WAEA+NT+C
Sbjct: 828 RLTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTAC 887
Query: 950 YIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLG 1009
YI NR+ +R T YE++KGR+P++ +FH FG CYIL ++ +K D K+ GIFLG
Sbjct: 888 YIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLG 947
Query: 1010 YSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDS 1069
YS S+AYRV+NS T+ V ES++V DD P K + + + DA
Sbjct: 948 YSTNSRAYRVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTSGDNVADA--------- 998
Query: 1070 EKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEE 1129
A+S AE S +ES + IQ H +E
Sbjct: 999 -----------------AKSGENAENSDSATDESNINQPDKRSSTRIQ------KMHPKE 1035
Query: 1130 LIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVW 1189
LIIG + TRS + S +S IEPK V+EAL+D+ WI AMQEEL QF+RN+VW
Sbjct: 1036 LIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVW 1095
Query: 1190 DLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEA 1249
+LVP+P N+IGTKW+F+NK NE+G +TRNKARLVAQGY+Q EG+D+ ETFAPVARLE+
Sbjct: 1096 ELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLES 1155
Query: 1250 IRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGL 1309
IRLLL A LYQMDVKSAFLNG + EEVYV+QP GF D HP+HVY+LKK+LYGL
Sbjct: 1156 IRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKALYGL 1215
Query: 1310 KQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKE 1369
KQAPRAWY+RL+ FL + + +G +D TLF + ++++I QIYVDDI+FG + + +
Sbjct: 1216 KQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLRH 1275
Query: 1370 FSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMH 1429
F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q++Y K ++KKF +E+ TP
Sbjct: 1276 FVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKRTPAP 1335
Query: 1430 PTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKR 1489
LSK++ GT VDQ LYR MIGSLLYLTASRPDI ++V +CAR+Q++P+ SHLT VKR
Sbjct: 1336 THLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLTQVKR 1395
Query: 1490 IFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQ 1549
I +Y+ GT++ G++Y + L+G+CDAD+AG +RKSTSG C +LG NLISW SKKQ
Sbjct: 1396 ILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSKKQ 1455
Query: 1550 ATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSR 1609
+++STAEAEYI+A + C+QL+WMK L++Y + + + +YCDN +AI +SKNP+ HSR
Sbjct: 1456 NCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQHSR 1515
Query: 1610 AKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
KHI+I+HH+IRD V ++ ++ +DTE Q ADIFTK L +F+ ++ L +
Sbjct: 1516 TKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFEKLRGKLGI 1568
>gb|AAO73523.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 1064 bits (2752), Expect = 0.0
Identities = 639/1675 (38%), Positives = 920/1675 (54%), Gaps = 170/1675 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK L + EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDACEILKSTHEGTSKVKMSRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K K+ D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKRSDVKPSHSKGIQC 302
Query: 315 FNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH I +CP K+ KG S+ S D +SE SD
Sbjct: 303 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 340
Query: 375 ADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A +G+ T +ED ++ S+I EL S ++L E +
Sbjct: 341 SDRDVNALIGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNK 494
LK+ DL ++++ E+ K E+ GF +++ E+ KS+ + +KGS
Sbjct: 389 AQLKKVIADLEAEKEAHKEEISELKGEV-GF--LNSKLENMTKSI-----KMLNKGSDT- 439
Query: 495 HEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKS 552
LD+ ++ G KN G +G+G++ + + ++ +
Sbjct: 440 ----LDEVLLLG--------------KNAGNQRGLGFNPKSAGRTTM------------T 469
Query: 553 TFVP-EGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKSD---PKSQKITILKRSET 608
FVP + T Q +S G Q K SK + + K P + T
Sbjct: 470 EFVPAKNRTGATMSQHRSRHHGMQQK-KSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGT 528
Query: 609 VHQNLIKPESKIPKQKDQKNKAATASEKTIPK-------------GVKPKVLNDQKTLSI 655
N K +PK K T+ + + GVK +LN + +
Sbjct: 529 QSSNSRKKMMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTS 588
Query: 656 HP--------KVQGRKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCT 707
+ K+ G + + S+N V LV GL NL+SISQ CD G++V F+K+ C
Sbjct: 589 YVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC- 647
Query: 708 LVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKI 767
LV + + KG R ++ + CL S D+ +WH+R GH + R + KI
Sbjct: 648 LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKI 707
Query: 768 SKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTAS 827
V+G+PN+ +CG CQ GK VK S + +TSR LELLH+DL GP+ S
Sbjct: 708 LDKSAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVES 767
Query: 828 LYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFE 887
L G +Y V+VDD+SR+TWV FI+ K EVF ++Q EK+ I ++RSDHG EFE
Sbjct: 768 LGGKRYAYVVVDDFSRFTWVNFIREKSGTFEVFKKLSLRLQREKDCVIKRIRSDHGREFE 827
Query: 888 NEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNT 947
N F FC GI HEFS+ TPQQNG+VERKNRTLQE AR M+H L + WAEA+NT
Sbjct: 828 NSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNT 887
Query: 948 SCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIF 1007
+CYI NR+ +R T YE++KGR+P++ +FH FG CYIL ++ +K D K+ GIF
Sbjct: 888 ACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIF 947
Query: 1008 LGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPS 1067
LGYS S+AYRV+NS T+ V ES++V DD P K + + + DA
Sbjct: 948 LGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTSGDNVADA------- 1000
Query: 1068 DSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHH 1127
A+S AE S +ES + IQ H
Sbjct: 1001 -------------------AKSGENAENSDSATDESNINQPDKRSSTRIQ------KMHP 1035
Query: 1128 EELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRND 1187
+ELIIG + TRS + S +S IEPK V+EAL+D+ WI AMQEEL QF+RN+
Sbjct: 1036 KELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNE 1095
Query: 1188 VWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARL 1247
VW+LVP+P N+IGTKW+F+NK NE+G +TRNKARLVAQGY+Q EG+D+ ETFAPVARL
Sbjct: 1096 VWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARL 1155
Query: 1248 EAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLY 1307
E+IRLLL A LYQMDVKSAFLNG + EEVYV+QP GF D HP+HVY+LKK+LY
Sbjct: 1156 ESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKALY 1215
Query: 1308 GLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLC 1367
GLKQAPRAWY+RL+ FL + + +G +D TLF + ++++I QIYVDDI+FG + +
Sbjct: 1216 GLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEML 1275
Query: 1368 KEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTP 1427
+ F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q++Y K ++KKF +E+ TP
Sbjct: 1276 RHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKRTP 1335
Query: 1428 MHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAV 1487
LSK++ GT VDQK YR MIGSLLYLTASRPDI ++V +CAR+Q++P+ SHL V
Sbjct: 1336 APTHLKLSKDEAGTSVDQKPYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLNQV 1395
Query: 1488 KRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASK 1547
KRI +Y+ GT++ G++Y L+G+CDAD+AG +RKSTSG C +LG NLISW SK
Sbjct: 1396 KRILKYVNGTSDYGIMYCHCSSSMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSK 1455
Query: 1548 KQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILH 1607
KQ +++STAEAEYI+A + C+QL+WMK L++Y + + + +YCDN +AI +SKNP+ H
Sbjct: 1456 KQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQH 1515
Query: 1608 SRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
SR KHI+I+HH+IRD V ++ ++ +DTE Q ADIFTK L +F+ ++ L +
Sbjct: 1516 SRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFEKLRGKLGI 1570
>gb|AAO73527.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 1062 bits (2746), Expect = 0.0
Identities = 638/1675 (38%), Positives = 914/1675 (54%), Gaps = 170/1675 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K K+ D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKRSDVKPSHSKGIQC 302
Query: 315 FNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH I +CP K+ KG S+ S D +SE SD
Sbjct: 303 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 340
Query: 375 ADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 341 SDRDVNALTGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNK 494
LK+ DL ++++ E+ K E+ N T + +K L NK
Sbjct: 389 AQLKKVIADLEAEKEAHEEEISELKGEVGFLNSKLETMKKSIKML-------------NK 435
Query: 495 HEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKS 552
LD+ ++ G KN G +G+G++ + + ++ +
Sbjct: 436 GSDTLDEVLLLG--------------KNAGNQRGLGFNPKFAGRTTM------------T 469
Query: 553 TFVP-EGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKSD---PKSQKITILKRSET 608
FVP + T Q S G+Q K SK + + K P + T
Sbjct: 470 EFVPAKNRTGTTMSQHLSRHHGTQQK-KSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGT 528
Query: 609 VHQNLIKPESKIPKQKDQKNKAATASEKTIPK-------------GVKPKVLNDQKTLSI 655
N K +PK K T+ + + GVK +LN + +
Sbjct: 529 QSSNSRKKMMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTS 588
Query: 656 HP--------KVQGRKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCT 707
+ K+ G + + S+N V LV GL NL+SISQ CD G++V F+K+ C
Sbjct: 589 YVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC- 647
Query: 708 LVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKI 767
LV + + KG R ++ + CL S D+ +WH+R GH + R + KI
Sbjct: 648 LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKI 707
Query: 768 SKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTAS 827
V+G+PN+ +CG CQ GK VK S + +TSR LELLH+DL GP+ S
Sbjct: 708 IDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLRHQTTSRVLELLHMDLMGPMQVES 767
Query: 828 LYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFE 887
L G +Y V+VDD+SR+TWV FI+ K EVF ++Q EK+ I ++RSDHG EFE
Sbjct: 768 LGGKRYAYVVVDDFSRFTWVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFE 827
Query: 888 NEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNT 947
N F FC GI HEFS+ TPQQNG+VERKNRTLQE AR M+H L + WAEA+NT
Sbjct: 828 NSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNT 887
Query: 948 SCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIF 1007
+CYI NR+ +R T YE++KGR+P++ +FH FG CYIL ++ +K D K+ GIF
Sbjct: 888 ACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIF 947
Query: 1008 LGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPS 1067
LGYS S+AYRV+NS T+ V ES++V DD P K + + + DA
Sbjct: 948 LGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTLGDNVADA------- 1000
Query: 1068 DSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHH 1127
A+S AE S +ES + IQ H
Sbjct: 1001 -------------------AKSGENAENSDSATDESNINQPDKRSSTRIQ------KMHP 1035
Query: 1128 EELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRND 1187
+ELIIG + TRS + S +S IEPK V+EAL+D+ WI AMQEEL QF+RN+
Sbjct: 1036 KELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNE 1095
Query: 1188 VWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARL 1247
VW+LVP+P N+IGTKW+F+NK NE+G +TRNKARLVAQGY+Q EG+D+ ETFAPVARL
Sbjct: 1096 VWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARL 1155
Query: 1248 EAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLY 1307
E+IRLLL A LYQMDVKSAFLNG + EEVYV+QP GF D HP+HVY+LKK+LY
Sbjct: 1156 ESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKALY 1215
Query: 1308 GLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLC 1367
GLKQAPRAWY+RL+ FL + + +G +D TLF + ++++I QIYVDDI+FG + +
Sbjct: 1216 GLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEML 1275
Query: 1368 KEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTP 1427
+ F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q++Y K ++KKF +E+ TP
Sbjct: 1276 RHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKRTP 1335
Query: 1428 MHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAV 1487
LSK++ GT VDQ LYR MIGSLLYLTASRPDI ++V +CAR+Q++P+ SHLT V
Sbjct: 1336 APTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLTQV 1395
Query: 1488 KRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASK 1547
KRI +Y+ GT++ G++Y + L+G+CDAD+AG +RKSTSG C +LG NLISW SK
Sbjct: 1396 KRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSK 1455
Query: 1548 KQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILH 1607
KQ +++STAEAEYI+A + C+QL+WMK L++Y + + + +YCDN +AI +SKNP+ H
Sbjct: 1456 KQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQH 1515
Query: 1608 SRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
SR KHI+I+HH+IRD V ++ ++ +DTE Q ADIFTK L +F+ ++ L +
Sbjct: 1516 SRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFEKLRGKLGI 1570
>gb|AAO73529.1| gag-pol polyprotein [Glycine max]
Length = 1577
Score = 1053 bits (2724), Expect = 0.0
