

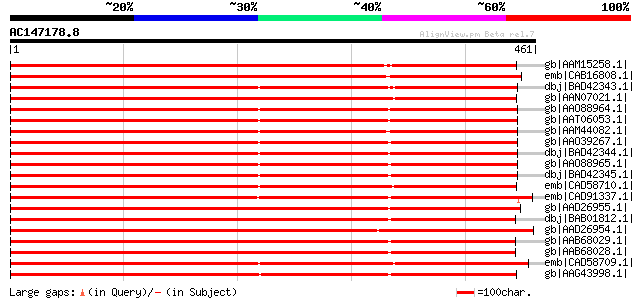
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.8 + phase: 0
(461 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM15258.1| putative sugar transporter [Arabidopsis thaliana]... 481 e-134
emb|CAB16808.1| sugar transporter like protein [Arabidopsis thal... 467 e-130
dbj|BAD42343.1| sorbitol transporter [Malus x domestica] 456 e-127
gb|AAN07021.1| putative mannitol transporter [Orobanche ramosa] 454 e-126
gb|AAO88964.1| sorbitol transporter [Malus x domestica] 452 e-125
gb|AAT06053.1| sorbitol transporter [Malus x domestica] 451 e-125
gb|AAM44082.1| putative sorbitol transporter [Prunus cerasus] 449 e-124
gb|AAO39267.1| sorbitol transporter [Prunus cerasus] 447 e-124
dbj|BAD42344.1| sorbitol transporter [Malus x domestica] 447 e-124
gb|AAO88965.1| sorbitol transporter [Malus x domestica] 445 e-123
dbj|BAD42345.1| sorbitol transporter [Malus x domestica] 445 e-123
emb|CAD58710.1| polyol transporter [Plantago major] 440 e-122
emb|CAD91337.1| sorbitol-like transporter [Glycine max] 439 e-121
gb|AAD26955.1| putative sugar transporter [Arabidopsis thaliana]... 434 e-120
dbj|BAB01812.1| sugar transporter protein [Arabidopsis thaliana]... 431 e-119
gb|AAD26954.1| putative sugar transporter [Arabidopsis thaliana]... 431 e-119
gb|AAB68029.1| putative sugar transporter; member of major facil... 430 e-119
gb|AAB68028.1| putative sugar transporter; member of major facil... 430 e-119
emb|CAD58709.1| polyol transporter [Plantago major] 429 e-118
gb|AAG43998.1| mannitol transporter [Apium graveolens var. dulce] 422 e-116
>gb|AAM15258.1| putative sugar transporter [Arabidopsis thaliana]
gi|4218010|gb|AAD12218.1| putative sugar transporter
[Arabidopsis thaliana] gi|15224183|ref|NP_179438.1|
mannitol transporter, putative [Arabidopsis thaliana]
gi|25308961|pir||G84564 probable sugar transporter
[imported] - Arabidopsis thaliana
Length = 508
Score = 481 bits (1239), Expect = e-134
Identities = 234/445 (52%), Positives = 324/445 (72%), Gaps = 3/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA +FI+++L+I+D Q+++LAGILN CAL + AG+ SD IGRRYTI LS++ F +G
Sbjct: 43 MSGAQIFIRDDLKINDTQIEVLAGILNLCALVGSLTAGKTSDVIGRRYTIALSAVIFLVG 102
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
S+LMGYG ++P+LM+GRCIAG GVGFAL+I VYSAEISS S+RGFLTSLP+L I++G L
Sbjct: 103 SVLMGYGPNYPVLMVGRCIAGVGVGFALMIAPVYSAEISSASHRGFLTSLPELCISLGIL 162
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY GKL+L+LGWR+ML I + PS+ L + ++ ESPRWLVMQGRL +AKK+++
Sbjct: 163 LGYVSNYCFGKLTLKLGWRLMLGIAAFPSLILAFGITRMPESPRWLVMQGRLEEAKKIMV 222
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
L+SN+++EAE+R ++I A +D + + KK G +E+ KP P V ILI
Sbjct: 223 LVSNTEEEAEERFRDILTAAEVDVTEIKEVGGGVKKKNHGKSVWRELVIKPRPAVRLILI 282
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H F++ G+E + LYSPRIF + G+ K LLLATVG+G+++ F +++ FLLDK
Sbjct: 283 AAVGIHFFEHATGIEAVVLYSPRIFKKAGVVSKDKLLLATVGVGLTKAFFIIIATFLLDK 342
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S+GG++F++ L V +V+ + G WA+ +I+ Y F +IG+G
Sbjct: 343 VGRRKLLLTSTGGMVFALTSLAVSLTMVQ--RFG-RLAWALSLSIVSTYAFVAFFSIGLG 399
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPLRLRAQG + V +NRI N V SF+S+ K IT GG FF+F G V
Sbjct: 400 PITWVYSSEIFPLRLRAQGASIGVAVNRIMNATVSMSFLSMTKAITTGGVFFVFAGIAVA 459
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG 445
WWF++ LPETKG LE+ME +FG
Sbjct: 460 AWWFFFFMLPETKGLPLEEMEKLFG 484
>emb|CAB16808.1| sugar transporter like protein [Arabidopsis thaliana]
gi|7270615|emb|CAB80333.1| sugar transporter like
protein [Arabidopsis thaliana]
gi|15234491|ref|NP_195385.1| mannitol transporter,
putative [Arabidopsis thaliana] gi|25308953|pir||A85433
sugar transporter like protein [imported] - Arabidopsis
thaliana
Length = 493
Score = 467 bits (1201), Expect = e-130
Identities = 221/449 (49%), Positives = 318/449 (70%), Gaps = 2/449 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA++FI+E+L+ +D+Q+++L GILN CAL ++AGR SD IGRRYTI+L+SI F LG
Sbjct: 38 MSGAMVFIEEDLKTNDVQIEVLTGILNLCALVGSLLAGRTSDIIGRRYTIVLASILFMLG 97
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
SILMG+G ++P+L+ GRC AG GVGFAL++ VYSAEI++ S+RG L SLP L I+IG L
Sbjct: 98 SILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYSAEIATASHRGLLASLPHLCISIGIL 157
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+ NYF KL + +GWR+ML I ++PS+ L +L++ ESPRWL+MQGRL + K++L
Sbjct: 158 LGYIVNYFFSKLPMHIGWRLMLGIAAVPSLVLAFGILKMPESPRWLIMQGRLKEGKEILE 217
