

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.14 + phase: 0
(465 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
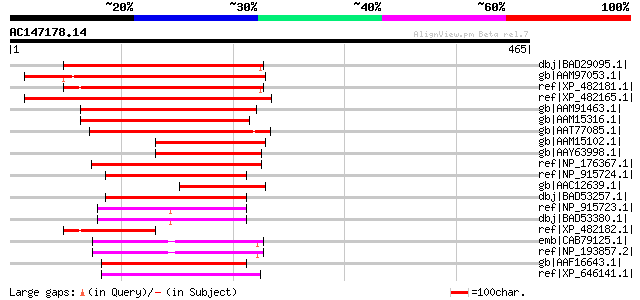
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD29095.1| membrane protein-like [Oryza sativa (japonica cu... 277 6e-73
gb|AAM97053.1| unknown protein [Arabidopsis thaliana] gi|2645052... 274 4e-72
ref|XP_482181.1| membrane protein-like [Oryza sativa (japonica c... 274 5e-72
ref|XP_482165.1| unknown protein [Oryza sativa (japonica cultiva... 259 1e-67
gb|AAM91463.1| At2g36630/F1O11.26 [Arabidopsis thaliana] gi|1870... 219 1e-55
gb|AAM15316.1| unknown protein [Arabidopsis thaliana] gi|4415908... 214 6e-54
gb|AAT77085.1| expressed protein [Oryza sativa (japonica cultiva... 207 7e-52
gb|AAM15102.1| hypothetical protein [Arabidopsis thaliana] gi|36... 150 1e-34
gb|AAY63998.1| unknown [Brassica napus] 149 1e-34
ref|NP_176367.1| expressed protein [Arabidopsis thaliana] gi|450... 120 7e-26
ref|NP_915724.1| P0415A04.10 [Oryza sativa (japonica cultivar-gr... 118 3e-25
gb|AAC12639.1| hypothetical protein [Mesembryanthemum crystallin... 118 3e-25
dbj|BAD53257.1| membrane protein-like [Oryza sativa (japonica cu... 118 3e-25
ref|NP_915723.1| P0415A04.9 [Oryza sativa (japonica cultivar-gro... 118 4e-25
dbj|BAD53380.1| membrane protein-like [Oryza sativa (japonica cu... 118 4e-25
ref|XP_482182.1| unknown protein [Oryza sativa (japonica cultiva... 117 1e-24
emb|CAB79125.1| hypothetical protein [Arabidopsis thaliana] gi|2... 116 2e-24
ref|NP_193857.2| expressed protein [Arabidopsis thaliana] 116 2e-24
gb|AAF16643.1| T23J18.20 [Arabidopsis thaliana] 116 2e-24
ref|XP_646141.1| hypothetical protein DDB0190426 [Dictyostelium ... 112 3e-23
>dbj|BAD29095.1| membrane protein-like [Oryza sativa (japonica cultivar-group)]
Length = 481
Score = 277 bits (708), Expect = 6e-73
Identities = 132/182 (72%), Positives = 159/182 (86%), Gaps = 3/182 (1%)
Query: 49 ETKGFLKAMVDFLWESGKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFV 108
E G L+ + +FLW++ +SY VWPEM+ W+I++GS+IGF GAA GSVGGVGGGGIFV
Sbjct: 53 EEVGILRKVANFLWQTDGNSYHHVWPEMELGWQIVLGSLIGFFGAAFGSVGGVGGGGIFV 112
Query: 109 PMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLML 168
PML LIIGFDPKSSTAISKCMIMGAA+STVYYN++L++PTLDMP+IDYDLALL QPMLML
Sbjct: 113 PMLTLIIGFDPKSSTAISKCMIMGAAVSTVYYNLKLKHPTLDMPVIDYDLALLIQPMLML 172
Query: 169 GISIGVICNVMFADWMVTVLLIILFIGTSTKALIKGINTWKKETMLKKETAKQLE---EE 225
GISIGVI NV+F DW+VTVLLIILF+GTSTKA +KG+ TWKKET++K+E AK+LE EE
Sbjct: 173 GISIGVIFNVLFPDWLVTVLLIILFLGTSTKAFLKGVETWKKETIIKREAAKRLEQTSEE 232
Query: 226 PK 227
P+
Sbjct: 233 PE 234
>gb|AAM97053.1| unknown protein [Arabidopsis thaliana] gi|26450521|dbj|BAC42374.1|
unknown protein [Arabidopsis thaliana]
gi|28951043|gb|AAO63445.1| At2g25737 [Arabidopsis
thaliana] gi|25083770|gb|AAN72117.1| unknown protein