Identities = 632/1672 (37%), Positives = 927/1672 (54%), Gaps = 163/1672 (9%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 17 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMTILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ T KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 272 DEDQSVKMAMLSNK----LEYLARKQKKFLS------KRGSYKNSKKEDQ----KG--CF 315
DE + + L + L + R+QK + ++GS K +++ KG C
Sbjct: 244 DEGLTNAVVFLGKQFNKVLNRMDRRQKPHVRNISLDIRKGSEYQRKSDEKPSHSKGIQCR 303
Query: 316 NCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEA 375
C+ GH +CP K++ KG S+ S +D +SE SD +
Sbjct: 304 GCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESD---S 342
Query: 376 DDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 435
D D A G + +ED ++ S+I EL +EL E +
Sbjct: 343 DRDVNALTGRFES------------AEDSSDTDSEITFDELAIFYRELCIKSEKILQQEA 390
Query: 436 DLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKH 495
LK+ +L ++++ E+ K E+ GF +++ E+ KS+ + +KGS
Sbjct: 391 QLKKVIANLEAEKEAHEEEISKLKGEV-GF--LNSKLENMTKSI-----KMLNKGSD--- 439
Query: 496 EIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 555
LD+ + G KV + +G+G++ + + ++ + FV
Sbjct: 440 --MLDZVLQLG---KKVGNQ---------RGLGFNHKSAGRTTM------------TEFV 473
Query: 556 P-EGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKSDPKSQKITIL--------KRS 606
P + + T Q +S G+Q K SK + + K L + S
Sbjct: 474 PAKNSTGATMSQHRSRHHGTQQK-RSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQGS 532
Query: 607 ETVHQNLIKPESKIPKQKDQKNKAATASEKTI--------PKGVKPKVLNDQKTLSIHP- 657
+ + + P+ KI + A+A E GVK ++N + + +
Sbjct: 533 SSGRKMMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVT 592
Query: 658 -------KVQGRKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVN 710
K+ G + S+N V LV GL NL+SISQ CD G++V F+K+ C LV
Sbjct: 593 FGDGSKGKITGMGKLVHEGLPSLNKVLLVKGLTVNLISISQLCDEGFNVNFTKSEC-LVT 651
Query: 711 KDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKL 770
+ + KG R ++ + + CL S D+ +WH+R GH + R + KI
Sbjct: 652 NEKSEVLMKGSRSKDNCYLWTPQESSHSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDK 711
Query: 771 QLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYG 830
V+G+PN+ +CG CQ GK VK S + +TSR LELLH+DL GP+ SL G
Sbjct: 712 GAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGG 771
Query: 831 SKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEP 890
+Y V+VDD+SR+TWV FI+ K EVF ++Q EK+ I ++RSDHG EFEN
Sbjct: 772 KRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSK 831
Query: 891 FELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCY 950
F FC GI HEFS+ TPQQNG+VERKNRTLQE AR M+H L + WAEA+NT+CY
Sbjct: 832 FTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACY 891
Query: 951 IQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGY 1010
I NR+ +R T YE++KGR+P + +FH FG CYIL ++ +K D K+ GIFLGY
Sbjct: 892 IHNRVTLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGY 951
Query: 1011 SERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSE 1070
S S+AYRV+NS T+ V ES++V DD P K + + + D +D+
Sbjct: 952 STNSRAYRVFNSRTRTVMESINVVVDDLTPARKKDVEEDVR---------TSGDNVADTA 1002
Query: 1071 KYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEEL 1130
K E AE++ A P + +P + + H +EL
Sbjct: 1003 KSAE-----------NAENSDSATDEPNINQPDK------------RPSIRIQKMHPKEL 1039
Query: 1131 IIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWD 1190
IIG + TRS + S +S IEPK V+EAL+D+ WI AMQEEL QF+RN+VW+
Sbjct: 1040 IIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWE 1099
Query: 1191 LVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAI 1250
LVP+P N+IGTKW+F+NK NE+G +TRNKARLVAQGY+Q EG+D+ ETFAPVARLE+I
Sbjct: 1100 LVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLESI 1159
Query: 1251 RLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLK 1310
RLLL A LYQMDVKSAFLNG + EE YV+QP GF D HP+HVY+LKK+LYGLK
Sbjct: 1160 RLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYGLK 1219
Query: 1311 QAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEF 1370
QAPRAWY+RL+ FL + + +G +D TLF + ++++I QIYVDDI+FG + + + F
Sbjct: 1220 QAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLRHF 1279
Query: 1371 SKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHP 1430
+ MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+KY K ++KKF +E+ TP
Sbjct: 1280 VQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASHKRTPAPT 1339
Query: 1431 TCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRI 1490
LSK++ GT VDQ LYR MIGSLLYLTASRPDI ++V +CAR+Q++P+ SHL VKRI
Sbjct: 1340 HLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLNQVKRI 1399
Query: 1491 FRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQA 1550
+Y+ GT++ G++Y L+G+CDAD+AG +RKSTSG C +LG NLISW SKKQ
Sbjct: 1400 LKYVNGTSDYGIMYCHCSGSMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSKKQN 1459
Query: 1551 TIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRA 1610
+++STAEAEYI+A + C+QL+WMK L++Y + + + +YCDN +AI +SKNP+ HSR
Sbjct: 1460 CVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQHSRT 1519
Query: 1611 KHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
KHI+I+HH+IR+ V ++ ++ +DTE Q ADIFTK L ++F+ ++ L +
Sbjct: 1520 KHIDIRHHYIRELVDDKVITLEHVDTEEQIADIFTKALDAKQFEKLRGKLGI 1571
>gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 1049 bits (2712), Expect = 0.0
Identities = 621/1665 (37%), Positives = 921/1665 (55%), Gaps = 88/1665 (5%)
Query: 41 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 89
+ WK M + I GL +E W E+G D L +++ T A++
Sbjct: 21 YGHWKVKMRALIRGLGKEAWIATSVGWKAPVVKGENGEDVLKTEDQW--------TDAEE 72
Query: 90 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 149
+ + +I S+ + ++ ++ + +AK + L +EG+ VK ++ ML Q+
Sbjct: 73 AKATANSRALSLIFNSVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 132
Query: 150 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAK 209
E M + E+IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 133 ENLTMDESENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 192
Query: 210 DLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEE--ESP 267
D +T+ E++V L+ +E+ + KG SK +SE E E
Sbjct: 193 DTDTIDFEEVVGMLQAYELEITS-------------GKGGYSKGVALAVSSEKNEIQELK 239
Query: 268 DGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDC 327
D S ++ AM + AR Q + + K + C C+ GH +C
Sbjct: 240 DSMSMMAKNFSRAMKRVEKRGFARNQGSDRDRDRDRDRNSKRSEIQCHECQGYGHIKAEC 299
Query: 328 PDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVA 387
P L+++ K + +KF KS +S+S SD E++++D K V V
Sbjct: 300 PSLKRKDLKCSECRG-IGHTKFDCIGSKSKPDRSYIAESDSDSDDEDSEEDVKGFVSFVG 358
Query: 388 TVSSEAVSEAESDSE---DENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDL 444
+ + VS SDSE ++ E+ + +D E L+E N L KEK + L
Sbjct: 359 IIEDDNVSSDSSDSEVGCEKEEISADDESDVEMDVDGEFRKLYE---NWLVLSKEKVIWL 415
Query: 445 MKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIM 504
++ K +++ K EL N I + + + +K +E S + +I + +
Sbjct: 416 EEKVK-VQEQIEQLKGELAVANQIKSEMILKYSAKEEKNRELSQDLSDTRKKIHMLNKGT 474
Query: 505 AGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGL---KSTFVPEGTNA 561
+D A + + G G S K+ K+ + KS VP
Sbjct: 475 KDLDSILAAGRVGKSNFGLGYHGGGSSTKTNFVRSKAAAPTQSQSVFRSKSNSVPARRKY 534
Query: 562 VTADQSKSE--ASGSQAKITSKPENL------------KIKVMTKSDP---KSQKITILK 604
+ S+ +G + + ++ K+K K P ++ K+ + +
Sbjct: 535 QNQNHYHSQRTVTGYECYYCGRHGHIQRYCYRYAARLSKLKRQGKLYPHQGRNSKMYVRR 594
Query: 605 RSETVH--QNLIKPESKIPKQKDQ-KNKAATASEKTIPKGVKPKVLNDQKTLSIHPKVQG 661
H I K P D ++ T S+ + K N +++G
Sbjct: 595 EDLYCHVAYTSIAEGVKKPWYFDSGASRHMTGSQANLNNYSSVKESNVMFGGGAKGRIKG 654
Query: 662 RKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGK 721
+ + NV+ V+GL NL+S+SQ CD G V+F+K C N+ +++ +
Sbjct: 655 KGDLTETEKPHLTNVYFVEGLTANLISVSQLCDEGLTVSFNKVKCWATNERNQNTLTGVR 714
Query: 722 RVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDY 781
N Y + ++ +CL + + +WH+RLGH N R +SK+ ++V+G+P + +
Sbjct: 715 TGNNCY------MWEEPKICLRAEKEDPVLWHQRLGHMNARSMSKLVNKEMVRGVPELKH 768
Query: 782 HSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDY 841
+CGAC +GK ++ K + + T++ L+L+H+DL GP+ T S+ G +Y V+VDD+
Sbjct: 769 IEKIVCGACNQGKQIRVQHKRVEGIQTTQVLDLIHMDLMGPMQTESIAGKRYVFVLVDDF 828
Query: 842 SRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGIL 901
SR+ WV+FI+ K F Q+++EK++ I ++RSD GGEF NE F FCE GI
Sbjct: 829 SRYAWVRFIREKSETANSFKILALQLKNEKKMGIKQIRSDRGGEFMNEAFNSFCESQGIF 888
Query: 902 HEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPML 961
H++S+PRTPQ NGVVERKNRTLQEMAR MIH N + + FWAEA++T+CY+ NR+Y+R
Sbjct: 889 HQYSAPRTPQSNGVVERKNRTLQEMARAMIHGNGVPEKFWAEAISTACYVINRVYVRLGS 948
Query: 962 EKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYN 1021
+KT YE++KG++PN+SYF FGC CYI+N KD L KFD++++ G FLGY+ S AYRV+N
Sbjct: 949 DKTPYEIWKGKKPNLSYFRVFGCVCYIMNDKDQLGKFDSRSEEGFFLGYATNSLAYRVWN 1008
Query: 1022 SETQCVEESMHVKFDDRE-PESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPE 1080
+ +EESM+V FDD PE + ++ + T+ S + E + +
Sbjct: 1009 KQRGKIEESMNVVFDDGSMPELQIIVRNRNEPQTSISNNHGEERNDNQFDN--------- 1059
Query: 1081 AEITPEAESNSEAEPSPKVQNESASEDF-QDNTQQVIQPKFKHKSSHHEELIIGSKDSPR 1139
+I E + E P +V + AS+D D + + + K + G K R
Sbjct: 1060 GDINKSGEESDEEVPPAQVHRDHASKDIIGDPSGERVTRGVKQDYRQ----LAGIKQKHR 1115
Query: 1140 RTRSHFRQEESLIG-LLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQK 1198
S EE + +SI+EPK V+EAL D WILAM+EEL +F R+ VWDLVP+P Q
Sbjct: 1116 VMASFACFEEIMFSCFVSIVEPKNVKEALEDHFWILAMEEELEEFSRHQVWDLVPRPPQV 1175
Query: 1199 NIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAI 1258
N+IGTKW+F+NK +E G +TRNKARLVAQGY+Q EG+D+ ETFAPVARLE IR LL A
Sbjct: 1176 NVIGTKWIFKNKFDEVGNITRNKARLVAQGYTQVEGLDFDETFAPVARLECIRFLLGTAC 1235
Query: 1259 NHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYD 1318
G L+QMDVK AFLNG+IEEEVYV+QP GFE+L+ P +VYKLKK+LYGLKQAPRAWY+
Sbjct: 1236 GMGFKLHQMDVKCAFLNGIIEEEVYVEQPKGFENLEFPEYVYKLKKALYGLKQAPRAWYE 1295
Query: 1319 RLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEF 1378
RL+ FLI + RG VD TLF + I+I+QIYVDDI+FG T+ L K F K M EF
Sbjct: 1296 RLTTFLIVQGYTRGSVDKTLFVKNDVHGIIIIQIYVDDIVFGGTSDKLVKTFVKTMTTEF 1355
Query: 1379 EMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKED 1438
MSM+GELK+FLG+QINQ+ EG+ + Q+ Y + L+K+F + K TPM T L K++
Sbjct: 1356 RMSMVGELKYFLGLQINQTDEGITISQSTYAQNLVKRFGMCSSKPAPTPMSTTTKLFKDE 1415
Query: 1439 TGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTT 1498
G VD+KLYRGMIGSLLYLTA+RPD+ SV LCAR+QS+P+ SHL AVKRI +Y+ GT
Sbjct: 1416 KGVKVDEKLYRGMIGSLLYLTATRPDLCLSVGLCARYQSNPKASHLLAVKRIIKYVSGTI 1475
Query: 1499 NLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAE 1558
N GL Y + L+G+CDAD+ G+ +R+ST+G FLG NLISW SKKQ +++S+ +
Sbjct: 1476 NYGLNYTRDTSLVLVGYCDADWGGNLDDRRSTTGGVFFLGSNLISWHSKKQNCVSLSSTQ 1535
Query: 1559 AEYISAANCCTQLLWMKHQLEDYQIN-ANSIPIYCDNTAAICLSKNPILHSRAKHIEIKH 1617
+EYI+ +CCTQLLWM+ DY + + + + CDN +AI +SKNP+ HS KHI I+H
Sbjct: 1536 SEYIALGSCCTQLLWMRQMGLDYGMTFPDPLLVKCDNESAIAISKNPVQHSVTKHIAIRH 1595
Query: 1618 HFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
HF+R+ V++ + ++ + TE Q DIFTKPL + F ++K+L +
Sbjct: 1596 HFVRELVEEKQITVEHVPTEIQLVDIFTKPLDLNTFVNLQKSLGI 1640
>gb|AAN40030.1| putative gag-pol polyprotein [Zea mays]
Length = 2319
Score = 1045 bits (2703), Expect = 0.0
Identities = 639/1694 (37%), Positives = 921/1694 (53%), Gaps = 153/1694 (9%)
Query: 44 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 103
W M S ++G+ LW+I+ G+ EE TP + ++ + II
Sbjct: 138 WADKMKSHLIGVHPSLWEIVNVGMYKPAQGEE--------MTPEMMQEVHRNAQAVSIIK 189
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEM 163
S+ EY K+ + A+ ++ L + EG K K + L + + ES++ +
Sbjct: 190 GSLCPEEYRKVQGREDARDIWNILRMSHEGDPKAKRHRVEALESELARYDWTKGESLQSL 249
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKI-LRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSS 222
+ R LV+ +++L + V+++ +R+ + + I + D ++ L +
Sbjct: 250 FDRLMVLVNKIRVLGSEDWSDSKVTRLFMRAYKEKDKSLARMIRDRDDYEDMTPHQLFAK 309
Query: 223 LKVHEMSLNEHETSKKSKSIALPSKGKTS-KSSKAYKASESEEESPDGDSDEDQSVKMAM 281
++ HE +E K S AL + + + K SK +KA + E S D DS D+ M +
Sbjct: 310 IQQHE---SEEAPIKTRDSHALITNEQDNLKKSKDHKAKKVVETSSDEDSSSDEDTAMFI 366
Query: 282 LSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMK 341
+ KKF+ K ++ +K ++ C+ C + GHFI DCP+ ++++ K + K
Sbjct: 367 ---------KTFKKFVRKNDKFQ--RKGKKRACYECGQTGHFIADCPNKKEQEAKKEYKK 415
Query: 342 SSFNSS-------KFRKQIKKSLMATWEDLDSESGSDKEEA------------------D 376
F K +K + + W D S S++EE
Sbjct: 416 DKFKKGGKTKGYFKKKKYGQAHIGEEWNSDDESSSSEEEEVVANVAIQSTSSAQLFTNLQ 475
Query: 377 DDAKAAVGLVAT---VSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEH--RT 431
DD+ L+A V+ + + D +D+ + +K+ ++ ++ + L E +
Sbjct: 476 DDSYIPTCLMAKGDKVTLFSNDFSNDDDDDQIAMKNKMIKEFGLNGYNVITKLMEKLDKR 535
Query: 432 NELTDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGS 491
D +E + L K++ L EL +K+ ++ NL T D L + L+ +
Sbjct: 536 KATLDAQEDLLILEKERNLELQELIHNKD-IEVINL--KTSIDNLANEKNALESSMLSLN 592
Query: 492 GNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLK 551
E+ + ++ K + +++K S + S ++ K + C
Sbjct: 593 VQNQELQVQ------LENCKNINAPTLVFESKSS----SNDNSCKHCAKYHASCCLTNHA 642