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
L+SNS +EAE R ++IK A GID C ++V + K G G KE+ +P+P V R+L+
Sbjct: 218 LVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEGKKTHGEGVWKELILRPTPAVRRVLL 277
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A+G+H FQ+ G+E + LY PRIF + GIT K L L T+G+GI +T F + LLDK
Sbjct: 278 TALGIHFFQHASGIEAVLLYGPRIFKKAGITTKDKLFLVTIGVGIMKTTFIFTATLLLDK 337
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S GG++ ++ L + +N+ G + WA++ +I+ Y F +IG+G
Sbjct: 338 VGRRKLLLTSVGGMVIALTMLGFGLTMAQNA--GGKLAWALVLSIVAAYSFVAFFSIGLG 395
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+E+FPL+LRAQG + V +NR+ N V SF+S+ IT GG FF+F G +
Sbjct: 396 PITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMSFLSLTSAITTGGAFFMFAGVAAV 455
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNSN 449
W F++ LPETKG+SLE++E +F ++ +
Sbjct: 456 AWNFFFFLLPETKGKSLEEIEALFQRDGD 484
>dbj|BAD42343.1| sorbitol transporter [Malus x domestica]
Length = 526
Score = 456 bits (1173), Expect = e-127
Identities = 226/446 (50%), Positives = 317/446 (70%), Gaps = 3/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA L+I++ L+ISD+QV++LAG LN +L AGR SD+IGR+YTI+L+ + F +G
Sbjct: 56 MSGASLYIQKNLKISDVQVEVLAGTLNIYSLLGSAFAGRTSDWIGRKYTIVLAGVIFLVG 115
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++ LM+GR +AG GVG+ ++I VY+AEIS S+RGFLTS P++ +N+G L
Sbjct: 116 ALLMGFATNYAFLMVGRFVAGVGVGYGMMIAPVYTAEISPASFRGFLTSFPEVFVNVGIL 175
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY++NY KL L LGWR ML + +P+I L + +L + ESPRWLVMQGRLGDAKKVL
Sbjct: 176 LGYIANYAFSKLPLHLGWRFMLGVGGVPAIFLTVGVLFMPESPRWLVMQGRLGDAKKVLQ 235
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S SK+E + R+ +IK A GI + +IV V+K + G G KE+ P+P V ILI
Sbjct: 236 RTSESKEECQLRLDDIKEAAGIPPHLNDDIVQVTKSSH-GEGVWKELILHPTPAVRHILI 294
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H F+ G++ + LYSPRIF + GIT LLATV +G ++T+F L++ F LDK
Sbjct: 295 AAVGIHFFEQASGIDTVVLYSPRIFAKAGITSSNHKLLATVAVGFTKTVFILVATFFLDK 354
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S GG++FS++ L V IV++ K G P WAI + ++Y F +IG+G
Sbjct: 355 FGRRPLLLTSVGGMVFSLMFLGVGLTIVDHHK-GSVP-WAIGLCMAMVYFNVAFFSIGLG 412
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPL+LRAQG+ + V NR+T+ V +FIS+YK IT+GG FFL+ G +
Sbjct: 413 PITWVYSSEIFPLKLRAQGVSIGVACNRVTSGVVSMTFISLYKAITIGGAFFLYAGISAA 472
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
W F+Y+ LPET+GR+LED E +FGK
Sbjct: 473 AWIFFYTMLPETQGRTLEDTEVLFGK 498
>gb|AAN07021.1| putative mannitol transporter [Orobanche ramosa]
Length = 519
Score = 454 bits (1167), Expect = e-126
Identities = 213/445 (47%), Positives = 318/445 (70%), Gaps = 1/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA L+IK++L+I+D+QV++L G++N +L +AGR SDY+GRR TI+++S+ FF+G
Sbjct: 53 MSGATLYIKKDLKITDVQVEILVGLINIYSLIGSAVAGRTSDYLGRRITIVIASVIFFVG 112
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ +MG +++ +LM+GR +AG GVG+AL+I VY+AE++ S RGFLTS P++ IN G L
Sbjct: 113 AAVMGLANNYAVLMVGRFVAGLGVGYALMIAPVYAAEVAPASSRGFLTSFPEVFINFGVL 172
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGYLSNY K SL+LGWR+ML + ++P+I + + ++ + ESPRWLVMQGRLGDAKKVL
Sbjct: 173 LGYLSNYAFAKFSLKLGWRLMLGVGALPAIFIGLAVIVMPESPRWLVMQGRLGDAKKVLD 232
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S QEA+ R+ +I A G+ E+C ++V V K+ R GGG KE+ P+ V I I
Sbjct: 233 RTSDSPQEAQLRLADIMEAAGLPEDCHDDVVPVLKQDRGGGGVWKELIVHPTKPVLHITI 292
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G FQ G++ + +YSPRI+ + GIT LLAT+ +G+ +T+F L++ F++D+
Sbjct: 293 AAVGCQFFQQASGIDAVVMYSPRIYEKAGITSDEKKLLATIAVGLCKTVFILVTTFMVDR 352
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
IGRR+LLL S GG++ SML L ++++ P W ++ ++ Y F ++G+G
Sbjct: 353 IGRRVLLLTSCGGLVLSMLTLATGLTVIDHYGADRFP-WVVVLCVLTTYSSVAFFSMGMG 411
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPL+LRAQG G+ V +NR TN ++ SFIS+Y IT+GG F+LF G ++
Sbjct: 412 PIAWVYSSEIFPLKLRAQGCGLGVAINRATNGVILMSFISLYNAITIGGAFYLFSGIGIV 471
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG 445
W F+++ LPET+GR+LEDME +FG
Sbjct: 472 TWIFFFTLLPETRGRTLEDMEVLFG 496
>gb|AAO88964.1| sorbitol transporter [Malus x domestica]
Length = 491
Score = 452 bits (1162), Expect = e-125
Identities = 221/446 (49%), Positives = 315/446 (70%), Gaps = 2/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++I+++L+++D Q++++ G++ +L +AG+ SD++GRRYTI++S FF+G
Sbjct: 13 MSGASIYIEKDLKVTDTQIEIMIGVIEIYSLIGSAMAGKTSDWVGRRYTIVISGAIFFIG 72