[Arabidopsis thaliana] gi|30682879|ref|NP_850068.1|
expressed protein [Arabidopsis thaliana]
Length = 476
Score = 274 bits (701), Expect = 4e-72
Identities = 137/225 (60%), Positives = 173/225 (76%), Gaps = 10/225 (4%)
Query: 14 IAATWL-IMCILVMICNVSLAERVLKEKEPAKFV--------EKETKGFLKAMVDFLWES 64
+ + WL + + + + N SLA + + K + E E+ FLKA ++FLWES
Sbjct: 4 LRSKWLGLRSVTIFLINFSLAFAFVSAERRGKSLRLSTDETRENESSFFLKA-INFLWES 62
Query: 65 GKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTA 124
+ Y VWPE +F+W+I++G+++GF GAA GSVGGVGGGGIFVPML+LIIGFDPKS+TA
Sbjct: 63 DQIGYRHVWPEFEFNWQIVLGTLVGFFGAAFGSVGGVGGGGIFVPMLSLIIGFDPKSATA 122
Query: 125 ISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWM 184
ISKCMIMGA++STVYYN+RLR+PTLDMP+IDYDLALL QPMLMLGISIGV NV+F DW+
Sbjct: 123 ISKCMIMGASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGVAFNVIFPDWL 182
Query: 185 VTVLLIILFIGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTG 229
VTVLLI+LF+GTSTKA +KG TW KET+ KKE AK+LE +G
Sbjct: 183 VTVLLIVLFLGTSTKAFLKGSETWNKETIEKKEAAKRLESNGVSG 227
>ref|XP_482181.1| membrane protein-like [Oryza sativa (japonica cultivar-group)]
gi|40253788|dbj|BAD05726.1| membrane protein-like [Oryza
sativa (japonica cultivar-group)]
gi|40253412|dbj|BAD05341.1| membrane protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 465
Score = 274 bits (700), Expect = 5e-72
Identities = 132/182 (72%), Positives = 160/182 (87%), Gaps = 4/182 (2%)
Query: 49 ETKGFLKAMVDFLWESGKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFV 108
E G+++ +V+FLW SG++SY VWP M+F WKI++G +IGF GAA GSVGGVGGGGIFV
Sbjct: 38 EEVGYMRKVVNFLW-SGEASYHHVWPPMEFGWKIVLGILIGFFGAAFGSVGGVGGGGIFV 96
Query: 109 PMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLML 168
PML LIIGFD KSSTAISKCMIMGAA+STVYYN++L++PTLDMP+IDYDLALL QPMLML
Sbjct: 97 PMLTLIIGFDAKSSTAISKCMIMGAAVSTVYYNLKLKHPTLDMPVIDYDLALLIQPMLML 156
Query: 169 GISIGVICNVMFADWMVTVLLIILFIGTSTKALIKGINTWKKETMLKKETAKQLE---EE 225
GISIGV+ NV+F DW++TVLLIILF+GTSTKA +KG+ TWKKET+LK+E AK+LE EE
Sbjct: 157 GISIGVLFNVIFPDWLITVLLIILFLGTSTKAFLKGVETWKKETILKREAAKRLEQIAEE 216
Query: 226 PK 227
P+
Sbjct: 217 PE 218
>ref|XP_482165.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|40253486|dbj|BAD05436.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 469
Score = 259 bits (663), Expect = 1e-67
Identities = 132/224 (58%), Positives = 167/224 (73%), Gaps = 3/224 (1%)
Query: 14 IAATWLIMCILVMICNVSLA-ERVLKEKEPAKFVEK-ETKGFLKAMVDFLWESG-KSSYE 70
+AA + C V+ A +R L A E+ E L+ + +FLW SG ++SY
Sbjct: 8 VAAALAVACAATAAATVAAAADRGLWSAAAAAVAEEGEEASHLRKVANFLWRSGGENSYH 67
Query: 71 PVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMI 130
VWP M+F W+I++GS +GF+GAA GS+GGVGGGG F+PML LIIGFD KSS AISKCMI
Sbjct: 68 HVWPPMEFGWQIVLGSFVGFIGAAFGSIGGVGGGGFFMPMLTLIIGFDAKSSVAISKCMI 127
Query: 131 MGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLI 190