Query: 552 STFVPEGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKSDPKSQKITILKRS-ETVH 610
P+ V ++ S + +P+ +K + + K + TVH
Sbjct: 643 RKHSPQ----VKVEEILKRCSSNDGLKKVEPKYKSLK--PNNGRRGLGFNSSKENPSTVH 696
Query: 611 QNLIKPE-----------SKIPKQKDQKNKAATA--SEKTIPKGVKPKVLNDQK---TLS 654
+ P+ +I D+ ++ ++ S + G + ++ TL
Sbjct: 697 KGWRSPKFIEGTTLYDALGRIHSSNDKSSQVYSSGGSSWVLDSGCTNHMTGEKDMFHTLQ 756
Query: 655 IHPKVQ-------GRKSTIGNSSI------SINNVWLVDGLKHNLLSISQFCDNGYDVTF 701
+ + Q G+ IG I S++NV LVD L +NLLS+SQ C GY+ F
Sbjct: 757 LTQEAQEIVFGDSGKSKVIGIGKIPISDQQSLSNVLLVDSLSYNLLSVSQLCGMGYNCLF 816
Query: 702 SKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANW 761
S + ++ ++D S+ F G+ +Y ++F+ CL++ +DK W+WH+RL H
Sbjct: 817 SDVDVKILRREDSSVAFTGRLKGKLYLVDFTTSKVTPETCLVAKSDKGWLWHRRLAHVGM 876
Query: 762 RLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFG 821
R ++K+ K + GL N+ + D +CGACQ GK +SK++V+T RPLELLH+DLFG
Sbjct: 877 RNLAKLQKDNHIIGLTNVVFEKDRVCGACQAGKQHGVPHQSKNVVTTKRPLELLHMDLFG 936
Query: 822 PVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSD 881
PV S+ GSKYGLVIVDD+SR+TWV F+ K E+ F + Q+E ELKI KVRSD
Sbjct: 937 PVAYISIGGSKYGLVIVDDFSRFTWVFFLSDKGETQEILKKFMRRAQNEFELKIKKVRSD 996
Query: 882 HGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFW 941
+G EF+N E F + GI HEFS P TPQQNGVVERKNRTL E ARTM+ E +FW
Sbjct: 997 NGTEFKNTGVEEFLGEEGIKHEFSVPYTPQQNGVVERKNRTLIEAARTMLDEYKTPDNFW 1056
Query: 942 AEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAK 1001
AEAVNT+C+ NR+Y+ + +KTAYEL G +P + YF FGC C+ILN K KF +
Sbjct: 1057 AEAVNTACHAINRLYLHKIYKKTAYELLTGNKPKVDYFRVFGCKCFILNKKVKSSKFAPR 1116
Query: 1002 AQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDAS 1061
G LGY+ + YRV+N+ T VE ++ V FD ES S+ SN + +
Sbjct: 1117 VDEGFLLGYASNAHGYRVFNNSTGLVEIAIDVTFD----ESNGSQGHVSNDTVGNEKLPC 1172
Query: 1062 ES----------DQPSDSEKYT-------------------EVESSPEAEITPEAESNSE 1092
ES Q D E+ T +++P P E N +
Sbjct: 1173 ESIKKLAIGEVRPQERDDEEGTLWMTNEVVDVGARVVSDNVSTQANPSTSSHPNHEENHQ 1232
Query: 1093 AEPSPKVQNESASEDFQ------DNTQQVIQ--PKFKHKSSHHE-------ELIIGSKDS 1137
P+ V++E + D + N ++ IQ P H HH + I+GS
Sbjct: 1233 RMPTV-VEDEQENIDGEVPLDQVTNEEEQIQRHPSVPHPRVHHTIQRDHPVDNILGSIRR 1291
Query: 1138 PRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQ 1197
TRS +S +EP VEEAL D WI AMQEELN F RN+VW LV +P Q
Sbjct: 1292 GVTTRSRLANFCEFYSFVSSLEPLKVEEALDDPDWITAMQEELNNFTRNEVWSLVQRPKQ 1351
Query: 1198 KNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYA 1257
N+IGTKWVFRNK +E G VT+NKARLVAQGY+Q EG+D+ ET+APVARLE+IR+L++YA
Sbjct: 1352 -NVIGTKWVFRNKQDENGVVTKNKARLVAQGYTQVEGLDFGETYAPVARLESIRILIAYA 1410
Query: 1258 INHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWY 1317
NH LYQMDVKSAFLNG ++E VYV+QPPGFED K PNHVY L K+LYGLKQAPRAWY
Sbjct: 1411 TNHDFKLYQMDVKSAFLNGPLQERVYVEQPPGFEDPKKPNHVYLLHKALYGLKQAPRAWY 1470
Query: 1318 DRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDE 1377
D L +FLIKN F G+ D+TLF R + ++ + QIYVDDIIFGSTN C+EFSK+M +
Sbjct: 1471 DCLKDFLIKNGFTIGKADSTLFTRKVDNELFVCQIYVDDIIFGSTNEKFCEEFSKVMTNR 1530
Query: 1378 FEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKE 1437
FEMSMMGELK+FLG Q+ Q KEG ++ QTKYT+++LKKF +E K TPM L
Sbjct: 1531 FEMSMMGELKYFLGFQVKQLKEGTFLCQTKYTQDMLKKFGMEKAKHAKTPMPSNGHLDLN 1590
Query: 1438 DTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGT 1497
+ G VDQKLYR MIGSLLYL ASRPDI+ SVC+CARFQ++P++ HL AVKRI RYL T
Sbjct: 1591 EEGKPVDQKLYRSMIGSLLYLCASRPDIMLSVCMCARFQANPKDCHLVAVKRILRYLVHT 1650
Query: 1498 TNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTA 1557
NLGL Y K + L+G+ D+DYAG +++RKST+G CQFLG +L+SW+SKKQ +A+STA
Sbjct: 1651 QNLGLWYPKGSFFDLLGYSDSDYAGCKVDRKSTTGTCQFLGRSLVSWSSKKQNCVALSTA 1710
Query: 1558 EAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKH 1617
EAEYI+A CC QLLWMK L D+ N IP+ CDN +AI L+ NP+ HSR KHI+I+
Sbjct: 1711 EAEYIAAGACCAQLLWMKQTLRDFGCEFNKIPLLCDNESAIKLANNPVQHSRTKHIDIRR 1770
Query: 1618 HFIRDYVQKGILDI 1631
HF+RD+ KG +++
Sbjct: 1771 HFLRDHEAKGDIEL 1784
>gb|AAO73525.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 1044 bits (2699), Expect = 0.0
Identities = 634/1679 (37%), Positives = 926/1679 (54%), Gaps = 168/1679 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 17 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAK-AMFASLCANFEGSKKVKEAKALMLVHQYE 150
+ K + + + + ++ + AK A L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDACGEILKTTHEGTSKVKMSRLQLLATKFE 136
Query: 151 LFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKD 210
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D
Sbjct: 137 NLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQD 196
Query: 211 LNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGD 270
+ + V++L+ SL+ E+ L++ KKSK++A S E EE+ D D
Sbjct: 197 ICNMRVDELIGSLQTFELGLSDRN-EKKSKNLAFVSN------------DEGEEDEYDLD 243
Query: 271 SDEDQSVKMAMLSNK----LEYLARKQKK------FLSKRGSYKNSKKEDQ----KG--C 314
+DE + + +L + L + R+QK F ++GS + K +++ KG C
Sbjct: 244 TDEGLTNAVGLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYHKKSDEKPSHSKGIQC 303
Query: 315 FNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH +CP K++ KG S+ S +D +SE SD
Sbjct: 304 HGCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESD--- 342
Query: 375 ADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G E++ DS D +I EL S ++L E +
Sbjct: 343 SDRDVNALTGRF---------ESDEDSSD-----IEITFDELAISYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNK 494
LK+ +L ++++ E+ K E+ GF +++ E+ KS+ + +KGS
Sbjct: 389 AQLKKVIANLEAEKEAHEEEISELKGEV-GF--LNSKLENMTKSI-----KMLNKGSD-- 438
Query: 495 HEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKS 552
LD+ + G KN G +G+G++ + + ++ + K+ +
Sbjct: 439 ---MLDEVLQLG--------------KNVGNQRGLGFNHKSACRITMTEFVPA-KNSTGA 480
Query: 553 TFVPEGTNAVTADQSKSEASGSQAKITSKPENLKIKVM---------TKSDPKSQKITIL 603
T + Q KS+ + K ++K T+S +K+ +
Sbjct: 481 TMSQHRSRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSGRKMMWV 540
Query: 604 KRSETVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKTLSIHP------ 657
+ + V ++ + ++D + + T GVK ++N + + +
Sbjct: 541 PKHKIVSL-VVHTSLRASAKEDWYLDSGCSRHMT---GVKEFLVNIEPCSTSYVTFGDGS 596
Query: 658 --KVQGRKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKS 715
K+ G + + S+N V LV GL NL+SISQ CD G++V F+K+ C LV +
Sbjct: 597 KGKITGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSE 655
Query: 716 ITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKG 775
+ KG R ++ + CL S D+ +WH+R GH + R + KI V+G
Sbjct: 656 VLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRG 715
Query: 776 LPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGL 835
+PN+ +CG CQ GK VK S + +TS LELLH+DL GP+ SL G +Y
Sbjct: 716 IPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSMVLELLHMDLMGPMQVESLGGKRYAY 775
Query: 836 VIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFC 895
V+VDD+SR+TWV FI+ K EVF ++Q EK+ I ++RSDHG EFEN F FC
Sbjct: 776 VVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFC 835
Query: 896 EKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRI 955
GI HEFS+ TPQQNG+VERKNRTLQE R M+H L + WAEA+NT+CYI NR+
Sbjct: 836 TSEGITHEFSAAITPQQNGIVERKNRTLQEATRVMLHAKELPYNLWAEAMNTACYIHNRV 895
Query: 956 YIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSK 1015
+R T YE++KGR+P + +FH FG CYIL ++ +K D K+ GIFLGYS S+
Sbjct: 896 TLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSR 955
Query: 1016 AYRVYNSETQCVEESMHVKFDDREPESKTSEQ-----SESNAGTT--DSEDASESDQPSD 1068
AYRV+NS T+ V ES++V DD P K + SE N T +E+A +SD +D
Sbjct: 956 AYRVFNSRTRTVMESINVVVDDLTPARKKDVEEDVRTSEDNVADTAKSAENAEKSDSTTD 1015
Query: 1069 SEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHE 1128
+ + SP I Q +QPK
Sbjct: 1016 EPNINQPDKSPFIRI------------------------------QKMQPK--------- 1036
Query: 1129 ELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDV 1188
ELIIG + TRS + S +S IEPK V+EAL+D+ WI AMQEEL QF+RN+V
Sbjct: 1037 ELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEV 1096
Query: 1189 WDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLE 1248
W+LVP+P N+IGTKW+F+NK NE+G +TRNKARLVAQGY+Q EG+D+ ETFAPVARLE
Sbjct: 1097 WELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLE 1156
Query: 1249 AIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYG 1308
+IRLLL A LYQMDVKSAFLNG + EE YV+QP GF D H +HVY+LKK+LYG
Sbjct: 1157 SIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHLDHVYRLKKALYG 1216
Query: 1309 LKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCK 1368
LKQAPRAWY+RL+ FL + + +G +D TLF + ++++I QIYVDDI+FG + + +
Sbjct: 1217 LKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLR 1276
Query: 1369 EFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPM 1428
F MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+KY K ++KKF +E+ TP
Sbjct: 1277 HFVPQMQSEFEMSLVGELHYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASHKRTPA 1336
Query: 1429 HPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVK 1488
LSK++ GT VDQ LYR MIGSLLYLTASRPDI F+V +CAR+Q++P+ SHL VK
Sbjct: 1337 PTHLKLSKDEAGTSVDQNLYRSMIGSLLYLTASRPDITFAVGVCARYQANPKISHLNQVK 1396
Query: 1489 RIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKK 1548
RI +Y+ GT++ G++Y D L+G+CDAD+AG +RK TSG C +LG NLISW SKK
Sbjct: 1397 RILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKCTSGGCFYLGTNLISWFSKK 1456
Query: 1549 QATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHS 1608
Q +++STAEAEYI+A + C+QL+WMK L++Y + + + +YCDN +AI +SKNP+ H+
Sbjct: 1457 QNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQHN 1516
Query: 1609 RAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNMHFVSD 1667
R KHI+I+HH+IRD V I+ ++ +DTE Q ADIFTK L +F+ ++ L + D
Sbjct: 1517 RTKHIDIRHHYIRDLVDDKIITLEHVDTEEQVADIFTKALDANQFEKLRGKLGTCLLED 1575
>gb|AAC64917.1| gag-pol polyprotein [Glycine max]
Length = 1550
Score = 1042 bits (2695), Expect = 0.0
Identities = 625/1661 (37%), Positives = 921/1661 (54%), Gaps = 153/1661 (9%)
Query: 48 MYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKLYKKHHKIRGII 102
M +F+ LD W + G + LD EG + T + +L + K +
Sbjct: 1 MVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDELALGNSKALNAL 60
Query: 103 VASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEE 162
+ + + ++ + AK + L EG+ KVK ++ +L ++E +MK++E I E
Sbjct: 61 FNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFENLKMKEEECIHE 120
Query: 163 MYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSS 222
+ + + L + V KILRSLP R+ KVTAIEEA+D+ + V++L+ S
Sbjct: 121 FHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDICNMRVDELIGS 180
Query: 223 LKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAML 282
L+ E+ L++ T KKSK++A S E EE+ D D+DE + + +L
Sbjct: 181 LQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDTDEGLTNAVVLL 227
Query: 283 SNK----LEYLARKQKK------FLSKRGSYKNSKKEDQKG-------CFNCKKPGHFIV 325
+ L + R+QK F ++GS + K+ D+K C C+ GH
Sbjct: 228 GKQFNKVLNRMDRRQKPHVRNIPFDIRKGS-EYQKRSDEKPSHSKGIQCHGCEGYGHIKA 286
Query: 326 DCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGL 385
+CP K++ KG S+ S +D +SE SD +D D A G
Sbjct: 287 ECPTHLKKQRKGLSVCRS------------------DDTESEQESD---SDRDVNALTGR 325
Query: 386 VATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLM 445
+ +ED ++ S+I EL S +EL E + LK+ +L
Sbjct: 326 FES------------AEDSSDTDSEITFDELAISYRELCIKSEKILQQEAQLKKVIANLE 373
Query: 446 KQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMA 505
++++ E+ K E+ GF +++ E+ KS+ + +KGS LD+ +
Sbjct: 374 AEKEAHEDEISELKGEI-GF--LNSKLENMTKSI-----KMLNKGSD-----LLDEVLQL 420
Query: 506 GIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVT 563
G KN G +G+G++ + + ++ + K+ +T +
Sbjct: 421 G--------------KNVGNQRGLGFNHKSAGRTTMTEFVPA-KNSTGATMSQHRSRHHG 465
Query: 564 ADQSKSEASGSQAKITSKPENLKIKVM---------TKSDPKSQKITILKRSETVHQNLI 614
Q KS+ + K ++K T+S +K+ + + +TV ++
Sbjct: 466 TQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSGRKMMWVPKHKTVSL-VV 524
Query: 615 KPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKTLSIHP--------KVQGRKSTI 666
+ ++D + + T GVK ++N + + + K+ G +
Sbjct: 525 HTSLRASAKEDWYLDSGCSRHMT---GVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLV 581
Query: 667 GNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKRVENV 726
+ S+N V LV GL NL+SISQ CD G++V F+K+ C LV + + KG R ++
Sbjct: 582 HDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDN 640
Query: 727 YKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDAL 786
+ CL S D+ +WH+R GH + R + KI V+G+PN+ +
Sbjct: 641 CYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRI 700
Query: 787 CGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTW 846
CG CQ GK VK S + +TSR LELLH+DL GP+ SL +Y V+VDD+SR+TW
Sbjct: 701 CGECQIGKQVKMSNQKLQHQTTSRVLELLHMDLMGPMQVESLGRKRYAYVVVDDFSRFTW 760
Query: 847 VKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSS 906
V FI+ K EVF ++Q EK+ I ++RSDHG EFEN F FC GI HEFS+