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG+ +++ LM GR +AG GVG+AL I VYSAE+S S RGFLTS P++ +NIG L
Sbjct: 73 AILMGFSTNYTFLMCGRFVAGIGVGYALTIAPVYSAEVSPTSSRGFLTSFPEVFVNIGIL 132
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGYLSNY L LGWR+ML + +IPS+GL + +L + ESPRWLVMQGRLG+AK+VL
Sbjct: 133 LGYLSNYAFSFCPLDLGWRLMLGVGAIPSVGLAVGVLAMPESPRWLVMQGRLGEAKRVLD 192
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+E+ R+ +IK A GI E C +IV VS + G G KE+ P+P V ILI
Sbjct: 193 RTSDSKEESMLRLADIKEAAGIPEECNDDIVQVSGHSH-GEGVWKELLVHPTPTVRHILI 251
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AAIG H FQ G++ + LYSPR+F + GIT LLL TVG+G+S+T+FTL++ F LD+
Sbjct: 252 AAIGFHFFQQASGIDALVLYSPRVFAKAGITSTNQLLLCTVGVGLSKTVFTLVATFFLDR 311
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S G++ +++ L IV+ + G WA+I + + GF + GIG
Sbjct: 312 VGRRPLLLTSMAGMVGALVCLGTSLTIVDQHE-GVRMTWAVILCLCCVLAYVGFFSSGIG 370
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPLRLRAQG G+ V +NR+ + + +FIS+YK IT+GGTFFL+ +
Sbjct: 371 PIAWVYSSEIFPLRLRAQGCGMGVAVNRLMSGILSMTFISLYKAITMGGTFFLYAAIGTV 430
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
GW F+++ LPET+GR+LEDME +FGK
Sbjct: 431 GWIFFFTMLPETQGRTLEDMEVLFGK 456
>gb|AAT06053.1| sorbitol transporter [Malus x domestica]
Length = 491
Score = 451 bits (1159), Expect = e-125
Identities = 220/446 (49%), Positives = 315/446 (70%), Gaps = 2/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++I+++L+++D Q++++ G++ +L +AG+ SD++GRRYTI++S FF+G
Sbjct: 13 MSGASIYIEKDLKVTDTQIEIMIGVIEIYSLIGSAMAGKTSDWVGRRYTIVISGAIFFIG 72
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG+ +++ LM GR +AG GVG+AL I VYSAE+S S RGFLTS P++ +NIG L
Sbjct: 73 AILMGFSTNYTFLMCGRFVAGIGVGYALTIAPVYSAEVSPTSSRGFLTSFPEVFVNIGIL 132
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGYLSNY L LGWR+ML + +IPS+GL + +L + ESPRWLVMQGRLG+AK+VL
Sbjct: 133 LGYLSNYAFSFCPLDLGWRLMLGVGAIPSVGLAVGVLAMPESPRWLVMQGRLGEAKRVLD 192
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+E+ R+ +IK A GI E C +IV VS + G G KE+ P+P V ILI
Sbjct: 193 RTSDSKEESMLRLADIKEAAGIPEECNDDIVQVSGHSH-GEGVWKELLVHPTPTVRHILI 251
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AAIG H FQ G++ + LYSPR+F + GIT LLL TVG+G+S+T+FTL++ F LD+
Sbjct: 252 AAIGFHFFQQASGIDALVLYSPRVFAKAGITSTNQLLLCTVGVGLSKTVFTLVATFFLDR 311
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S G++ +++ L +V+ + G WA+I + + GF + GIG
Sbjct: 312 VGRRPLLLTSMAGMVGALVCLGTSLTMVDQHE-GVRMTWAVILCLCCVLAYVGFFSSGIG 370
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPLRLRAQG G+ V +NR+ + + +FIS+YK IT+GGTFFL+ +
Sbjct: 371 PIAWVYSSEIFPLRLRAQGCGMGVAVNRLMSGILSMTFISLYKAITMGGTFFLYAAIGTV 430
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
GW F+++ LPET+GR+LEDME +FGK
Sbjct: 431 GWIFFFTMLPETQGRTLEDMEVLFGK 456
>gb|AAM44082.1| putative sorbitol transporter [Prunus cerasus]
Length = 538
Score = 449 bits (1154), Expect = e-124
Identities = 220/446 (49%), Positives = 320/446 (71%), Gaps = 3/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA+++IK++L++SD+++++L GILN +L AGR SD+IGRRYTI+L+ FF G
Sbjct: 56 MSGAVIYIKKDLKVSDVEIEVLVGILNLYSLIGSAAAGRTSDWIGRRYTIVLAGAIFFAG 115
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ ++ LM GR +AG GVG+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 116 ALLMGFAPNYAFLMFGRFVAGIGVGYALMIAPVYTAEVSPASSRGFLTSFPEVFINAGIL 175
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
GY+SNY KL LGWR+ML + +IPSI L I +L + ESPRWLVMQGRLGDA+KVL
Sbjct: 176 FGYVSNYGFSKLPTHLGWRLMLGVGAIPSIFLAIGVLAMPESPRWLVMQGRLGDARKVLD 235
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S +E++ R+ EIK A GI E+C +IV V K+++ G K++ +P+P V IL+
Sbjct: 236 KTSDSLEESKLRLGEIKEAAGIPEHCNDDIVEVKKRSQ-GQEVWKQLLLRPTPAVRHILM 294
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A+G+H FQ G++ + LYSPRIF + GIT+ +LL TV +G +T+F L++ F+LD+
Sbjct: 295 CAVGLHFFQQASGIDAVVLYSPRIFEKAGITNPDHVLLCTVAVGFVKTVFILVATFMLDR 354
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
IGRR LLL S G++F++ L + I+++S GE+ +WAI ++ ++ F +IG+G
Sbjct: 355 IGRRPLLLTSVAGMVFTLACLGLGLTIIDHS--GEKIMWAIALSLTMVLAYVAFFSIGMG 412
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPL+LRAQG + V +NR+ + + +FIS+YK IT+GG FFLF +
Sbjct: 413 PITWVYSSEIFPLQLRAQGCSIGVAVNRVVSGVLSMTFISLYKAITIGGAFFLFAAIAAV 472
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
GW F+++ LPET+GR+LEDME +FGK
Sbjct: 473 GWTFFFTMLPETQGRTLEDMEVLFGK 498
>gb|AAO39267.1| sorbitol transporter [Prunus cerasus]
Length = 509
Score = 447 bits (1151), Expect = e-124