MGAA+STVY N++ ++PTLDMP+IDYDLALL QPMLMLGISIGVI NV+F DW+VTVLLI
Sbjct: 128 MGAAVSTVYCNLKRKHPTLDMPVIDYDLALLIQPMLMLGISIGVIFNVIFPDWLVTVLLI 187
Query: 191 ILFIGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTGAYQRV 234
ILF+GTSTKA +KGI TWKKET++K+E K+ E+ + Y+ V
Sbjct: 188 ILFLGTSTKAFLKGIETWKKETIIKREAEKRSEQTSEELEYRPV 231
>gb|AAM91463.1| At2g36630/F1O11.26 [Arabidopsis thaliana]
gi|18700129|gb|AAL77676.1| At2g36630/F1O11.26
[Arabidopsis thaliana] gi|30686851|ref|NP_850267.1|
expressed protein [Arabidopsis thaliana]
Length = 459
Score = 219 bits (558), Expect = 1e-55
Identities = 97/158 (61%), Positives = 135/158 (85%)
Query: 64 SGKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSST 123
S S+ E +WP++KF WK+++ ++I FLG+A G+VGGVGGGGIFVPML LI+GFD KS+
Sbjct: 42 SSLSATEKIWPDLKFSWKLVLATVIAFLGSACGTVGGVGGGGIFVPMLTLILGFDTKSAA 101
Query: 124 AISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADW 183
AISKCMIMGA+ S+V+YN+R+R+PT ++P++DYDLALLFQPML+LGI++GV +V+F W
Sbjct: 102 AISKCMIMGASASSVWYNVRVRHPTKEVPILDYDLALLFQPMLLLGITVGVSLSVVFPYW 161
Query: 184 MVTVLLIILFIGTSTKALIKGINTWKKETMLKKETAKQ 221
++TVL+IILF+GTS+++ KGI WK+ET+LK E A+Q
Sbjct: 162 LITVLIIILFVGTSSRSFFKGIEMWKEETLLKNEMAQQ 199
>gb|AAM15316.1| unknown protein [Arabidopsis thaliana] gi|4415908|gb|AAD20139.1|
unknown protein [Arabidopsis thaliana]
gi|25408492|pir||G84782 hypothetical protein At2g36630
[imported] - Arabidopsis thaliana
Length = 288
Score = 214 bits (544), Expect = 6e-54
Identities = 94/152 (61%), Positives = 131/152 (85%)
Query: 64 SGKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSST 123
S S+ E +WP++KF WK+++ ++I FLG+A G+VGGVGGGGIFVPML LI+GFD KS+
Sbjct: 42 SSLSATEKIWPDLKFSWKLVLATVIAFLGSACGTVGGVGGGGIFVPMLTLILGFDTKSAA 101
Query: 124 AISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADW 183
AISKCMIMGA+ S+V+YN+R+R+PT ++P++DYDLALLFQPML+LGI++GV +V+F W
Sbjct: 102 AISKCMIMGASASSVWYNVRVRHPTKEVPILDYDLALLFQPMLLLGITVGVSLSVVFPYW 161
Query: 184 MVTVLLIILFIGTSTKALIKGINTWKKETMLK 215
++TVL+IILF+GTS+++ KGI WK+ET+LK
Sbjct: 162 LITVLIIILFVGTSSRSFFKGIEMWKEETLLK 193
>gb|AAT77085.1| expressed protein [Oryza sativa (japonica cultivar-group)]
gi|41469272|gb|AAS07154.1| expressed protein [Oryza
sativa (japonica cultivar-group)]
Length = 475
Score = 207 bits (526), Expect = 7e-52
Identities = 95/162 (58%), Positives = 133/162 (81%), Gaps = 1/162 (0%)
Query: 72 VWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIM 131
VWP++ F+W+++V +++GFLG+A G+VGGVGGGGIFVPML L++GFD KS+ A+SKCMIM
Sbjct: 61 VWPDLAFNWRVVVATVVGFLGSAFGTVGGVGGGGIFVPMLNLLVGFDTKSAAALSKCMIM 120
Query: 132 GAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLII 191
GA+ S+V+YN+++ +PT + P+IDY LALLFQPMLMLGI+IGV +V+F W++TVL+II
Sbjct: 121 GASASSVWYNLQVSHPTKEAPVIDYKLALLFQPMLMLGITIGVELSVIFPYWLITVLIII 180
Query: 192 LFIGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTGAYQR 233
LFIGTS+++ KGI WK ET ++ ET ++ EEE K+ R