Sbjct: 761 VNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSA 820
Query: 907 PRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAY 966
TPQQNG+VERKNRTLQE AR M+H L + WAEA+NT+CYI NR+ +R T Y
Sbjct: 821 AITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLY 880
Query: 967 ELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQC 1026
E++KGR+P + +FH G CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+
Sbjct: 881 EIWKGRKPTVKHFHICGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRT 940
Query: 1027 VEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEITPE 1086
V ES++V DD P K + + + D +D+ K E
Sbjct: 941 VMESINVVVDDLTPARKKDVEEDVR---------TSGDNVADTAKSAE-----------N 980
Query: 1087 AESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSHFR 1146
AE++ A P + +P + + H +ELIIG + TRS
Sbjct: 981 AENSDSATDEPNINQPDK------------RPSIRIQKMHPKELIIGDPNRGVTTRSREI 1028
Query: 1147 QEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKWV 1206
+ S +S IEPK V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+IGTKW+
Sbjct: 1029 EIISNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWI 1088
Query: 1207 FRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQ 1266
F+NK NE+G +TRNKARLVAQGY+Q EG+D+ ETFAP ARLE+IRLLL A LYQ
Sbjct: 1089 FKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPGARLESIRLLLGVACILKFKLYQ 1148
Query: 1267 MDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLIK 1326
MDVKSAFLNG + EE YV+QP GF D HP+HVY+LKK+LYGLKQAPRAWY+RL+ FL +
Sbjct: 1149 MDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQ 1208
Query: 1327 NDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGEL 1386
+ +G +D TLF + ++++I QIYVDDI+FG + + + F + MQ EFEMS++GEL
Sbjct: 1209 QGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGEL 1268
Query: 1387 KFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQK 1446
+FLG+Q+ Q ++ +++ Q+KY K ++KKF +E+ TP LSK++ GT VDQ
Sbjct: 1269 TYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQS 1328
Query: 1447 LYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRK 1506
LYR MIGSLLYLTASRPDI ++V CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y
Sbjct: 1329 LYRSMIGSLLYLTASRPDITYAVGGCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCH 1388
Query: 1507 SLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAAN 1566
D L+G+CDAD+AG +RKST G C +LG N ISW SKKQ +++STAEAEYI+A +
Sbjct: 1389 CSDSMLVGYCDADWAGSVDDRKSTFGGCFYLGTNFISWFSKKQNCVSLSTAEAEYIAAGS 1448
Query: 1567 CCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQK 1626
C+QL+WMK L++Y + + + +YCDN +AI +SKNP+ HSR KHI+I+HH+IRD V
Sbjct: 1449 SCSQLVWMKQMLKEYNVEQDVMTLYCDNLSAINISKNPVQHSRTKHIDIRHHYIRDLVDD 1508
Query: 1627 GILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNMHFVSD 1667
++ ++ +DTE Q ADIFTK L +F+ ++ L + + D
Sbjct: 1509 KVITLEHVDTEEQIADIFTKALDANQFEKLRGKLGICLLED 1549
>gb|AAD20307.1| copia-type pol polyprotein [Zea mays]
Length = 1063
Score = 1018 bits (2631), Expect = 0.0
Identities = 533/1049 (50%), Positives = 694/1049 (65%), Gaps = 47/1049 (4%)
Query: 650 QKTLSIHPKVQGRKSTIGNSSIS----INNVWLVDGLKHNLLSISQFCDNGYDVTFSKTN 705
Q+ ++ QG +G +IS I+NV+LVD L +NLLS+SQ C GY+ F+
Sbjct: 18 QRAITFGDGNQGLVKGLGKIAISPDHSISNVFLVDSLDYNLLSVSQLCQMGYNCLFTDIG 77
Query: 706 CTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLIS 765
T+ + D SI FKG +Y ++F D A+ CL++ + W+WH+RL H + +
Sbjct: 78 VTVFRRSDDSIAFKGVLEGQLYLVDF-DRAELDT-CLIAKTNMGWLWHRRLAHVGMKNLH 135
Query: 766 KISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNT 825
K+ K + + GL N+ + D +C ACQ GK V + K+I++T RPLELLH+DLFGP+
Sbjct: 136 KLLKGEHILGLTNVHFEKDRICSACQAGKQVGTHHPHKNIMTTDRPLELLHMDLFGPIAY 195
Query: 826 ASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGE 885
S+ GSKY LVIVDDYSR+TWV F++ K E F + Q+E L+I K+RSD+G E
Sbjct: 196 ISIGGSKYCLVIVDDYSRFTWVFFLQEKSQTQETLKGFLRRAQNEFGLRIKKIRSDNGTE 255
Query: 886 FENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAV 945
F+N E F E+ GI HEFSSP TPQQNGVVERKNRTL +MARTM+ E FWAEAV
Sbjct: 256 FKNSQIESFLEEEGIKHEFSSPYTPQQNGVVERKNRTLLDMARTMLDEYKTPDRFWAEAV 315
Query: 946 NTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRG 1005
NT+CY NR+Y+ +L+KT+YEL G++PNISYF FG C+IL + KF K G
Sbjct: 316 NTACYAINRLYLHRILKKTSYELLTGKKPNISYFRVFGSKCFILIKRGRKSKFAPKTVEG 375
Query: 1006 IFLGYSERSKAYRVYNSETQCVEESMHVKFDD-----------------REPESKTSEQS 1048
LGY ++AYRV+N T VE S V FD+ + P S
Sbjct: 376 FLLGYDSNTRAYRVFNKSTGLVEVSCDVVFDETNGSQVEQVDLDEIGEEQAPCIALRNMS 435
Query: 1049 ESNAGTTDSEDA-SESDQPSDSEKYTEVESSPEAEITPEA----ESNSEAEPSPKVQNES 1103
+ +SE+ S DQPS S +++SP + EA E N E EP N+
Sbjct: 436 IGDVCPKESEEPPSTQDQPSSS-----MQASPPTQNEDEAQNDEEQNQEDEPPQDDSNDQ 490
Query: 1104 ASEDFQDNTQQVIQPKFKH-------KSSHHEELIIGSKDSPRRTRS---HFRQEESLIG 1153
+ + +P+ H + H + I+G TRS HF + S +
Sbjct: 491 GGDTNDQEKEDEEEPRPPHPRVHQAIQRDHPVDTILGDIHKGVTTRSRVAHFCEHYSFV- 549
Query: 1154 LLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKWVFRNKLNE 1213
S IEP VEEAL D W++AMQEELN F RN+VW LVP+P Q N++GTKWVFRNK +E
Sbjct: 550 --SSIEPHRVEEALQDSDWVVAMQEELNNFTRNEVWHLVPRPNQ-NVVGTKWVFRNKQDE 606
Query: 1214 QGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAF 1273
G VTRNKARLVA+GYSQ EG+D+ ET+APVARLE+IR+LL+YA HG LYQMDVKSAF
Sbjct: 607 HGVVTRNKARLVAKGYSQVEGLDFGETYAPVARLESIRILLAYATYHGFKLYQMDVKSAF 666
Query: 1274 LNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQ 1333
LNG I+EEVYV+QPPGFED ++PNHVY+L K+LYGLKQAPRAWY+ L +FLI N F+ G+
Sbjct: 667 LNGPIKEEVYVEQPPGFEDSEYPNHVYRLSKALYGLKQAPRAWYECLRDFLIANGFKVGK 726
Query: 1334 VDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQ 1393
D TLF +TL+ D+ + QIYVDDIIFGSTN S C+EFS++M +FEMSMMGELK+FLG Q
Sbjct: 727 ADPTLFTKTLENDLFVCQIYVDDIIFGSTNKSTCEEFSRIMTQKFEMSMMGELKYFLGFQ 786
Query: 1394 INQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIG 1453
+ Q +EG ++ QTKYT+++L KF ++D K + TPM L + G VDQK+YR MIG
Sbjct: 787 VKQLQEGTFICQTKYTQDILTKFGMKDAKPIKTPMGTNGHLDLDTGGKSVDQKVYRSMIG 846
Query: 1454 SLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLI 1513
SLLYL ASRPDI+ SVC+CARFQSDP+ESHLTAVKRI RYL T GL Y + + LI
Sbjct: 847 SLLYLCASRPDIMLSVCMCARFQSDPKESHLTAVKRILRYLAYTPKFGLWYPRGSTFDLI 906
Query: 1514 GFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANCCTQLLW 1573
G+ DAD+AG +I RKSTSG CQFLG +L+SWASKKQ ++A+STAEAEYI+A +CC QLLW
Sbjct: 907 GYSDADWAGCKINRKSTSGTCQFLGRSLVSWASKKQNSVALSTAEAEYIAAGHCCAQLLW 966
Query: 1574 MKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGILDIQF 1633
M+ L DY +P+ CDN +AI ++ NP+ HSR KHI I++HF+RD+ QKG ++I +
Sbjct: 967 MRQTLLDYGYKLTKVPLLCDNESAIKMADNPVEHSRTKHIAIRYHFLRDHQQKGDIEISY 1026
Query: 1634 IDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
I+T+ Q ADIFTKPL + F ++ LN+
Sbjct: 1027 INTKDQLADIFTKPLDEQSFTRLRHELNI 1055
>gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902324|ref|NP_912508.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2011
Score = 1011 bits (2615), Expect = 0.0
Identities = 630/1669 (37%), Positives = 898/1669 (53%), Gaps = 131/1669 (7%)
Query: 29 GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQ 88
GKAP FNG +S WK M + + + +W I++ G AI T
Sbjct: 9 GKAPMFNGT--NYSTWKIKMSTHLKAMSFHIWSIVDVGF----------AITGTPLTEID 56
Query: 89 KKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQ 148
+ K + + ++ S+ + E+ ++S+ TA ++ L E + + K+AK L Q
Sbjct: 57 HRNLKLNAQAMNVLFNSLSQEEFDRVSNLETAYEIWNKLAEIHESTSEYKDAKLHFLKIQ 116
Query: 149 YELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEA 208
YE F M ES+ +MY R +V+ L+ L +Y + K+LR+LP ++ VT + +
Sbjct: 117 YETFSMLPHESVNDMYGRLNVIVNDLKGLGANYTNLEVAQKMLRALPEKYETLVTMLINS 176
Query: 209 KDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPD 268
D++ ++ L+ + ++M ++ KK A PSK + ++ S+S+ +
Sbjct: 177 -DMSRMTPASLLGKINTNDM----YKLKKKEMEEASPSKKCIALQAEVEDKSKSKVNEVN 231
Query: 269 GDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKG-------CFNCKKPG 321
D +E+ + +L+ + L ++K+ RGS N ++ + CF C
Sbjct: 232 KDLEEE----IVLLARRFNDLLGRRKE--RGRGSNSNRRRNRRPNKTLSNLRCFEC---- 281
Query: 322 HFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKA 381
D +E + K+ K +++A E A
Sbjct: 282 -------DSNEESSASSGSEEEGGDDASSKKKKMAVVAIKEA-------------PSLFA 321
Query: 382 AVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKY 441
+ L+A S+ S ++S+S+D+ + S EL+S+FE EL EK
Sbjct: 322 PLCLMAKSPSKVTSLSDSESDDDCDDVS----------YDELVSMFE----ELHAYSEKE 367
Query: 442 VDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIA--- 498
+ K K L+ EELK + T ++LK L G+ I+
Sbjct: 368 IIKFKTLKKDHASLEVLYEELKTSHERLTISHEKLKEAHDNLLSTTQHGALIDVGISCDL 427
Query: 499 LDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKS-----------KEYSLKSYCDCIK 547
LDD I + VAS ST + + S S + LK +
Sbjct: 428 LDDSATCHI--AHVASSSISTSCDDLMDMPNSSSSSCVSICDASLVVENNELKEQVAKLN 485
Query: 548 DGLKSTFVPEGT-NAVTADQS---KSEASGSQAKITSKPENLK---IKVMTKSDPKSQKI 600
L+ F + T + + ++Q E G K KP + +K K K +++
Sbjct: 486 KSLERCFKGKNTLDKILSEQQCILNKEGLGFILKKGKKPSHRATRFVKSNGKYCSKCREV 545
Query: 601 TILKRSETVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKTLSIHPKVQ 660
L T + + + Q A T G N+Q+ ++ +
Sbjct: 546 GHLVNYRTGGSHWV-----LDSGCTQHMTGDRAMFTTFEVGG-----NEQEKVTFGDNSK 595
Query: 661 GRKSTIG----NSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSI 716
G+ +G ++ +SI+NV LV L NLLS++Q CD F + + DKS
Sbjct: 596 GKVIGLGKIAISNDLSIDNVSLVKSLNFNLLSVAQICDLVLSCAFFPQEVIVSSLLDKSC 655
Query: 717 TFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGL 776
FKG R N+Y +F+ CL++ W+WH+RL H +SK+SK LV GL
Sbjct: 656 VFKGFRYGNLYLGDFNSSEANLKTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGL 715
Query: 777 PNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLV 836
++ + D LC ACQ GK V S +K I+STSRPLELLH++ FGP S+ G+ + LV
Sbjct: 716 KDVKFEKDKLCSACQAGKQVACSHPTKSIMSTSRPLELLHMEFFGPTTYKSIGGNSHCLV 775
Query: 837 IVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCE 896
IVDDYSR+TW+ F+ K E+F F + Q+E ++K+RSD+G +F+N E +C+
Sbjct: 776 IVDDYSRYTWMFFLHDKSIVAELFKKFAKRGQNEFNCTLVKIRSDNGSKFKNTNIEDYCD 835
Query: 897 KHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIY 956
GI HE S+ +PQQNGVVE KNRTL EMARTM+ E ++ FWAEA+NT+C+ NR+Y
Sbjct: 836 DLGIKHELSATYSPQQNGVVEMKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLY 895
Query: 957 IRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKA 1016
+ +L+KT+YEL GR+PN++YF FGC CYI L KF+++ G LGY+ SKA
Sbjct: 896 LHRLLKKTSYELIVGRKPNVAYFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKA 955
Query: 1017 YRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVE 1076
YRVYN VEE+ V+FD E+ S++ N E + + +E
Sbjct: 956 YRVYNKNKGIVEETADVQFD----ETNGSQEGHENLDDVGDEGLMRAMKNMSIGDVKPIE 1011
Query: 1077 SSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKH---KSSHHEELIIG 1133
+ + + E ++ A PS ++ E ++ Q + P H H + ++G
Sbjct: 1012 VEDKPSTSTQDEPSTSATPS-----QAQVEVEEEKAQDLPMPPRIHTALSKDHPIDQVLG 1066
Query: 1134 SKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVP 1193
+TRS +S +EPK V+EAL D WI AM +ELN F RN VW LV
Sbjct: 1067 DISKGVQTRSRVASICEHYSFVSCLEPKHVDEALCDPDWINAMHKELNNFARNKVWTLVE 1126
Query: 1194 KPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLL 1253
+ N+IGTKWVFRNK +E G V RNKAR VAQG++Q EG+D+ ETFAPV RLEAI +L
Sbjct: 1127 RLRDHNVIGTKWVFRNKQDENGLVVRNKARFVAQGFTQVEGLDFGETFAPVTRLEAICIL 1186
Query: 1254 LSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAP 1313
L++A I L+QMDVKSAFLNG I E V+V+QPPGFED K+PNHVYKL K+LYGLKQAP
Sbjct: 1187 LAFASCFNIKLFQMDVKSAFLNGEIAELVFVEQPPGFEDPKYPNHVYKLSKALYGLKQAP 1246
Query: 1314 RAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKL 1373
RAWY+RL +FL+ DF+ G+VDTTLF + + D + QIYVDDIIFG TN CKEF +
Sbjct: 1247 RAWYERLRDFLLSKDFKIGKVDTTLFTKIIGDDFFVCQIYVDDIIFGCTNEVFCKEFGDM 1306
Query: 1374 MQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCT 1433
M EFEMSM+GEL FF G+QI Q K+G F LED K + TPM
Sbjct: 1307 MSREFEMSMIGELSFFHGLQIKQLKDGT--------------FGLEDAKPIKTPMATNGH 1352
Query: 1434 LSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRY 1493
L ++ G VD KLYR MIGSLLYLTASRPDI+FSVC+CARFQ+ P+E HL AVKRI RY
Sbjct: 1353 LDLDEGGKPVDLKLYRSMIGSLLYLTASRPDIMFSVCMCARFQAAPKECHLVAVKRILRY 1412
Query: 1494 LKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIA 1553
LK ++ +GL Y K +KL+G+ D+DYAG +++RKSTSG+CQ LG +L+SW+SKKQ +A
Sbjct: 1413 LKHSSTIGLWYPKGAKFKLVGYSDSDYAGCKVDRKSTSGSCQMLGRSLVSWSSKKQNFVA 1472
Query: 1554 MSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHI 1613
+ AEAEY+SA +CC QLLWMK L DY I+ P+ C+N +AI ++ NP+ HSR KHI
Sbjct: 1473 LFIAEAEYVSAGSCCAQLLWMKQILLDYGISFTKTPLLCENDSAIKIANNPVQHSRTKHI 1532
Query: 1614 EIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
+I+HHF+RD+V K + I I TE Q ADIFTKPL RF ++ LN+
Sbjct: 1533 DIRHHFLRDHVAKCDIVISHIRTEDQLADIFTKPLDETRFCKLRNELNL 1581
>gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein homolog - maize
retrotransposon Opie-2
Length = 1068
Score = 990 bits (2559), Expect = 0.0
Identities = 516/1057 (48%), Positives = 686/1057 (64%), Gaps = 43/1057 (4%)