Identities = 224/446 (50%), Positives = 316/446 (70%), Gaps = 3/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++I+++L+ISD++V++L GILN +L AGR SD+IGRRYTI+ + FF G
Sbjct: 48 MSGASIYIQKDLKISDVEVEILIGILNLYSLIGSAAAGRTSDWIGRRYTIVFAGAIFFTG 107
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG +++ LM+GR +AG GVG+AL+I VY+AE+S S RG LTS P++ +NIG L
Sbjct: 108 ALLMGLATNYAFLMVGRFVAGIGVGYALMIAPVYNAEVSPASSRGALTSFPEVFVNIGIL 167
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY++NY L + LGWR+ML + PS+ L + +L + ESPRWLVMQGRLG+AK+VL
Sbjct: 168 LGYVANYAFSGLPIDLGWRLMLGVGVFPSVILAVGVLSMPESPRWLVMQGRLGEAKQVLD 227
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S +EA+ R+ +IK A GI E+C +++V V K + G KE+ P+P V ILI
Sbjct: 228 KTSDSLEEAQLRLADIKEAAGIPEHCVEDVVQVPKHSH-GEEVWKELLLHPTPPVRHILI 286
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AAIG H FQ + G++ + LYSPRIF + GITD TLLLATV +G S+T+FTL++ LD+
Sbjct: 287 AAIGFHFFQQLSGIDALVLYSPRIFEKAGITDSSTLLLATVAVGFSKTIFTLVAIGFLDR 346
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S G+I S+L L IV++ E+ +WA + + ++ GF +IG+G
Sbjct: 347 VGRRPLLLTSVAGMIASLLCLGTSLTIVDHET--EKMMWASVLCLTMVLAYVGFFSIGMG 404
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPL+LRAQG + +NRI + + SFIS+YK IT+GGTFFL+ G +
Sbjct: 405 PIAWVYSSEIFPLKLRAQGCSMGTAVNRIMSGVLSMSFISLYKAITMGGTFFLYAGIATV 464
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
GW F+Y+ LPET+GR+LEDME +FGK
Sbjct: 465 GWVFFYTMLPETQGRTLEDMEVLFGK 490
>dbj|BAD42344.1| sorbitol transporter [Malus x domestica]
Length = 491
Score = 447 bits (1150), Expect = e-124
Identities = 217/446 (48%), Positives = 315/446 (69%), Gaps = 2/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++I+++L+++D Q+++L GILN +L +AGR SD++GRRYTI++S FF G
Sbjct: 13 MSGAAIYIEKDLKVTDTQIEILLGILNLYSLIGSAMAGRTSDWVGRRYTIVISGAIFFTG 72
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG +++ LM GR +AG GVG+AL I VY+AE+S S RGFLTS P++ +N+G L
Sbjct: 73 AILMGLSTNYTFLMCGRFVAGLGVGYALTIAPVYAAEVSPASSRGFLTSFPEVFVNVGIL 132
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY +L +GWR+ML + +IPS+ L I +L + ESPRWLVMQGRLGDA++VL
Sbjct: 133 LGYISNYAFSFCALDVGWRLMLGVGAIPSVILAIGVLAMPESPRWLVMQGRLGDARQVLD 192
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+E+ R+ +IK A GI E+C +IV V+ + G G KE+F P+P V I+I
Sbjct: 193 KTSDSKEESMLRLADIKEAAGIPEDCNDDIVQVTGHSH-GEGVWKELFVHPTPTVLHIVI 251
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G H FQ G++ + LYSPR+F + GIT+ LLL TVG+G+S+T+FT ++ F LD+
Sbjct: 252 AALGFHFFQQASGIDALVLYSPRVFEKAGITNSNQLLLCTVGVGLSKTVFTFVATFFLDR 311
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR L+L S G++ S++ L IV+ + G WA++ + + GF + GIG
Sbjct: 312 VGRRPLVLTSMAGMVASLVCLGTSLTIVDQHE-GARMTWAVVLCLCCVLAFVGFFSTGIG 370
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPLRLRAQG G+ V +NR+ + + +FIS+YK IT+GG FFL+ +
Sbjct: 371 PIAWVYSSEIFPLRLRAQGCGMGVAVNRVMSGVLSMTFISLYKAITMGGAFFLYAAIGAV 430
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
GW F+++ LPET+GR+LEDME +FGK
Sbjct: 431 GWIFFFTMLPETQGRTLEDMEVLFGK 456
>gb|AAO88965.1| sorbitol transporter [Malus x domestica]
Length = 481
Score = 445 bits (1145), Expect = e-123
Identities = 223/446 (50%), Positives = 310/446 (69%), Gaps = 3/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA LFIKE L+ISD+QV+++ G LN +L +AGR SD+IGRRYTI+L+ FF+G
Sbjct: 13 MSGASLFIKENLKISDVQVEIMNGTLNLYSLIGSALAGRTSDWIGRRYTIVLAGTIFFIG 72
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ ++ LM GR +AG GVG+AL+I VY+AEIS S+RGFLTS P++ +NIG L
Sbjct: 73 ALLMGFAPNYAFLMFGRFVAGVGVGYALMIAPVYTAEISPASFRGFLTSFPEVFVNIGIL 132
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY KL + L WRIML + + PS+ L + +L + ESPRWLVMQGRLGDAK+VL
Sbjct: 133 LGYVSNYAFSKLPIHLNWRIMLGVGAFPSVILAVGVLAMPESPRWLVMQGRLGDAKRVLQ 192
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S S +E + R+ +IK A GI + ++V VSK++ G G KE+ P+P V ILI
Sbjct: 193 KTSESIEECQLRLDDIKEAAGIPKESNDDVVQVSKRSH-GEGVWKELLLHPTPAVRHILI 251
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H F+ G++ + LYSPRIF + GIT LLATV +G+ +T+ L++ LDK
Sbjct: 252 AALGIHFFEQSSGIDSVVLYSPRIFEKAGITSYDHKLLATVAVGVVKTICILVATVFLDK 311
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S G++FS+ L IV+ + +WAI+ I ++ + F +IG+G
Sbjct: 312 FGRRPLLLTSVAGMVFSLSCLGASLTIVDQQH--GKIMWAIVLCITMVLLNVAFFSIGLG 369
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPL+LRAQG + V +NR+T+ + +FIS+YK IT+GG FFL+ G +
Sbjct: 370 PITWVYSSEIFPLQLRAQGCSMGVAVNRVTSGVISMTFISLYKAITIGGAFFLYAGIAAV 429