Sbjct: 181 LFIGTSSRSFYKGILMWKDETRIQMET-REREEESKSSCAAR 221
>gb|AAM15102.1| hypothetical protein [Arabidopsis thaliana]
gi|3643608|gb|AAC42255.1| hypothetical protein
[Arabidopsis thaliana] gi|25412342|pir||B84652
hypothetical protein At2g25740 [imported] - Arabidopsis
thaliana
Length = 902
Score = 150 bits (378), Expect = 1e-34
Identities = 72/99 (72%), Positives = 85/99 (85%)
Query: 131 MGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLI 190
MGA++STVYYN+RLR+PTLDMP+IDYDLALL QPMLMLGISIGV NV+F DW+VTVLLI
Sbjct: 1 MGASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGVAFNVIFPDWLVTVLLI 60
Query: 191 ILFIGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTG 229
+LF+GTSTKA +KG TW KET+ KKE AK+LE +G
Sbjct: 61 VLFLGTSTKAFLKGSETWNKETIEKKEAAKRLESNGVSG 99
>gb|AAY63998.1| unknown [Brassica napus]
Length = 348
Score = 149 bits (377), Expect = 1e-34
Identities = 72/95 (75%), Positives = 83/95 (86%)
Query: 131 MGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLI 190
MGA++STVYYN+RLR+PTLDMP+IDYDLALL QPMLMLGIS+GV NVMF DWMVTVLLI
Sbjct: 1 MGASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISVGVAFNVMFPDWMVTVLLI 60
Query: 191 ILFIGTSTKALIKGINTWKKETMLKKETAKQLEEE 225
ILF+GTSTKA +KG TW KET+ K E AK+LE +
Sbjct: 61 ILFLGTSTKAFLKGRETWNKETIEKMEAAKRLESD 95
>ref|NP_176367.1| expressed protein [Arabidopsis thaliana] gi|4508075|gb|AAD21419.1|
50259 gi|13272465|gb|AAK17171.1| unknown protein
[Arabidopsis thaliana] gi|25404344|pir||B96643
hypothetical protein T13M11.10 [imported] - Arabidopsis
thaliana
Length = 458
Score = 120 bits (302), Expect = 7e-26
Identities = 54/154 (35%), Positives = 100/154 (64%), Gaps = 2/154 (1%)
Query: 74 PEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGA 133
P ++ I+ ++ FL +++ S GG+GGGG++VP++ ++ G D K++++ S M+ G
Sbjct: 51 PRIELTTSTIIAGLLSFLASSISSAGGIGGGGLYVPIMTIVAGLDLKTASSFSAFMVTGG 110
Query: 134 ALSTVYYNMRLRNPTLD-MPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIIL 192
+++ V N+ +RNP LID+DLALL +P ++LG+SIGVICN++F +W++T L +
Sbjct: 111 SIANVGCNLFVRNPKSGGKTLIDFDLALLLEPCMLLGVSIGVICNLVFPNWLITSLFAVF 170
Query: 193 FIGTSTKALIKGINTWKKET-MLKKETAKQLEEE 225
++ K G+ W+ E+ M+K + ++EE+
Sbjct: 171 LAWSTLKTFGNGLYYWRLESEMVKIRESNRIEED 204
>ref|NP_915724.1| P0415A04.10 [Oryza sativa (japonica cultivar-group)]
Length = 488
Score = 118 bits (296), Expect = 3e-25
Identities = 57/127 (44%), Positives = 80/127 (62%), Gaps = 1/127 (0%)
Query: 87 IIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMRLRN 146
++ FL A++ S GGVGGG +F+P+L L+ G K +TA S M+ G A S V YN+
Sbjct: 96 LLSFLAASVSSAGGVGGGSLFLPILNLVAGLSLKRATAYSSFMVTGGAASNVLYNLLCTG 155
Query: 147 -PTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGTSTKALIKGI 205
+IDYD+ALLFQP L+LG+SIGV+CNVMF +W++T L + +TK L G+
Sbjct: 156 CGRRAAAVIDYDIALLFQPCLLLGVSIGVVCNVMFPEWLITALFALFLAFCTTKTLRAGL 215
Query: 206 NTWKKET 212