Query: 632 TASEKTIPKGVKPKVLNDQKTLSIHPKVQGRKSTIGNSSIS----INNVWLVDGLKHNLL 687
T +K VK K + Q ++ QG+ +G +IS I+NV+LV+ L +NLL
Sbjct: 22 TGEKKMFTSYVKNK--DSQDSIIFGDGNQGKVKGLGKIAISNEHSISNVFLVESLGYNLL 79
Query: 688 SISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMND 747
S+SQ C+ GY+ F+ + ++ + D S+ FKG +Y ++F+ CL++
Sbjct: 80 SVSQLCNMGYNCLFTNIDVSVFRRCDGSLAFKGVLDGKLYLVDFAKEEAGLDACLIAKTS 139
Query: 748 KKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVS 807
W+WH+RL H + + K+ K + V GL N+ + D C ACQ GK V S +K++++
Sbjct: 140 MGWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFEKDRPCAACQAGKQVGGSHHTKNVMT 199
Query: 808 TSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQI 867
TSRPLE+LH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F++ K F +
Sbjct: 200 TSRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQEKSETQGTLKRFLRRA 259
Query: 868 QSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMA 927
Q+E ELK+ K+RSD+G EF+N E F E+ GI HEFS+P TPQQNGVVERKNRTL +MA
Sbjct: 260 QNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVERKNRTLIDMA 319
Query: 928 RTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCY 987
RTM+ E + FW EAVNT+C+ NR+Y+ +L+ T+YEL G +PN+SYF FG CY
Sbjct: 320 RTMLGEFKTPECFWTEAVNTACHAINRVYLHRILKNTSYELLTGNKPNVSYFRVFGSKCY 379
Query: 988 ILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQ 1047
IL K KF KA G LGY +KAYRV+N + VE S V FD E EQ
Sbjct: 380 ILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSGDVVFD--ETNGSPREQ 437
Query: 1048 SESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASED 1107
D +D E D P+ + + + E+ P+ + E +PSP ++D
Sbjct: 438 ------VVDCDDVDEEDIPTAA-----IRTMAIGEVRPQEQDERE-QPSPSTMVHPPTQD 485
Query: 1108 FQDNTQQVI----------------------QPKFKHKSSHHEELIIGSKDSPRRTRSHF 1145
+ QQ + Q + + H + I+G TRS
Sbjct: 486 DEQVHQQEVCDQGGAQDDHVLEEEAQPAPPTQVRAMIQRDHPVDQILGDISKGVTTRSRL 545
Query: 1146 RQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKW 1205
+S IEP VEEAL D W+LAMQEELN F+RN+VW LVP+P Q N++GTKW
Sbjct: 546 VNFCEHNSFVSSIEPFRVEEALLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVVGTKW 604
Query: 1206 VFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILY 1265
VFRNK +E+G VTRNKARLVA+GY+Q G+D+ ETFAPVARLE+IR+LL+YA +H LY
Sbjct: 605 VFRNKQDERGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLAYAAHHSFRLY 664
Query: 1266 QMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLI 1325
QMDVKSAFLNG I+EEVYV+QPPGFED ++P+HV KL K+LYGLKQAPRAWY+ L +FLI
Sbjct: 665 QMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLSKALYGLKQAPRAWYECLRDFLI 724
Query: 1326 KNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGE 1385
N F+ G+ D TLF +T D+ + QIYVDDIIFGSTN C+EFS++M +FEMSMMGE
Sbjct: 725 ANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQKSCEEFSRVMTQKFEMSMMGE 784
Query: 1386 LKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQ 1445
L +FLG Q+ Q K+G ++ QTKYT++LLK+F ++D K TPM G VDQ
Sbjct: 785 LNYFLGFQVKQLKDGTFISQTKYTQDLLKRFGMKDAKPAKTPMGTDGHTDLNKGGKSVDQ 844
Query: 1446 KLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYR 1505
K YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+E HL AVKRI RYL T GL Y
Sbjct: 845 KAYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKECHLVAVKRILRYLVATPCFGLWYP 904
Query: 1506 KSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAA 1565
K + L+G+ D+DYAG +++RKSTSG CQFLG +L+SW SKKQ ++A+STAEAEY++A
Sbjct: 905 KGSTFDLVGYSDSDYAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTSVALSTAEAEYVAAG 964
Query: 1566 NCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQ 1625
CC QLLWM+ L D+ N + +P+ CDN +AI +++NP+ HSR KHI+I+HHF+RD+ Q
Sbjct: 965 QCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVEHSRTKHIDIRHHFLRDHQQ 1024
Query: 1626 KGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
KG +++ + TE+Q ADIFTKPL + F ++ LN+
Sbjct: 1025 KGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1061
>gb|AAN40025.1| putative gag-pol polyprotein [Zea mays]
Length = 1892
Score = 984 bits (2545), Expect = 0.0
Identities = 540/1143 (47%), Positives = 707/1143 (61%), Gaps = 61/1143 (5%)
Query: 573 GSQAKITSKPENLKIKVMTKSD-PKSQKITIL-------KRSETVHQNLIKPESKIPKQK 624
G KI + P+NL V K+ P+ + IL +S+ +H K S
Sbjct: 750 GDNVKINAPPKNLSNFVKGKAPMPQDNEGYILYPAGYPENKSKKIHSR--KSHSGPNYAF 807
Query: 625 DQKNKAATASEKTIPKGVKPKVLNDQKTLSIHPKVQGRK----STIGNSSISINNVWLVD 680
K + + + + T K + K +N +I K S G S W++D
Sbjct: 808 MYKGETSRSRQPTHAKLPRKKTINASNDHAISFKTFDASYVLTSKCGKVYASGGTSWILD 867
Query: 681 --------GLKHNLLSISQFCDNGYDVTFSKTN------CTLVNKDDKSITFKGKRVENV 726
G K S + D +TF N T+ + D SI FKG +
Sbjct: 868 SGCTNHMTGEKKMFSSYEKNKDPQRAITFGDGNQGLVKGVTVFRRSDDSIAFKGVLEGQL 927
Query: 727 YKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDAL 786
Y + F D A+ CL++ + W+WH+RL H + + K+ K + + GL N+ + D +
Sbjct: 928 YLVVF-DRAELDT-CLIAKTNMGWLWHRRLAHVGMKNLHKLLKGEHILGLTNVHFEKDRI 985
Query: 787 CGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTW 846
C ACQ GK V + K+I++T RPLELLH+DLFGP+ S+ GSKY LVIVDDYSR+TW
Sbjct: 986 CSACQAGKQVGTHHPHKNIMTTDRPLELLHMDLFGPIAYISIGGSKYCLVIVDDYSRFTW 1045
Query: 847 VKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSS 906
V F++ K E F + Q+E L+I K+RSD+G EF+N E F E+ GI HEFSS
Sbjct: 1046 VFFLQEKSQTQETLKGFLRRAQNEFGLRIKKIRSDNGTEFKNSQIESFLEEEGIKHEFSS 1105
Query: 907 PRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAY 966
P TPQQNGVVERKNRTL +MARTM+ E FWAEAVNT+CY NR+Y+ +L+KT+Y
Sbjct: 1106 PYTPQQNGVVERKNRTLLDMARTMLDEYKTPDRFWAEAVNTACYAINRLYLHRILKKTSY 1165
Query: 967 ELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQC 1026
EL G++PNISYF FG C+IL + KF K G LGY ++AYRV+N T
Sbjct: 1166 ELLTGKKPNISYFRVFGSKCFILIKRGRKSKFAPKTVEGFLLGYDSNTRAYRVFNKSTGL 1225
Query: 1027 VEESMHVKFDDRE----PESKTSEQSESNAGTTDSEDASESDQ-PSDSEKYTEVESSP-- 1079
VE S V FD+ + E E A + S D P +SE+ + P
Sbjct: 1226 VEVSCDVVFDETNGSQVEQVDLDEIGEEQAPCIALRNMSIGDVCPKESEEPPSTQDQPPS 1285
Query: 1080 ----------EAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKH------- 1122
E E + E N E +P N+ + + +P+ H
Sbjct: 1286 SMQASPPTQNEDEAQNDEEQNQEVKPPQDKSNDQGGDTNDQEKEDEEEPRPPHPRVHQAI 1345
Query: 1123 KSSHHEELIIGSKDSPRRTRS---HFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEE 1179
+ H + I+G TRS HF + S + S IEP VEEAL D W++AMQEE
Sbjct: 1346 QRDHPVDTILGDIHKGVTTRSRVAHFCEHYSFV---SSIEPHRVEEALQDSDWVVAMQEE 1402
Query: 1180 LNQFQRNDVWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTE 1239
LN F RN+VW LVP+P Q N++GTKWVFRNK +E G VTRNKARLVA+GYSQ EG+D+ E
Sbjct: 1403 LNNFTRNEVWHLVPRPNQ-NVVGTKWVFRNKQDEHGVVTRNKARLVAKGYSQVEGLDFDE 1461
Query: 1240 TFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHV 1299
T+APVARLE+IR+LL+YA HG LYQMDVKSAFLNG I+EEVYV+QPPGFED ++PNHV
Sbjct: 1462 TYAPVARLESIRILLAYATYHGFKLYQMDVKSAFLNGPIKEEVYVEQPPGFEDSEYPNHV 1521
Query: 1300 YKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIF 1359
Y+L K+LYGLKQAPRAWY+ L +FLI N F+ G+ D TLF +TL+ D+ + QIYVDDIIF
Sbjct: 1522 YRLSKALYGLKQAPRAWYECLRDFLIANGFKVGKADPTLFTKTLENDLFVCQIYVDDIIF 1581
Query: 1360 GSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLE 1419
GSTN S C+EFS++M +FEMSMMGELK+FLG Q+ Q +EG ++ QTKYT+++L KF ++
Sbjct: 1582 GSTNESTCEEFSRIMTQKFEMSMMGELKYFLGFQVKQLREGTFISQTKYTQDILAKFGMK 1641
Query: 1420 DCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDP 1479
D K + TPM L + G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQSDP
Sbjct: 1642 DAKPIKTPMGTNGHLDLDTGGKSVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQSDP 1701
Query: 1480 RESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGE 1539
+ESHLTAVKRI RYL T GL Y + + LIG+ DAD+AG +I RKSTSG CQFLG
Sbjct: 1702 KESHLTAVKRILRYLAYTPKFGLWYPRGSTFDLIGYSDADWAGCKINRKSTSGTCQFLGR 1761
Query: 1540 NLISWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAIC 1599
+L+SWASKKQ ++A+STAEAEYI+A +CC QLLWM+ L DY +P+ CDN +AI
Sbjct: 1762 SLVSWASKKQNSVALSTAEAEYIAAGHCCAQLLWMRQTLRDYGYKLTKVPLLCDNESAIK 1821
Query: 1600 LSKNPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKN 1659
++ NP+ HSR KHI I++HF+RD+ QKG ++I +I+T+ Q ADIFTKPL + F ++
Sbjct: 1822 MADNPVEHSRTKHIAIRYHFLRDHQQKGDIEISYINTKDQLADIFTKPLDEQSFTRLRHE 1881
Query: 1660 LNM 1662
LN+
Sbjct: 1882 LNI 1884
Score = 113 bits (282), Expect = 6e-23
Identities = 124/509 (24%), Positives = 216/509 (42%), Gaps = 50/509 (9%)
Query: 4 DEESVTTK------YTSVKHDYDTADKKTDS-----GKAPKFNGDPEEFSWWKTNMYSFI 52
D SVT+K Y+ + Y K T GK P FNG E+++ W M +
Sbjct: 95 DAASVTSKRQERKKYSKIPLRYPRVPKHTPLLSVPLGKPPTFNG--EDYAMWSNLMRFHL 152
Query: 53 MGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYM 112
L + +WD++E GV + +E D ++ + + I++AS+ + EY
Sbjct: 153 TSLHKRIWDVVEYGVQVPSIGDEDYDTDE------VAQIEHFNSQAATILLASLSKEEYN 206
Query: 113 KMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVS 172
K+ AK ++ L EG + K K + + F ++ E ++MY+R +TLV+
Sbjct: 207 KVQGLKNAKEIWDLLKTAHEGDELTKITKRETIEGELGRFHLRQGEEPQDMYNRLKTLVN 266
Query: 173 GLQILKKSYVASDHVSK-ILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLN 231
++ L + V K ILR+L +V I ++ E+++ + E +
Sbjct: 267 QVRNLGSTKWDDHEVVKVILRALIFLNPTQVQLIRGNPRYPLMTPEEVIGNFVSFECMI- 325
Query: 232 EHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLAR 291
+ SKK + PS + A+KA+E ++E + Q + + L N+ L
Sbjct: 326 --KGSKKINELDEPSTSEAQPV--AFKATEEKKEE---STPSRQPIDASKLDNEEMALII 378
Query: 292 KQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRK 351
K + + K+ K+ K +K C+ C KPGHFI CP + + +G K K
Sbjct: 379 KSFRQILKQRKGKDYKPRSKKVCYKCGKPGHFIAKCP-MSSDSDRGDDKKGRRKEKKRYY 437
Query: 352 QIKKSLMATWEDLDS-ESGSDKEEADDDAKAA---------VGLVATVSSEAVSEAESDS 401
+ K + DS ES SD + +D A A VG ++ + + +S S
Sbjct: 438 KKKGGDAHVCREWDSDESSSDSSDDEDAANIAVTKGLLFPNVGHKCLMAKDGKKKVKSKS 497
Query: 402 EDENEVYSKIPRQELVDSLKELLSLF----EHRTNELTDLKEKYVDLMKQQKSTLLELKA 457
+ E S + D+L+ L + + + NEL + DL+ Q+ L +K
Sbjct: 498 STKYETSSDEDDKNEEDNLRILFANLNMEQKEKLNELISAIHEKDDLLDSQEDFL--IKE 555
Query: 458 SKEELKGFNLISTTYEDRLKSLCQKLQEK 486
+K+ +K N + E C+KL +
Sbjct: 556 NKKHVKVKNAYALEREK-----CEKLSSE 579
>ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (japonica cultivar-group)]
gi|28269414|gb|AAO37957.1| putative gag-pol polyprotein
[Oryza sativa (japonica cultivar-group)]
Length = 1969
Score = 972 bits (2512), Expect = 0.0
Identities = 554/1298 (42%), Positives = 760/1298 (57%), Gaps = 87/1298 (6%)
Query: 433 ELTDLKEKYV-----------------DLMKQQ------KSTLLELKASKEELKGFNLIS 469
EL +LKEK+V D ++QQ K+ LLE A+KE +
Sbjct: 510 ELKELKEKFVHDRIGRCENCPILTSDNDELRQQVAMLKTKNDLLESFATKEPIHSSCANC 569
Query: 470 TTYEDRLK-------------SLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMI 516
E LK S + K D S K L + K +MI
Sbjct: 570 AILETELKDAKTVIDSIKSIDSCSSCISLKVDLESAKKENSYLQQSLERFAQGKKKLNMI 629
Query: 517 YSTYKNK--GKGIGYSEEKSKEYSLKSYCDCIKDGLKS------TFVPEGTNAVTADQSK 568
K +GIGY +S + DG+ TF G T+ +
Sbjct: 630 LDQSKVSINNQGIGYDFAESLRIGTHEILG-VTDGMIELAQKPITFKSAGFIGNTSSSTP 688
Query: 569 SEASGSQAKITSKPENLKIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKDQKN 628
+ +TSK + +++ PK+ K K++ Q KP K +
Sbjct: 689 KTSEPKMVPMTSKSKPVELP-----RPKNPKQVEHKQN----QRQTKPVEKTKYECTYCG 739
Query: 629 KAA-------TASEKTIPKGVKPKVLNDQK------------TLSIHPKVQGR---KSTI 666
KA ++ + G + + K T++ GR K TI
Sbjct: 740 KAGHLDFGVGRSNSWLVDSGCSRHMTGEAKWFTSLTRASGDETITFGDASSGRVMAKGTI 799
Query: 667 G-NSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKRVEN 725
N + +V LV LK+NLLS+SQ CD +V F K +++ + + F RV
Sbjct: 800 KVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGR 858
Query: 726 VYKINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSD 784
V+ NF A CL+ S N + WH+RLGH + +S+IS + L++GLP + D
Sbjct: 859 VFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVQKD 918
Query: 785 ALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRW 844
+C C+ GK+ SS K +V T P +LLH+D GP S+ G Y LV+VDD+SR+
Sbjct: 919 LVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRY 978
Query: 845 TWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEF 904
+WV F++SK+ F S + E + +RSD+G EF+N FE FC+ G+ H+F
Sbjct: 979 SWVYFLESKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQF 1038
Query: 905 SSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKT 964
SSP PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT
Sbjct: 1039 SSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKT 1098
Query: 965 AYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSET 1024
YEL GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T
Sbjct: 1099 PYELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLST 1157
Query: 1025 QCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEIT 1084
+ E+ V FD+ P ++ + ED+ + D D ++S+P + T
Sbjct: 1158 NKIVETCEVTFDEASPGARPEISGVPDESIFVDEDSDDDD---DDSIPPPLDSTPPVQET 1214
Query: 1085 PEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSH 1144
+ S + +P + SA+E+ T P+ + +I G + R RS+
Sbjct: 1215 GSPSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSY 1273