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
GW F+Y PET+GR+LEDME +FGK
Sbjct: 430 GWVFFYMLYPETQGRTLEDMEVLFGK 455
>dbj|BAD42345.1| sorbitol transporter [Malus x domestica]
Length = 535
Score = 445 bits (1144), Expect = e-123
Identities = 220/446 (49%), Positives = 314/446 (70%), Gaps = 3/446 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++IK++L+ISD++V++L GILN +L AGR SD++GRRYTI+L+ FF+G
Sbjct: 55 MSGAAIYIKDDLKISDVEVEVLLGILNLYSLIGSAAAGRTSDWVGRRYTIVLAGAIFFVG 114
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++ LM GR +AG GVG+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 115 ALLMGFATNYSFLMFGRFVAGIGVGYALMIAPVYTAEVSPASSRGFLTSFPEVFINSGIL 174
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY KL LGWR+ML + +IPSI L + +L + ESPRWLVMQGRLGDA +VL
Sbjct: 175 LGYVSNYAFSKLPTHLGWRLMLGVGAIPSIFLAVGVLAMPESPRWLVMQGRLGDATRVLD 234
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+E+ R+ +IK A GI E+CT ++V V K+++ G KE+ P+P V ILI
Sbjct: 235 KTSDSKEESMLRLADIKEAAGIPEHCTDDVVQVPKRSQ-GQDVWKELLLHPTPAVRHILI 293
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AIG+H FQ G++ + LYSPRIF + GIT+ LL TV +G +T+F L++ F +DK
Sbjct: 294 CAIGIHFFQQASGIDAVVLYSPRIFEKAGITNSDKKLLCTVAVGFVKTVFILVATFFVDK 353
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S G+I S+ GL + I++ + E LWA + + ++ + F +IG+G
Sbjct: 354 VGRRPLLLASVAGMILSLTGLGLGLTIIDQNH--ERILWAAVLCLTMVLLYVAFFSIGMG 411
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPL+LRAQG + V MNR+ + + +FIS+Y+ IT+GG FFL+ +
Sbjct: 412 PITWVYSSEIFPLKLRAQGCSLGVAMNRVVSGVLSMTFISLYEAITIGGAFFLYAAIASV 471
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGK 446
W F+++ LPET GR+LEDME +FGK
Sbjct: 472 AWVFFFTMLPETHGRTLEDMEVLFGK 497
>emb|CAD58710.1| polyol transporter [Plantago major]
Length = 530
Score = 440 bits (1131), Expect = e-122
Identities = 213/445 (47%), Positives = 313/445 (69%), Gaps = 2/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA L+IK++L+ISD+QV+LL G +N +L +AGR SD++GRRYT + +S+ FF+G
Sbjct: 65 MSGATLYIKDDLKISDVQVELLVGTINIYSLVGSAVAGRTSDWVGRRYTTVFASVVFFVG 124
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG +++ LM GR +AG GVG+AL+I VY+AE++ S RGFLTS P++ IN G L
Sbjct: 125 AILMGIATNYVFLMAGRFVAGIGVGYALMIAPVYAAEVAPASCRGFLTSFPEVFINFGVL 184
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LG++SNY K L+LGWR+ML + +IP++ L I ++ + ESPRWLV+QGRLGDA++VL
Sbjct: 185 LGFVSNYAFAKFPLKLGWRMMLGVGAIPAVFLAIGVIYMPESPRWLVLQGRLGDARRVLD 244
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S +E++ R+ +IK A GI E+C + V V K ++ G G +E+ P+ V ILI
Sbjct: 245 KTSDSLEESKLRLADIKEAAGIPEDCNDDFVQVQKHSQ-GQGVWRELLLHPTKPVLHILI 303
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
+G+H FQ G++ + LYSPRI+ R GITD LLAT+ +GIS+T F L++ F +D+
Sbjct: 304 CGVGIHFFQQGIGIDSVVLYSPRIYDRAGITDTSDKLLATIAVGISKTFFILITTFYVDR 363
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLLVS GV SM L I++ + ++ ++ +++ ++ GF ++G+G
Sbjct: 364 FGRRFLLLVSCAGVALSMFALGTVLTIIDRNPDAKQ-TGVLVLVVLLTMVIVGFFSMGLG 422
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPL+LRAQG G+ V MNR + ++ SFIS+YK IT+GG+FFLF G L
Sbjct: 423 PIAWVYSSEIFPLKLRAQGCGLGVAMNRFMSGVILMSFISLYKEITIGGSFFLFGGITTL 482
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG 445
GW F+++ PET+GR+LE+ME +FG
Sbjct: 483 GWIFFFTLFPETRGRTLEEMEGLFG 507
>emb|CAD91337.1| sorbitol-like transporter [Glycine max]
Length = 523
Score = 439 bits (1128), Expect = e-121
Identities = 224/472 (47%), Positives = 325/472 (68%), Gaps = 16/472 (3%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++IK +L++SD Q+++L GI+N +L +AGR SD+IGRRYTI L F +G
Sbjct: 52 MSGAAIYIKRDLKVSDEQIEILLGIINLYSLIGSCLAGRTSDWIGRRYTIGLGGAIFLVG 111
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
S LMG+ + LM GR +AG G+G+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 112 STLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGIL 171
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY KL+L++GWR+ML + +IPS+ L +L + ESPRWLVM+GRLG+A+KVL
Sbjct: 172 LGYISNYGFSKLTLKVGWRMMLGVGAIPSVVLTEGVLAMPESPRWLVMRGRLGEARKVLN 231
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+EA+ R+ EIK A GI E+C ++V V+K++ +G G KE+F P+P + I+I
Sbjct: 232 KTSDSKEEAQLRLAEIKQAAGIPESCNDDVVQVNKQS-NGEGVWKELFLYPTPAIRHIVI 290
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H FQ GV+ + LYSPRIF + GIT+ LLATV +G +T+F L + F LD+
Sbjct: 291 AALGIHFFQQASGVDAVVLYSPRIFEKAGITNDTHKLLATVAVGFVKTVFILAATFTLDR 350
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S GG++ S+L L + ++++S+ + +WA+ +I ++ +IG G
Sbjct: 351 VGRRPLLLSSVGGMVLSLLTLAISLTVIDHSE--RKLMWAVGSSIAMVLAYVATFSIGAG 408
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPLRLRAQG V +NR T+ V +F+S+ + IT+GG FFL+ G +
Sbjct: 409 PITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVSMTFLSLTRAITIGGAFFLYCGIATV 468
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG------------KNSNSEV-QMKYGSN 459
GW F+Y+ LPET+G++LEDME FG +N N +V Q++ G+N
Sbjct: 469 GWIFFYTVLPETRGKTLEDMEGSFGTFRSKSNASKAVENENGQVAQVQLGTN 520
>gb|AAD26955.1| putative sugar transporter [Arabidopsis thaliana]
gi|15226696|ref|NP_179210.1| mannitol transporter,
putative [Arabidopsis thaliana] gi|25308959|pir||A84537
probable sugar transporter [imported] - Arabidopsis
thaliana
Length = 511
Score = 434 bits (1115), Expect = e-120
Identities = 210/448 (46%), Positives = 309/448 (68%), Gaps = 1/448 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA +FIK++L++SD+Q+++L GILN +L AGR SD+IGRRYTI+L+ FFF G
Sbjct: 47 MSGAAIFIKDDLKLSDVQLEILMGILNIYSLIGSGAAGRTSDWIGRRYTIVLAGFFFFCG 106
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++P +M+GR +AG GVG+A++I VY+ E++ S RGFL+S P++ INIG L
Sbjct: 107 ALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTTEVAPASSRGFLSSFPEIFINIGIL 166
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNYF KL +GWR ML I ++PS+ L I +L + ESPRWLVMQGRLGDA KVL
Sbjct: 167 LGYVSNYFFAKLPEHIGWRFMLGIGAVPSVFLAIGVLAMPESPRWLVMQGRLGDAFKVLD 226
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
SN+K+EA R+ +IK AVGI ++ T +++ V K +G G K++ +P+P V ILI
Sbjct: 227 KTSNTKEEAISRLNDIKRAVGIPDDMTDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILI 286
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A +G+H Q G++ + LYSP IF R G+ K LLATV +G+ +TLF ++ L+D+
Sbjct: 287 ACLGIHFSQQASGIDAVVLYSPTIFSRAGLKSKNDQLLATVAVGVVKTLFIVVGTCLVDR 346
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S GG+ FS+ L +++ + G+ WAI + + ++G G
Sbjct: 347 FGRRALLLTSMGGMFFSLTALGTSLTVIDRNP-GQTLKWAIGLAVTTVMTFVATFSLGAG 405
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
VTWVY++EIFP+RLRAQG + V++NR+ + + +F+S+ K +T+GG F LF G V
Sbjct: 406 PVTWVYASEIFPVRLRAQGASLGVMLNRLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAVA 465
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNS 448
W F+++FLPET+G LE++E++FG S
Sbjct: 466 AWVFFFTFLPETRGVPLEEIESLFGSYS 493
>dbj|BAB01812.1| sugar transporter protein [Arabidopsis thaliana]
gi|15230212|ref|NP_188513.1| mannitol transporter,
putative [Arabidopsis thaliana]
Length = 539
Score = 431 bits (1108), Expect = e-119
Identities = 213/444 (47%), Positives = 307/444 (68%), Gaps = 2/444 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA+++IK +L+I+D+Q+ +LAG LN +L AGR SD+IGRRYTI+L+ FF G
Sbjct: 57 MSGAMIYIKRDLKINDLQIGILAGSLNIYSLIGSCAAGRTSDWIGRRYTIVLAGAIFFAG 116
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG ++ LM GR IAG GVG+AL+I VY+AE+S S RGFL S P++ IN G +
Sbjct: 117 AILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTAEVSPASSRGFLNSFPEVFINAGIM 176
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN L L++GWR+ML I ++PS+ L I +L + ESPRWLVMQGRLGDAK+VL
Sbjct: 177 LGYVSNLAFSNLPLKVGWRLMLGIGAVPSVILAIGVLAMPESPRWLVMQGRLGDAKRVLD 236
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S EA R+++IK+A GI +C ++V VS++ G G +E+ +P+P V R++I
Sbjct: 237 KTSDSPTEATLRLEDIKHAAGIPADCHDDVVQVSRRNSHGEGVWRELLIRPTPAVRRVMI 296
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AAIG+H FQ G++ + L+SPRIF G+ LLATV +G+ +T F L++ FLLD+
Sbjct: 297 AAIGIHFFQQASGIDAVVLFSPRIFKTAGLKTDHQQLLATVAVGVVKTSFILVATFLLDR 356
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
IGRR LLL S GG++ S+ L I++ S+ ++ +WA++ I + +IG G
Sbjct: 357 IGRRPLLLTSVGGMVLSLAALGTSLTIIDQSE--KKVMWAVVVAIATVMTYVATFSIGAG 414
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPLRLR+QG + V++NR+T+ + SF+ + K +T GG F+LF G +
Sbjct: 415 PITWVYSSEIFPLRLRSQGSSMGVVVNRVTSGVISISFLPMSKAMTTGGAFYLFGGIATV 474
Query: 421 GWWFYYSFLPETKGRSLEDMETIF 444
W F+Y+FLPET+GR LEDM+ +F
Sbjct: 475 AWVFFYTFLPETQGRMLEDMDELF 498
>gb|AAD26954.1| putative sugar transporter [Arabidopsis thaliana]
gi|15226682|ref|NP_179209.1| mannitol transporter,
putative [Arabidopsis thaliana] gi|25308957|pir||H84536
probable sugar transporter [imported] - Arabidopsis
thaliana
Length = 511
Score = 431 bits (1108), Expect = e-119
Identities = 212/460 (46%), Positives = 310/460 (67%), Gaps = 1/460 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA +FIK++L++SD+Q+++L GILN +L AGR SD++GRRYTI+L+ FFF G