Query: 1145 FRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTK 1204
+ + + EPK V ALSD+ W+ AM EEL F+RN VW LV P N+IGTK
Sbjct: 1274 ELVNSAFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTK 1330
Query: 1205 WVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIIL 1264
WVF+NKL E G + RNKARLVAQG++Q EG+D+ ETFAPVARLEAIR+LL++A + G L
Sbjct: 1331 WVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKL 1390
Query: 1265 YQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFL 1324
+QMDVKSAFLNGVIEEEVYVKQPPGFE+ K PNHV+KL+K+LYGLKQAPRAWY+RL FL
Sbjct: 1391 FQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFL 1450
Query: 1325 IKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMG 1384
++N FE G VD TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMG
Sbjct: 1451 LQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMG 1510
Query: 1385 ELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVD 1444
EL FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VD
Sbjct: 1511 ELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVD 1570
Query: 1445 QKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLY 1504
Q+ YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKRIFRY+K T G+ Y
Sbjct: 1571 QREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWY 1630
Query: 1505 RKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISA 1564
S + F DAD+AG +I+RKSTSG C FLG +L+SW+S+KQ+++A STAEAEY++A
Sbjct: 1631 SCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAA 1690
Query: 1565 ANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYV 1624
A+ C+Q+LWM L+DY ++ + +P+ CDNT+AI ++KNP+ HSR KHIEI++HF+RD V
Sbjct: 1691 ASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNV 1750
Query: 1625 QKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
+KG + ++F+++E Q ADIFTKPL RF+F++ L +
Sbjct: 1751 EKGTIVLEFVESEKQLADIFTKPLDRSRFEFLRSELGV 1788
Score = 117 bits (294), Expect = 3e-24
Identities = 116/499 (23%), Positives = 207/499 (41%), Gaps = 55/499 (11%)
Query: 31 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 87
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 88 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH 147
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 148 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 206
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVYGGFSLAA 234
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVD 326
++E + + L ARK + K K + CF C +P H V+
Sbjct: 235 LHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 327 CPDLQK------EKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAK 380
CP L+K +K +G+ K +K I K L A E S+ SD ++ + K
Sbjct: 292 CPKLKKKSDKTTKKPEGQGRKG--KKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDK 349
Query: 381 AAVGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKEL 423
G+ ++E A+ + + SE IP + DS+ +
Sbjct: 350 DFSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDE 409
Query: 424 LSLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASKEELKGFNLIST--TYED 474
LS E + + ++ +KY ++ +K + + LE+ + + + ST +Y
Sbjct: 410 LSK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLS 468
Query: 475 RLKSLCQKLQEKCDKGSGN 493
+ L KL + C G+GN
Sbjct: 469 EINLLKDKL-KSCALGAGN 486
>gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902378|ref|NP_912535.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2145
Score = 970 bits (2507), Expect = 0.0
Identities = 553/1298 (42%), Positives = 760/1298 (57%), Gaps = 87/1298 (6%)
Query: 433 ELTDLKEKYV-----------------DLMKQQ------KSTLLELKASKEELKGFNLIS 469
EL +LKEK+V D ++QQ K+ LLE A+KE +
Sbjct: 510 ELKELKEKFVHDRIGRCENCPILTSDNDELRQQVAMLRTKNDLLESFATKEPIHSSCANC 569
Query: 470 TTYEDRLK-------------SLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMI 516
E LK S + K D S K L + K +MI
Sbjct: 570 AILETELKDAKTVIDSIKSIDSCSSCISLKVDLESAKKENSYLQQSLERFAQGKKKLNMI 629
Query: 517 YSTYKNK--GKGIGYSEEKSKEYSLKSYCDCIKDGLKS------TFVPEGTNAVTADQSK 568
K +GIGY +S + DG+ TF G T+ +
Sbjct: 630 LDQSKVSINNQGIGYDFAESLRIGTHEILG-VTDGMIELAQKPITFKSAGFIGNTSSSTP 688
Query: 569 SEASGSQAKITSKPENLKIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKDQKN 628
+ +TSK + +++ PK+ K K++ Q KP K +
Sbjct: 689 KTSEPKMVPMTSKSKPVELP-----RPKNPKQVEHKQN----QRQTKPVEKTKYECTYCG 739
Query: 629 KAA-------TASEKTIPKGVKPKVLNDQK------------TLSIHPKVQGR---KSTI 666
KA ++ + G + + K T++ GR K TI
Sbjct: 740 KAGHLDFGVGRSNSWLVDSGCSRHMTGEAKWFTSLTRASSDETITFGDASSGRVMAKGTI 799
Query: 667 G-NSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKRVEN 725
N + +V LV LK+NLLS+SQ CD +V F K +++ + + F RV
Sbjct: 800 KVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGR 858
Query: 726 VYKINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSD 784
V+ NF A CL+ S N + WH+RLGH + +S+IS + L++GLP + D
Sbjct: 859 VFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKD 918
Query: 785 ALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRW 844
+C C+ GK+ SS K +V T P +LLH+D GP S+ G Y LV+VDD+SR+
Sbjct: 919 LVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRY 978
Query: 845 TWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEF 904
+WV F++SK+ F S + E + +RSD+G EF+N FE FC+ G+ H+F
Sbjct: 979 SWVYFLESKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQF 1038
Query: 905 SSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKT 964
SSP PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT
Sbjct: 1039 SSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKT 1098
Query: 965 AYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSET 1024
YEL GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T
Sbjct: 1099 PYELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLST 1157
Query: 1025 QCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEIT 1084
+ E+ V FD+ P ++ + ED+ + D D ++S+P + T
Sbjct: 1158 NKIVETCEVTFDEASPGARPEISGVPDESIFVDEDSDDDD---DDSIPPPLDSTPPVQET 1214
Query: 1085 PEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSH 1144
+ S + +P + SA+E+ T P+ + +I G + R RS+
Sbjct: 1215 GSPSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSY 1273
Query: 1145 FRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTK 1204
+ + + EPK V ALSD+ W+ AM EEL F+RN VW LV P N+IGTK
Sbjct: 1274 ELVNSAFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTK 1330
Query: 1205 WVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIIL 1264
WVF+NKL E G + RNKARLVAQG++Q EG+D+ ETFAPVARLEAIR+LL++A + G L
Sbjct: 1331 WVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKL 1390
Query: 1265 YQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFL 1324
+QMDVKSAFLNGVIEEEVYVKQPPGFE+ K PNHV+KL+K+LYGLKQAPRAWY+RL FL
Sbjct: 1391 FQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFL 1450
Query: 1325 IKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMG 1384
++N FE G VD TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMG
Sbjct: 1451 LQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMG 1510
Query: 1385 ELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVD 1444
EL FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VD
Sbjct: 1511 ELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVD 1570
Query: 1445 QKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLY 1504
Q+ YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKR+FRY+K T G+ Y
Sbjct: 1571 QREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWY 1630
Query: 1505 RKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISA 1564
S + F DAD+AG +I+RKSTSG C FLG +L+SW+S+KQ+++A STAEAEY++A
Sbjct: 1631 SCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAA 1690
Query: 1565 ANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYV 1624
A+ C+Q+LWM L+DY ++ + +P+ CDNT+AI ++KNP+ HSR KHIEI++HF+RD V
Sbjct: 1691 ASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNV 1750
Query: 1625 QKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
+KG + ++F+++E Q ADIFTKPL RF+F++ L +
Sbjct: 1751 EKGTIVLEFVESEKQLADIFTKPLDRSRFEFLRSELGV 1788
Score = 117 bits (294), Expect = 3e-24
Identities = 116/499 (23%), Positives = 207/499 (41%), Gaps = 55/499 (11%)
Query: 31 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 87
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 88 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH 147
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 148 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 206
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVYGGFSLAA 234
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVD 326
++E + + L ARK + K K + CF C +P H V+
Sbjct: 235 LHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 327 CPDLQK------EKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAK 380
CP L+K +K +G+ K +K I K L A E S+ SD ++ + K
Sbjct: 292 CPKLKKKSDKTTKKPEGQGRKG--KKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDK 349
Query: 381 AAVGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKEL 423
G+ ++E A+ + + SE IP + DS+ +
Sbjct: 350 DFSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDE 409
Query: 424 LSLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASKEELKGFNLIST--TYED 474
LS E + + ++ +KY ++ +K + + LE+ + + + ST +Y
Sbjct: 410 LSK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLS 468
Query: 475 RLKSLCQKLQEKCDKGSGN 493
+ L KL + C G+GN
Sbjct: 469 EINLLKDKL-KSCALGAGN 486
>ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultivar-group)]
gi|32488723|emb|CAE03600.1| OSJNBb0004A17.2 [Oryza sativa
(japonica cultivar-group)]
Length = 1877
Score = 969 bits (2506), Expect = 0.0
Identities = 497/1020 (48%), Positives = 675/1020 (65%), Gaps = 14/1020 (1%)
Query: 648 NDQKTLSIHPKVQGR---KSTIG-NSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSK 703
+ +T++ GR K TI N + +V LV LK+NLLS+SQ CD +V F K
Sbjct: 860 SSDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKK 919
Query: 704 TNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWR 762
+++ + + F RV V+ NF A CL+ S N + WH+RLGH +
Sbjct: 920 DRSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIGFD 978
Query: 763 LISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGP 822
+S+IS + L++GLP + D +C C+ GK+ SS K +V T P +LLH+D GP
Sbjct: 979 HLSRISGMDLIRGLPKLKVPKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGP 1038
Query: 823 VNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDH 882
S+ G Y LV+VDD+SR++WV F++SK+ F S + E + +RSD+
Sbjct: 1039 ARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRSDN 1098
Query: 883 GGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWA 942
G EF+N FE FC+ G+ H+FSSP PQQNGVVERKNRTL EMARTM+ E + FW
Sbjct: 1099 GSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWT 1158
Query: 943 EAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKA 1002
EA++ +C+I NR+++R +L KT YEL GRRP +S+ FGC C++L + + L KF++++
Sbjct: 1159 EAISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRS 1217
Query: 1003 QRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASE 1062
GIFLGY+ S+AYRVY T + E+ V FD+ P ++ + ED+ +
Sbjct: 1218 LDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVPDESIFVDEDSDD 1277
Query: 1063 SDQPSDSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKH 1122
D D ++S+P + T + S + +P + SA+E+ T P+
Sbjct: 1278 DD---DDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQ 1333
Query: 1123 KSSHHEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQ 1182
+ +I G + R RS+ + + + EPK V ALSD+ W+ AM EEL
Sbjct: 1334 NRHPPDSMIGGLGERVTRNRSYELVNSAFV---ASFEPKNVCHALSDENWVNAMHEELEN 1390
Query: 1183 FQRNDVWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFA 1242
F+RN VW LV P N+IGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ETFA
Sbjct: 1391 FERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFA 1450
Query: 1243 PVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKL 1302
PVARLEAIR+LL++A + G L+QMDVKSAFLNGVIEEEVYVKQPPGFE+ K PNHV+KL
Sbjct: 1451 PVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKL 1510
Query: 1303 KKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGST 1362
+K+LYGLKQAPRAWY+RL FL++N FE G VD TLF D L+VQIYVDDIIFG +
Sbjct: 1511 EKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGS 1570
Query: 1363 NASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCK 1422
+ +L +FS +M EFEMSMMGEL FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK
Sbjct: 1571 SHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCK 1630
Query: 1423 VMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRES 1482
+ TPM T +L ++ G VDQ+ YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR S
Sbjct: 1631 PIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTS 1690
Query: 1483 HLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLI 1542
H AVKR+FRY+K T G+ Y S + F DAD+AG +I+RKSTSG C FLG +L+
Sbjct: 1691 HRQAVKRMFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLV 1750
Query: 1543 SWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSK 1602
SW+S+KQ+++A STAEAEY++AA+ C+Q+LWM L+DY ++ + +P+ CDNT+AI ++K
Sbjct: 1751 SWSSRKQSSVAQSTAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAK 1810
Query: 1603 NPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
NP+ HSR KHIEI++HF+RD V+KG + ++F+++E Q ADIFTKPL RF+F++ L +
Sbjct: 1811 NPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESEKQLADIFTKPLDRSRFEFLRSELGV 1870
Score = 117 bits (294), Expect = 3e-24
Identities = 116/499 (23%), Positives = 207/499 (41%), Gaps = 55/499 (11%)
Query: 31 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 87
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 88 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH 147
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 148 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 206
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVYGGFSLAA 234
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVD 326
++E + + L ARK + K K + CF C +P H V+
Sbjct: 235 LHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 327 CPDLQK------EKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAK 380
CP L+K +K +G+ K +K I K L A E S+ SD ++ + K
Sbjct: 292 CPKLKKKSDKTTKKPEGQGRKG--KKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDK 349
Query: 381 AAVGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKEL 423
G+ ++E A+ + + SE IP + DS+ +
Sbjct: 350 DFSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDE 409
Query: 424 LSLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASKEELKGFNLIST--TYED 474
LS E + + ++ +KY ++ +K + + LE+ + + + ST +Y
Sbjct: 410 LSK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLS 468
Query: 475 RLKSLCQKLQEKCDKGSGN 493
+ L KL + C G+GN
Sbjct: 469 EINLLKDKL-KSCALGAGN 486
>gb|AAL35396.1| Opie2a pol [Zea mays]
Length = 1048
Score = 969 bits (2504), Expect = 0.0
Identities = 493/1021 (48%), Positives = 665/1021 (64%), Gaps = 25/1021 (2%)
Query: 658 KVQGRKSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSIT 717
KV+G +S SI+NV+LV+ L +N LS+SQ C+ GY+ F+ + ++ + D S+
Sbjct: 30 KVKGLGKIAISSEHSISNVFLVESLGYNFLSVSQLCNMGYNCLFTNVDVSVFRRSDGSLA 89
Query: 718 FKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLP 777
FKG +Y ++F+ CL++ W+WH+RL H + + K+ K + V GL
Sbjct: 90 FKGVLDGKLYLVDFAKEEAGLDACLIAKTSMGWLWHRRLAHVGMKNLHKLLKGEHVIGLT 149
Query: 778 NIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVI 837
N+ + D C ACQ GK V+ S +K++++TSRPLE+LH+DLFGPV S+ GSKYGLVI
Sbjct: 150 NVQFKKDRPCVACQAGKQVRGSHHTKNVMTTSRPLEMLHMDLFGPVAYLSIGGSKYGLVI 209
Query: 838 VDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEK 897
VDD+SR+TWV F++ K + + Q+E ELK+ K+RSD+G EF+N E F +
Sbjct: 210 VDDFSRFTWVFFLQDKSETQGTLKRYLRRAQNEFELKVKKIRSDNGSEFKNLQVEEFLVE 269
Query: 898 HGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYI 957
GI HEFS+P TPQQNGVVERKNRTL +MARTM+ E + FW+EAVNT+C+ NR+Y+
Sbjct: 270 EGIKHEFSAPYTPQQNGVVERKNRTLMDMARTMLGEFKTPERFWSEAVNTACHSINRVYL 329
Query: 958 RPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAY 1017
+L+ T+YEL G +PN+SYF FG CYIL K KF KA G LGY +KAY
Sbjct: 330 HRLLKNTSYELLTGNKPNVSYFRVFGSKCYILVKKGRTSKFAPKAVEGFLLGYDSNTKAY 389
Query: 1018 RVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDS-------- 1069
RV+N + VE S V FD E EQ + +D E D P+ +
Sbjct: 390 RVFNKSSGLVEVSSDVVFD--ETNGSLREQ------VVNLDDVDEEDVPTAAMRTMAIGD 441
Query: 1070 ---EKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQV-----IQPKFK 1121
+++ E + + + E P ++ ++D Q ++ Q +
Sbjct: 442 VRPQEHLEQDQPSSTTMVHPPTQDDEQAPQVGAHDQGGAQDVQVEEEEAPQAPPTQVRAT 501
Query: 1122 HKSSHHEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELN 1181
+ +H I+G TRS +S IEP VEE L D W+LAMQEELN
Sbjct: 502 IQRNHPVNQILGDISKGVTTRSRLVNFCEHYSFVSSIEPFRVEEVLLDPDWVLAMQEELN 561
Query: 1182 QFQRNDVWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETF 1241
F+RN+VW LVP+P Q N++GTKWVFRNK +E G VTRNKARLVA+GY+Q G+D+ ETF
Sbjct: 562 NFKRNEVWTLVPRPKQ-NVVGTKWVFRNKQDEHGVVTRNKARLVAKGYAQVAGLDFEETF 620
Query: 1242 APVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYK 1301
APVARL++IR+ L+YA +H LYQMDVKSAFLNG I+EEVYV+QPPGFED + P+HV K
Sbjct: 621 APVARLKSIRIWLAYAAHHSFRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERFPDHVCK 680
Query: 1302 LKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGS 1361
L K+LYGLKQAPRAWY+ L +FLI N F+ G+ D TLF +T D+ + QIYVDDIIFGS
Sbjct: 681 LSKALYGLKQAPRAWYECLRDFLIANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGS 740
Query: 1362 TNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDC 1421
TN + C+EFS++M +FEMSMMGEL +FLG Q+ Q K+G ++ QTKYT++L+K+F ++D
Sbjct: 741 TNQNSCEEFSRVMTQKFEMSMMGELSYFLGFQVRQLKDGTFISQTKYTQDLIKRFGMKDA 800
Query: 1422 KVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRE 1481
K TPM G VDQK YR IGSLLYL ASRPDI+ SVC+CARFQSDPRE
Sbjct: 801 KPAKTPMGTDGHTDLNKGGKSVDQKAYRSTIGSLLYLCASRPDIMLSVCMCARFQSDPRE 860
Query: 1482 SHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENL 1541
HL AVKRI RYL T G+ Y K + LIG+ D+DYA +++RKSTS CQFLG +L
Sbjct: 861 CHLVAVKRILRYLVATPCFGIWYPKGSTFDLIGYSDSDYARCKVDRKSTSRMCQFLGRSL 920
Query: 1542 ISWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLS 1601
+SW SKKQ ++A+STAEAEY++ CC QLLWM+ L D+ N + +P+ CDN +AI ++
Sbjct: 921 VSWNSKKQTSVALSTAEAEYVAVGQCCAQLLWMRQTLRDFGYNLSEVPLLCDNESAIRIA 980
Query: 1602 KNPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLN 1661
+NP+ HSR KHI+I+HHF+RD+ QKG++++ + +E+ ADIFTKPL + F + LN
Sbjct: 981 ENPVEHSRTKHIDIRHHFLRDHQQKGVIEVFHVSSENHLADIFTKPLDEQTFCKLHSELN 1040
Query: 1662 M 1662
+
Sbjct: 1041 V 1041
>ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 1410
Score = 966 bits (2498), Expect = 0.0
Identities = 491/996 (49%), Positives = 664/996 (66%), Gaps = 10/996 (1%)
Query: 668 NSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKRVENVY 727
N + +V LV LK+NLLS+SQ CD +V F K +++ + + F RV V+
Sbjct: 417 NDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVF 475
Query: 728 KINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDAL 786
NF A CL+ S N + WH+RLGH + +S+IS + L++GLP + D +
Sbjct: 476 FANFDSSAPGPSRCLVASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKAPKDLV 535
Query: 787 CGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTW 846
C C+ GK+ SS K +V T P +LLH++ GP S+ G Y LV+VDD+SR++W
Sbjct: 536 CAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMNTVGPARVQSVGGKWYVLVVVDDFSRYSW 595
Query: 847 VKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSS 906
V F++SK+ F S + E + +RSD+G EF+N FE FC+ G+ H+FSS
Sbjct: 596 VYFLESKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSS 655
Query: 907 PRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAY 966
P PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT Y
Sbjct: 656 PYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPY 715
Query: 967 ELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQC 1026
EL GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T
Sbjct: 716 ELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNK 774
Query: 1027 VEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEITPE 1086
+ E+ V FD+ P ++ + ED+ + D D ++S+P + T
Sbjct: 775 IVETCEVTFDEASPGARPEISGVLDESIFVDEDSDDDD---DDSIPPPLDSTPPVQETGS 831
Query: 1087 AESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSHFR 1146
+ S + +P + SA+E+ T P+ + +I G + R RS+
Sbjct: 832 PSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYDL 890
Query: 1147 QEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKWV 1206
+ + + EPK V ALSD+ W+ AM EEL F+RN VW LV P N+IGTKWV
Sbjct: 891 VNSAFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWV 947
Query: 1207 FRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQ 1266
F+NKL E G + RNKARLVAQG++Q EG+D+ ETFAPVARLEAIR+LL++A + G L+Q
Sbjct: 948 FKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQ 1007
Query: 1267 MDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLIK 1326
MDVKSAFLNGVIEEEVYVKQPPGFE+ K PNHV+KL K+LYGLKQAPRAWY+RL FL++
Sbjct: 1008 MDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLDKALYGLKQAPRAWYERLKTFLLQ 1067
Query: 1327 NDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGEL 1386
N FE G VD TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMGEL
Sbjct: 1068 NGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGEL 1127
Query: 1387 KFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQK 1446
FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+
Sbjct: 1128 TFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQR 1187
Query: 1447 LYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRK 1506
YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKRIFRY+K T G+ Y
Sbjct: 1188 EYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSC 1247
Query: 1507 SLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAAN 1566
S + F DAD+AG +I+RKSTSG C FLG +L+SW+S+KQ+++A STAEAEY++AA+
Sbjct: 1248 SSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAAS 1307
Query: 1567 CCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQK 1626
C+Q+LWM L+DY ++ + +P+ CDNT+AI ++KNP+ HSR KHIEI++HF+RD V+K
Sbjct: 1308 ACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEK 1367
Query: 1627 GILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
G + ++F+++E Q ADIFTKPL RF+F++ L +
Sbjct: 1368 GTIVLEFVESEKQLADIFTKPLDRSRFEFLRSELGV 1403
Score = 110 bits (275), Expect = 4e-22
Identities = 82/305 (26%), Positives = 139/305 (44%), Gaps = 17/305 (5%)
Query: 85 TPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALM 144
TPA +++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 29 TPANMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQ 88
Query: 145 LVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVT 203
+Y+ F M ESI+ + RF +VS L+ + K + +D+ +L L W KVT
Sbjct: 89 FHKEYQRFEMHPGESIDSYFKRFGEIVSKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVT 148
Query: 204 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESE 263
+I E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+
Sbjct: 149 SITESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVCGGFS 208
Query: 264 EESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHF 323
+ ++E + + L ARK + + K K + CF C +P H
Sbjct: 209 LAALHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRNKKRGKTNEPFVCFECGEPNHI 265
Query: 324 IVDCPDLQK------EKFKGKSMKSSFNSSKFRKQIKKSLMATWE----DLDSESGSDKE 373
V+CP L+K +K +G+ K + +K I K L A E D+DS+ D+E
Sbjct: 266 RVNCPKLKKKSDKTTKKPEGQGRKG--KNDLMKKAIHKVLAALEEVQLSDIDSDD-DDQE 322
Query: 374 EADDD 378
+ D D
Sbjct: 323 KGDKD 327
>gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1577
Score = 937 bits (2423), Expect = 0.0
Identities = 492/1024 (48%), Positives = 650/1024 (63%), Gaps = 36/1024 (3%)
Query: 648 NDQKTLSIHPKVQGRKSTIG----NSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSK 703
N+Q+ ++ +G+ +G ++ +SI+NV LV L NLLS++Q CD F
Sbjct: 574 NEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVSLVKSLNFNLLSVAQICDLSLSCAFFP 633
Query: 704 TNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRL 763
+ + DKS FKG R N+Y ++F+ CL++ W+WH+RL H
Sbjct: 634 QEVIVSSLLDKSCVFKGFRYGNLYLVDFNSSEANLKTCLVAKTSLGWLWHRRLAHVGMNQ 693
Query: 764 ISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPV 823
+SK SK LV GL ++ + D LC ACQ GK V S +K I+STS+PLELLH+DLF P
Sbjct: 694 LSKFSKRDLVMGLKDVKFEKDKLCSACQAGKQVACSHPTKSIMSTSKPLELLHMDLFDPT 753
Query: 824 NTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHG 883
S+ G+ + LVIVDDYSR+TWV F+ K ++F F + Q+E ++K+RS+ G
Sbjct: 754 TYKSIGGNSHCLVIVDDYSRYTWVFFLHDKSIVADLFKKFAKRAQNEFSCTLVKIRSNIG 813
Query: 884 GEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAE 943
EF+N E +C+ GI HE + +PQQNGVVERKNRTL EMARTM+ E ++ FWAE
Sbjct: 814 SEFKNTNIEDYCDDLGIKHELFATYSPQQNGVVERKNRTLIEMARTMLDEYGVSDSFWAE 873
Query: 944 AVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQ 1003
A+NT+C+ NR+Y+ +L+KT+YE+ GR+PNI+YF FGC CYI L KF+++
Sbjct: 874 AINTACHATNRLYLHRVLKKTSYEVIVGRKPNIAYFRVFGCKCYIHRKGVRLTKFESRCD 933
Query: 1004 RGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASE- 1062
G LGY+ +SKAYRVYN VEE+ V+FD E+ S++ N E
Sbjct: 934 EGFLLGYASKSKAYRVYNKNKGIVEETADVQFD----ETNGSQEGHENLDDVGDEGLMRV 989
Query: 1063 -SDQPSDSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFK 1121
+ K EVE P + + E ++ A PS ++ E ++ Q+ P
Sbjct: 990 MKNMSIGDVKPIEVEDKPST--STQDEPSTSAMPS-----QAQVEVEEEKAQEPPMPPRI 1042
Query: 1122 HKS---SHHEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQE 1178
H + H + ++G +TRS +S +E K V+EAL D W+ AM E
Sbjct: 1043 HTALSKDHPIDQVLGDISKGVQTRSRVTSICEHYSFVSCLERKHVDEALCDPDWMNAMHE 1102
Query: 1179 ELNQFQRNDVWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYT 1238
EL F RN VW LV +P N+IGTKWVFRNK +E G V RNKARLVAQG++Q EG+D+
Sbjct: 1103 ELKNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENGLVVRNKARLVAQGFTQVEGLDFG 1162
Query: 1239 ETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNH 1298