Sbjct: 47 MSGASIFIKDDLKLSDVQLEILMGILNIYSLVGSGAAGRTSDWLGRRYTIVLAGAFFFCG 106
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++P +M+GR +AG GVG+A++I VY+AE++ S RGFLTS P++ INIG L
Sbjct: 107 ALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTAEVAPASSRGFLTSFPEIFINIGIL 166
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNYF KL LGWR ML + ++PS+ L I +L + ESPRWLV+QGRLGDA KVL
Sbjct: 167 LGYVSNYFFSKLPEHLGWRFMLGVGAVPSVFLAIGVLAMPESPRWLVLQGRLGDAFKVLD 226
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
SN+K+EA R+ +IK AVGI ++ T +++ V K +G G K++ +P+P V ILI
Sbjct: 227 KTSNTKEEAISRLDDIKRAVGIPDDMTDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILI 286
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A +G+H Q G++ + LYSP IF + G+ K LLATV +G+ +TLF ++ ++D+
Sbjct: 287 ACLGIHFAQQASGIDAVVLYSPTIFSKAGLKSKNDQLLATVAVGVVKTLFIVVGTCVVDR 346
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S GG+ S+ L S V N G+ WAI + + +IG G
Sbjct: 347 FGRRALLLTSMGGMFLSLTALGT-SLTVINRNPGQTLKWAIGLAVTTVMTFVATFSIGAG 405
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
VTWVY +EIFP+RLRAQG + V++NR+ + + +F+S+ K +T+GG F LF G
Sbjct: 406 PVTWVYCSEIFPVRLRAQGASLGVMLNRLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAAA 465
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQMKYGSNN 460
W F+++FLPET+G LE+MET+FG + ++ +N
Sbjct: 466 AWVFFFTFLPETRGIPLEEMETLFGSYTANKKNNSMSKDN 505
>gb|AAB68029.1| putative sugar transporter; member of major facilitative
superfamily; integral membrane protein [Beta vulgaris]
Length = 545
Score = 430 bits (1106), Expect = e-119
Identities = 218/445 (48%), Positives = 302/445 (66%), Gaps = 3/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA++++KE+ ISD Q+ +L GILN L AGR SD+IGRRYTI+L+ FF+G
Sbjct: 58 MSGAIIYLKEDWHISDTQIGVLVGILNIYCLFGSFAAGRTSDWIGRRYTIVLAGAIFFVG 117
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++ LM+GR + G GVG+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 118 ALLMGFATNYAFLMVGRFVTGIGVGYALMIAPVYTAEVSPASSRGFLTSFPEVFINAGIL 177
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN L L WR ML I +IPSI L I +L + ESPRWLVMQGRLGDAKKVL
Sbjct: 178 LGYISNLAFSSLPTHLSWRFMLGIGAIPSIFLAIGVLAMPESPRWLVMQGRLGDAKKVLN 237
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSK-KTRSGGGALKEMFYKPSPHVYRIL 239
IS+S +EA+ R+ EIK GI C ++I V K K +SG KE+F+ P+P V R +
Sbjct: 238 RISDSPEEAQLRLSEIKQTAGIPAECDEDIYKVEKTKIKSGNAVWKELFFNPTPAVRRAV 297
Query: 240 IAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLD 299
IA IG+H FQ G++ + LYSPRIF GIT+ LLATV +G+ +TLF L++ F LD
Sbjct: 298 IAGIGIHFFQQASGIDAVVLYSPRIFQSAGITNARKQLLATVAVGVVKTLFILVATFQLD 357
Query: 300 KIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGI 359
K GRR LLL S GG+I ++L L + ++++S + WAI I ++ + +IG+
Sbjct: 358 KYGRRPLLLTSVGGMIIAILTLAMSLTVIDHSH--HKITWAIALCITMVCAVVASFSIGL 415
Query: 360 GAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNV 419
G +TWVYS+E+FPLRLRAQG + V +NR+ + + F+ + IT GG FFLF G +
Sbjct: 416 GPITWVYSSEVFPLRLRAQGTSMGVAVNRVVSGVISIFFLPLSHKITTGGAFFLFGGIAI 475
Query: 420 LGWWFYYSFLPETKGRSLEDMETIF 444
+ W+F+ +FLPET+GR+LE+M +F
Sbjct: 476 IAWFFFLTFLPETRGRTLENMHELF 500
>gb|AAB68028.1| putative sugar transporter; member of major facilitative
superfamily; integral membrane protein [Beta vulgaris]
gi|7484647|pir||T14606 probable sugar transport protein
205 - beet
Length = 549
Score = 430 bits (1106), Expect = e-119
Identities = 218/445 (48%), Positives = 302/445 (66%), Gaps = 3/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA++++KE+ ISD Q+ +L GILN L AGR SD+IGRRYTI+L+ FF+G
Sbjct: 58 MSGAIIYLKEDWHISDTQIGVLVGILNIYCLFGSFAAGRTSDWIGRRYTIVLAGAIFFVG 117
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++ LM+GR + G GVG+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 118 ALLMGFATNYAFLMVGRFVTGIGVGYALMIAPVYTAEVSPASSRGFLTSFPEVFINAGIL 177
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN L L WR ML I +IPSI L I +L + ESPRWLVMQGRLGDAKKVL
Sbjct: 178 LGYISNLAFSSLPTHLSWRFMLGIGAIPSIFLAIGVLAMPESPRWLVMQGRLGDAKKVLN 237
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSK-KTRSGGGALKEMFYKPSPHVYRIL 239
IS+S +EA+ R+ EIK GI C ++I V K K +SG KE+F+ P+P V R +
Sbjct: 238 RISDSPEEAQLRLSEIKQTAGIPAECDEDIYKVEKTKIKSGNAVWKELFFNPTPAVRRAV 297
Query: 240 IAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLD 299
IA IG+H FQ G++ + LYSPRIF GIT+ LLATV +G+ +TLF L++ F LD
Sbjct: 298 IAGIGIHFFQQASGIDAVVLYSPRIFQSAGITNARKQLLATVAVGVVKTLFILVATFQLD 357
Query: 300 KIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGI 359
K GRR LLL S GG+I ++L L + ++++S + WAI I ++ + +IG+
Sbjct: 358 KYGRRPLLLTSVGGMIIAILTLAMSLTVIDHSH--HKITWAIALCITMVCAVVASFSIGL 415
Query: 360 GAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNV 419
G +TWVYS+E+FPLRLRAQG + V +NR+ + + F+ + IT GG FFLF G +
Sbjct: 416 GPITWVYSSEVFPLRLRAQGTSMGVAVNRVVSGVISIFFLPLSHKITTGGAFFLFGGIAI 475
Query: 420 LGWWFYYSFLPETKGRSLEDMETIF 444
+ W+F+ +FLPET+GR+LE+M +F
Sbjct: 476 IAWFFFLTFLPETRGRTLENMHELF 500
>emb|CAD58709.1| polyol transporter [Plantago major]
Length = 529
Score = 429 bits (1102), Expect = e-118
Identities = 215/455 (47%), Positives = 311/455 (68%), Gaps = 2/455 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA FI+E+L I+D+QV+LL G +N +L +AGR SD++GRRYTI+ +S FFLG
Sbjct: 64 MSGATQFIQEDLIITDVQVELLVGTINIYSLVGSAVAGRTSDWVGRRYTIVFASTIFFLG 123
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG+ +++ LM+GR +AG GVG+AL+I VY+AE++ S RGFLTS P++ IN G L
Sbjct: 124 AILMGFATNYAFLMVGRFVAGIGVGYALMIAPVYAAEVAPASCRGFLTSFPEVFINFGVL 183
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN+ KL L LGWR+ML + ++PS+ L + +L + ESPRWLV+QGRLGDAKKVL
Sbjct: 184 LGYVSNFAFAKLPLTLGWRMMLGVGAVPSVLLGVGVLYMPESPRWLVLQGRLGDAKKVLD 243
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S +E++ R+ +IK A G+ +C IV V K+++ G G KE+ P+ V ILI
Sbjct: 244 KTSDSLEESKLRLADIKEAAGVPLDCHDEIVQVQKRSQ-GQGVWKELLLHPTKPVLHILI 302
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
+G+H FQ G++ + LYSPRI+ + GI + LLAT+ +G+S+T F L++ F +D+
Sbjct: 303 CGVGIHFFQQGIGIDSVVLYSPRIYEKAGIKNTSDKLLATIAVGVSKTFFILITTFFVDR 362
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S GV SM L I++ + G +IF +I+ + GF ++G+G
Sbjct: 363 FGRRPLLLTSCAGVALSMFALGTSLTIIDRNPDGNIK-GLLIFAVILTMAIVGFFSMGLG 421
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+EIFPL+LRAQG + V MNR + ++ SFIS+YK IT+GG FFLF G +
Sbjct: 422 PIAWVYSSEIFPLKLRAQGCSMGVAMNRFMSGVILMSFISLYKAITIGGAFFLFGGITTV 481
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQMK 455
+ F+Y+ PET+GR+LE+ME +FG + +MK
Sbjct: 482 AFIFFYTLFPETQGRTLEEMEELFGTFFSWRTRMK 516
>gb|AAG43998.1| mannitol transporter [Apium graveolens var. dulce]
Length = 513
Score = 422 bits (1084), Expect = e-116
Identities = 209/445 (46%), Positives = 296/445 (65%), Gaps = 3/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
++GA ++IKE+L SD+Q++++ GI+N +L IAGR SD+IGRRYT++L+ I FFLG
Sbjct: 45 LSGASIYIKEDLHFSDVQIEIIIGIINIYSLLGSAIAGRTSDWIGRRYTMVLAGIIFFLG 104
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+I MG ++F LM GR +AG GVG+A++I VY+AE++ S RGFLTS P++ IN G L
Sbjct: 105 AIFMGLATNFAFLMFGRFVAGIGVGYAMMIAPVYTAEVAPSSSRGFLTSFPEVFINSGVL 164
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN+ K L LGWRIML I + PS+ L I++L + ESPRWLVMQGRLG+A+ VL
Sbjct: 165 LGYVSNFAFAKCPLWLGWRIMLGIGAFPSVALAIIVLYMPESPRWLVMQGRLGEARTVLE 224
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S SK+EA QR+ +IK A GID++C ++V V K+T+ KE+ P+ V I
Sbjct: 225 KTSTSKEEAHQRLSDIKEAAGIDKDCNDDVVQVPKRTKD-EAVWKELILHPTKPVRHAAI 283
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
IG+H FQ CG++ + LYSPRIF + GI LLAT+ +G+ +T+F L+S F LDK
Sbjct: 284 TGIGIHFFQQACGIDAVVLYSPRIFEKAGIKSNSKKLLATIAVGVCKTVFILISTFQLDK 343
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
IGRR L+L S GG++ ++ L ++ S WA I +Y + G+G
Sbjct: 344 IGRRPLMLTSMGGMVIALFVLAGSLTVINKSH--HTGHWAGGLAIFTVYAFVSIFSSGMG 401
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+ WVYS+E+FPLRLRAQG + V +NR + + +FIS+YK +T+GG F LF +
Sbjct: 402 PIAWVYSSEVFPLRLRAQGCSIGVAVNRGMSGIIGMTFISMYKAMTIGGAFLLFAVVASI 461
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG 445
GW F Y+ PET+GR+LE++E +FG
Sbjct: 462 GWVFMYTMFPETQGRNLEEIELLFG 486
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.327 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 725,463,840
Number of Sequences: 2540612
Number of extensions: 30054619
Number of successful extensions: 120311
Number of sequences better than 10.0: 5579
Number of HSP's better than 10.0 without gapping: 3080
Number of HSP's successfully gapped in prelim test: 2500
Number of HSP's that attempted gapping in prelim test: 107308
Number of HSP's gapped (non-prelim): 8695
length of query: 461
length of database: 863,360,394
effective HSP length: 131
effective length of query: 330
effective length of database: 530,540,222
effective search space: 175078273260
effective search space used: 175078273260
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC147178.8