ETFAPVARLEAI +LL++A I L+QMDVKSAFLN D K+PNH
Sbjct: 1163 ETFAPVARLEAICILLAFASCFDIKLFQMDVKSAFLN----------------DTKYPNH 1206
Query: 1299 VYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDII 1358
VYKL K+LYGL+QAPRAWY+RL +FL+ DF+ G+VD TLF + + D + QIYVDDII
Sbjct: 1207 VYKLSKALYGLRQAPRAWYERLRDFLLSKDFKIGKVDITLFTKIIGDDFFVYQIYVDDII 1266
Query: 1359 FGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKL 1418
FGSTN CKEF +M EFEMSM+GEL FFLG+QI Q K G +V QTKY K+LLK+F L
Sbjct: 1267 FGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIKQLKNGTFVSQTKYIKDLLKRFGL 1326
Query: 1419 EDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSD 1478
ED K + TPM L ++ G VD KLYR MIGSLLYLT SRPDI+FSVC+CARFQ+
Sbjct: 1327 EDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTVSRPDIMFSVCMCARFQAA 1386
Query: 1479 PRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLG 1538
P+E HL AVKRI RYLK ++ +GL Y K +KL+G+ D DYAG +++RKSTS +CQ LG
Sbjct: 1387 PKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVGYSDPDYAGCKVDRKSTSSSCQMLG 1446
Query: 1539 ENLISWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAI 1598
+L+SW+SKKQ ++A+STAE EY+SA +CC QLLWMK L DY I+ P+ CDN AI
Sbjct: 1447 RSLVSWSSKKQNSVALSTAETEYVSAGSCCAQLLWMKQTLLDYGISFTKTPLLCDNDGAI 1506
Query: 1599 CLSKNPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKK 1658
++ NP+ HSR KHI+I+HHF+RD+V K + I I TE Q ADIFTKPL RF ++
Sbjct: 1507 KIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHIRTEDQLADIFTKPLDETRFCKLRN 1566
Query: 1659 NLNM 1662
LN+
Sbjct: 1567 ELNI 1570
Score = 88.2 bits (217), Expect = 2e-15
Identities = 115/463 (24%), Positives = 190/463 (40%), Gaps = 83/463 (17%)
Query: 29 GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDG--VDDLDLDEEGAAIDRR---I 83
GKAP FNG +S WK M + + + +W I++ G + L E ID R +
Sbjct: 9 GKAPMFNGT--NYSTWKIKMSTHLKAMSFHIWSIVDVGFAITGTPLME----IDHRNLQL 62
Query: 84 HTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKAL 143
+ A L+ S+ + E+ ++S+ TA ++ L EG+ + K+AK
Sbjct: 63 NAQAMNALFN-----------SLSQEEFDRVSNLETAYEIWNKLAEIHEGTSEYKDAKLH 111
Query: 144 MLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVT 203
L QYE F M ES+ +MY R +V+ L+ L +Y + K+LR+LP ++ VT
Sbjct: 112 FLKIQYETFSMLPHESVNDMYGRLNVIVNDLKGLGANYTDLEVAQKMLRALPEKYETLVT 171
Query: 204 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSK--GKTSKSSKAYKASE 261
+ + D++ ++ L+ + ++M E K S S ++ KT + + ++ E
Sbjct: 172 MLINS-DMSRMTPASLLGKINTNDM---RKERGKGSNSNRRRNRRPNKTLSNLRCFECGE 227
Query: 262 S---EEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCK 318
+ P D D D+S K K G YK KK ++G K
Sbjct: 228 KGHFASKCPSKDDDGDKSSK-------------------KKSGGYKLMKKLKKEG----K 264
Query: 319 KPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDD 378
K FI + S+ SS KK +M G +
Sbjct: 265 KIEAFI-------------EEWDSNEESSPHPGPRKKMVMM------QAPGRRRWPLLPS 305
Query: 379 AKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLK 438
+ + VS + +ESD + ++ Y ELV +EL + E + LK
Sbjct: 306 RRLHHSSLHFVSWQKAPLSESDDDCDDVSY-----DELVSMFEELHAYSEKEIVKFKALK 360
Query: 439 EKYVD---LMKQQKSTLLELKASKEELKGF--NLISTTYEDRL 476
+ + L ++ K++ L S E+LK NL+STT L
Sbjct: 361 KDHASLEVLYEELKTSHERLTISHEKLKEAHDNLLSTTQHGAL 403
>emb|CAE04884.2| OSJNBa0042I15.6 [Oryza sativa (japonica cultivar-group)]
Length = 1510
Score = 936 bits (2418), Expect = 0.0
Identities = 488/981 (49%), Positives = 638/981 (64%), Gaps = 34/981 (3%)
Query: 711 KDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKL 770
+DD SI FKG ++Y ++F CL++ + W+WH+RL H R ++ + K
Sbjct: 521 RDDSSIAFKGVLKGDLYLVDFDVDRVNPEACLIAKSSMGWLWHRRLAHVGMRNLASLLKG 580
Query: 771 QLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYG 830
+ + GL N+ + D +C ACQ GK V S K+I++T+RPLELLH+DLFGPV S+ G
Sbjct: 581 EHILGLSNVSFEMDRVCSACQAGKQVGSPHPIKNIMTTTRPLELLHMDLFGPVAYISIGG 640
Query: 831 SKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEP 890
+KYG VIVDD+S +TWV F+ K A +VF F Q Q+ +L I +VRSD+GGEF+N
Sbjct: 641 NKYGFVIVDDFSCFTWVYFLHDKSEAQDVFKRFTKQAQNLYDLTIKRVRSDNGGEFKNTQ 700
Query: 891 FELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCY 950
E F ++ GI HEFS+P P QNG+VERKNRTL E AR M+ E + FWAEAV+T+C+
Sbjct: 701 VEEFLDEEGIKHEFSAPYDPPQNGIVERKNRTLIEAARAMLDEYKTSDVFWAEAVSTACH 760
Query: 951 IQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGY 1010
NR+Y+ +L+KT+YEL G++PN+SYF FG +IL+ KF K G LGY
Sbjct: 761 AINRLYLHKILKKTSYELLSGKKPNVSYFRVFGSKFFILSKMPRSSKFSPKVDEGFLLGY 820
Query: 1011 SERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASES-DQP--S 1067
+ AYRV+N +T + +HV D +P ++ + +++D E DQP S
Sbjct: 821 ESNAHAYRVFN-KTSGEQVVIHV-VRDVDPSQAIGTKAIGDIRPVETQDDQEDRDQPPSS 878
Query: 1068 DSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNE-------SASEDFQD---------- 1110
S T V S+ E E+ + N P P+V S SED
Sbjct: 879 TSNSPTSVVSA-EPEVPGPIDRNLRTSPGPEVPGSTVRNLRTSGSEDVPTAQVDGIDAAG 937
Query: 1111 ---NTQQVIQPKFKHKSSHHE-------ELIIGSKDSPRRTRSHFRQEESLIGLLSIIEP 1160
+T Q P H HH + I+G TRS +S +EP
Sbjct: 938 TLGHTDQAQVPLVHHPRIHHTVQRDHPVDNILGDIRKGVTTRSRVASFCQHYSFVSSLEP 997
Query: 1161 KTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRN 1220
VE+AL D W++AMQEELN F RN VW+LV +P Q N+IGTKW+FRNK +E V RN
Sbjct: 998 TRVEDALGDSDWVMAMQEELNNFARNQVWNLVERPKQ-NVIGTKWIFRNKQDEHVVVVRN 1056
Query: 1221 KARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEE 1280
KARLV QG++Q EG+D+ ETFAPVARLE+IR+LL+YA +H L+QMDVKSAFLNG I E
Sbjct: 1057 KARLVTQGFTQVEGLDFGETFAPVARLESIRILLAYAAHHDFRLFQMDVKSAFLNGPISE 1116
Query: 1281 EVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFR 1340
VYV+QPPGFED K PNHVYKL K+LYGLKQAPRAWY+ L +FL+KN FE G DTTLF
Sbjct: 1117 LVYVEQPPGFEDPKLPNHVYKLHKALYGLKQAPRAWYECLRDFLLKNGFEIGNADTTLFT 1176
Query: 1341 RTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEG 1400
+ K D+ I QIYVDDIIFGSTNAS C+EFS +M FEMSMMGEL FFL +Q+ Q++EG
Sbjct: 1177 KKFKSDLFICQIYVDDIIFGSTNASFCEEFSSIMTKRFEMSMMGELTFFLWLQVKQAQEG 1236
Query: 1401 VYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTA 1460
++ QTKY K++LKKF +ED K + TPM L +D G VDQK+YR MIGSLLYL A
Sbjct: 1237 TFISQTKYVKDILKKFGMEDAKPIKTPMPTNGHLDLDDNGKCVDQKVYRSMIGSLLYLCA 1296
Query: 1461 SRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADY 1520
SRPDI+ SVC+CARFQ++P+E HL AVKRI RYL T NLGL Y K D++L+G+ D+DY
Sbjct: 1297 SRPDIMLSVCMCARFQAEPKECHLIAVKRIQRYLVHTPNLGLWYPKGCDFELLGYSDSDY 1356
Query: 1521 AGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLED 1580
AG +++RKS +G CQFLG +L+SW KKQ +I +ST EAEY++A +CC QLLWMK L+D
Sbjct: 1357 AGCKVDRKSITGTCQFLGPSLVSWFPKKQNSIVLSTTEAEYVAAGSCCAQLLWMKQTLKD 1416
Query: 1581 YQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQW 1640
+ N P+ CDN +AI ++ NP+ HS+ KHI+I HHF+RD+ KG + + + TE Q
Sbjct: 1417 FGYNFTKTPLLCDNESAIKIANNPVQHSKTKHIDIHHHFLRDHETKGDICLTHVRTETQL 1476
Query: 1641 ADIFTKPLSVERFDFIKKNLN 1661
ADIFTKPL +RF ++ LN
Sbjct: 1477 ADIFTKPLDEKRFCELRSELN 1497
Score = 92.8 bits (229), Expect = 9e-17
Identities = 106/436 (24%), Positives = 175/436 (39%), Gaps = 87/436 (19%)
Query: 52 IMGLDEELWDILEDGVD----DLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIP 107
++ L +W ++ GVD D++L E Q++L ++ + I++++
Sbjct: 5 LISLHPSIWKVVCTGVDVPHDDMELTSE------------QEQLIHRNAQASNAILSTLS 52
Query: 108 RTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRF 167
E+ K+ AK + +L EGS V+EAK +L + F M D E+ +EMY R
Sbjct: 53 LEEFNKVDGLEEAKEICDTLQLAHEGSPAVREAKIELLEGRLGRFVMDDKETPQEMYDRM 112
Query: 168 QTLVSGLQILKKSYVASDHVSK-ILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVH 226
LV+ ++ L + + V K +LR R V+ I E KD L++ D++ + H
Sbjct: 113 MILVNKIKGLGSEDMTNHFVVKRLLREFGPRNPTLVSMIREKKDFKRLTLSDILGRIVSH 172
Query: 227 EMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKL 286
EM E KT + K +K + G D+D+
Sbjct: 173 EMQEEEAR--------------KTRRRVKRFKHFLRKSGYGKGRKDDDK----------- 207
Query: 287 EYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNS 346
KK+ ++ CFNC + GHFI D P + K KG K
Sbjct: 208 -------------------GKKQSKRACFNCGEYGHFIADFPKSNEAKAKGGKKKPE--- 245
Query: 347 SKFRKQIKKSLM-ATWEDLDSESGSDKEEADDDAKA-AVGLVATVSSEAVSEAE------ 398
R + ++ M W D E K + K G VATV+ ++ S ++
Sbjct: 246 ---RAHVAEAHMPEVWYSGDEEDPEVKPKPKPKDKVDGEGGVATVTFKSSSSSKERLFNN 302
Query: 399 -SDSEDENEVYS-------KIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKS 450
SD +D++ YS K+ Q+ + ++ S E NEL D+ + + Q +
Sbjct: 303 LSDDDDDSYHYSCFMAQGRKVMTQKPSHTSLDVDSSDEESDNELDDVLKSFSKPAMQHLA 362
Query: 451 TLLE----LKASKEEL 462
L+ LK E L
Sbjct: 363 KLMRALDTLKKENERL 378
>ref|XP_468929.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|28209450|gb|AAO37468.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1128
Score = 920 bits (2378), Expect = 0.0
Identities = 477/948 (50%), Positives = 626/948 (65%), Gaps = 15/948 (1%)
Query: 719 KGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPN 778
+GK E K+ F D + + V+ L++ W+WH+RL H +SK+SK LV GL +
Sbjct: 185 EGKEQE---KVTFGDNSKRNVIGLVAKTSFGWLWHRRLAHVGMNQLSKLSKRDLVVGLKD 241
Query: 779 IDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIV 838
+ + D LC ACQ K V S +K I+STSRPLELLH+DLFGP S+ G+ + LVIV
Sbjct: 242 VKFEKDKLCSACQASKQVACSHPTKSIMSTSRPLELLHMDLFGPTTYKSIGGNSHCLVIV 301
Query: 839 DDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKH 898
DDYS +TWV F+ K E+F F + Q+E ++K+RSD+G +F+N E +C+
Sbjct: 302 DDYSCYTWVFFLHDKCIVAELFKKFAKRAQNEFSCTLVKIRSDNGSKFKNTNIEDYCDDL 361
Query: 899 GILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIR 958
I HE S+ +PQQNGVVERKNRTL EMARTM+ E ++ FWAEA+NT+C+ NR+Y+
Sbjct: 362 SIKHELSATYSPQQNGVVERKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLH 421
Query: 959 PMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYR 1018
+L+KT+YEL GR+PN++YF FGC CYI L KF+++ G LGY+ SKAYR
Sbjct: 422 RLLKKTSYELIVGRKPNVAYFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYR 481
Query: 1019 VYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESS 1078
VYN VEE+ V+FD E+ S++ N E + + +E
Sbjct: 482 VYNKNKGIVEETADVQFD----ETNGSQEGHENLDDVGDEGLMRAMKNMSIGDVKPIEVE 537
Query: 1079 PEAEITPEAESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSS--HHEELIIGSKD 1136
+ + + E ++ A PS + Q E E QD + P+ S H + ++G
Sbjct: 538 DKPSTSTQDEPSTSASPS-QAQVEVEKEKAQDPP---MPPRIYTALSKDHPIDQVLGDIS 593
Query: 1137 SPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPF 1196
+TRS +S +EPK V+EAL D W+ A+ EELN F RN VW LV +P
Sbjct: 594 KGVQTRSPVASICEHYSFVSCLEPKHVDEALYDPDWMNAIHEELNNFARNKVWTLVERPR 653
Query: 1197 QKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSY 1256
N+IGTKWVFRNK +E V RNKARLVAQG++Q E +D+ ETF PVARLEAIR+LL++
Sbjct: 654 DHNVIGTKWVFRNKQDENRLVVRNKARLVAQGFTQVEDLDFGETFGPVARLEAIRILLAF 713
Query: 1257 AINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAW 1316
A I L+QMDVKSAFLNG I E V+V+QPPGF+D K+PNHVYKL K+LYGLKQAPRAW
Sbjct: 714 ASCFDIKLFQMDVKSAFLNGEIAELVFVEQPPGFDDPKYPNHVYKLSKALYGLKQAPRAW 773
Query: 1317 YDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQD 1376
Y+RL +FL+ DF+ G+VDTTLF + + D + QIYVDDIIFGSTN CKEF +M
Sbjct: 774 YERLRDFLLSKDFKIGKVDTTLFTKIIGDDFFVCQIYVDDIIFGSTNEVFCKEFGDMMSR 833
Query: 1377 EFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSK 1436
EFEMSM+ EL FFLG+QI Q K+G +V QTKY K+LLK+F LED K + TPM L
Sbjct: 834 EFEMSMIEELSFFLGLQIKQLKDGTFVSQTKYIKDLLKRFGLEDAKPIKTPMATNWHLDL 893
Query: 1437 EDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKG 1496
++ G VD KLYR MIGSLLYLTASRPDI+FSVC+ ARFQ+ P+E HL AVKRI RYLK
Sbjct: 894 DEGGKPVDLKLYRSMIGSLLYLTASRPDIMFSVCMYARFQAAPKECHLVAVKRILRYLKH 953
Query: 1497 TTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMST 1556
++ + L Y K +KL+G+ D+DYAG +++RKSTSG+CQ LG +L+SW+SKKQ ++A+ST
Sbjct: 954 SSTISLWYPKGAKFKLVGYSDSDYAGYKVDRKSTSGSCQMLGRSLVSWSSKKQNSVALST 1013
Query: 1557 AEAEYISAANCCTQLLWMKHQLEDYQIN--ANSIPIYCDNTAAICLSKNPILHSRAKHIE 1614
AEAEYISA +CC QLLWMK L DY I+ P+ C+N + I ++ NP+ H R KHI+
Sbjct: 1014 AEAEYISAGSCCAQLLWMKQILLDYGISFTETQTPLLCNNDSTIKIANNPVQHFRTKHID 1073
Query: 1615 IKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
I+HHF+ D+V K + I I TE Q ADIFTKPL RF ++ LN+
Sbjct: 1074 IRHHFLTDHVAKCDIVISHIRTEDQLADIFTKPLDETRFCKLRNELNV 1121
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.131 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,732,729,465
Number of Sequences: 2540612
Number of extensions: 117196508
Number of successful extensions: 476521
Number of sequences better than 10.0: 7532
Number of HSP's better than 10.0 without gapping: 3679
Number of HSP's successfully gapped in prelim test: 4056
Number of HSP's that attempted gapping in prelim test: 426488
Number of HSP's gapped (non-prelim): 34313
length of query: 1667
length of database: 863,360,394
effective HSP length: 142
effective length of query: 1525
effective length of database: 502,593,490
effective search space: 766455072250
effective search space used: 766455072250
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 82 (36.2 bits)
Medicago: description of AC147435.7