W E+
Sbjct: 216 RIWSSES 222
>gb|AAC12639.1| hypothetical protein [Mesembryanthemum crystallinum]
gi|7484617|pir||T12229 hypothetical protein - common ice
plant (fragment)
Length = 132
Score = 118 bits (296), Expect = 3e-25
Identities = 56/77 (72%), Positives = 67/77 (86%)
Query: 153 LIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGTSTKALIKGINTWKKET 212
+IDYDLALLFQPMLMLGISIGV NV+FADWMVTVLLI+LF+GTSTKA ++GI+TWKKET
Sbjct: 1 IIDYDLALLFQPMLMLGISIGVAFNVIFADWMVTVLLIVLFVGTSTKAFMRGIDTWKKET 60
Query: 213 MLKKETAKQLEEEPKTG 229
+++KE AK+ E G
Sbjct: 61 LMQKEAAKRAESNGADG 77
>dbj|BAD53257.1| membrane protein-like [Oryza sativa (japonica cultivar-group)]
Length = 461
Score = 118 bits (296), Expect = 3e-25
Identities = 57/127 (44%), Positives = 80/127 (62%), Gaps = 1/127 (0%)
Query: 87 IIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMRLRN 146
++ FL A++ S GGVGGG +F+P+L L+ G K +TA S M+ G A S V YN+
Sbjct: 69 LLSFLAASVSSAGGVGGGSLFLPILNLVAGLSLKRATAYSSFMVTGGAASNVLYNLLCTG 128
Query: 147 -PTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGTSTKALIKGI 205
+IDYD+ALLFQP L+LG+SIGV+CNVMF +W++T L + +TK L G+
Sbjct: 129 CGRRAAAVIDYDIALLFQPCLLLGVSIGVVCNVMFPEWLITALFALFLAFCTTKTLRAGL 188
Query: 206 NTWKKET 212
W E+
Sbjct: 189 RIWSSES 195
>ref|NP_915723.1| P0415A04.9 [Oryza sativa (japonica cultivar-group)]
Length = 590
Score = 118 bits (295), Expect = 4e-25
Identities = 57/137 (41%), Positives = 82/137 (59%), Gaps = 3/137 (2%)
Query: 79 DWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTV 138
D ++ I+ FL AA S GGVGGG ++VP+L ++ G K++TA S M+ G LS V
Sbjct: 72 DLGTVLACILSFLAAAFSSAGGVGGGSLYVPILNIVAGLSLKTATAFSTFMVTGGTLSNV 131
Query: 139 YYNM---RLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIG 195
Y + R PLIDYD+A++ QP L+LG+S+GVICNVMF +W++T L +
Sbjct: 132 LYTLIVLRGHEKGGHQPLIDYDIAVVSQPCLLLGVSVGVICNVMFPEWLITALFAVFLAS 191
Query: 196 TSTKALIKGINTWKKET 212
+ K G+ W+ ET
Sbjct: 192 ATFKTYGTGMKRWRAET 208
>dbj|BAD53380.1| membrane protein-like [Oryza sativa (japonica cultivar-group)]
gi|53792418|dbj|BAD53256.1| membrane protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 397
Score = 118 bits (295), Expect = 4e-25
Identities = 57/137 (41%), Positives = 82/137 (59%), Gaps = 3/137 (2%)
Query: 79 DWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTV 138
D ++ I+ FL AA S GGVGGG ++VP+L ++ G K++TA S M+ G LS V
Sbjct: 2 DLGTVLACILSFLAAAFSSAGGVGGGSLYVPILNIVAGLSLKTATAFSTFMVTGGTLSNV 61
Query: 139 YYNM---RLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIG 195
Y + R PLIDYD+A++ QP L+LG+S+GVICNVMF +W++T L +
Sbjct: 62 LYTLIVLRGHEKGGHQPLIDYDIAVVSQPCLLLGVSVGVICNVMFPEWLITALFAVFLAS 121
Query: 196 TSTKALIKGINTWKKET 212
+ K G+ W+ ET
Sbjct: 122 ATFKTYGTGMKRWRAET 138
>ref|XP_482182.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|40253789|dbj|BAD05727.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|40253413|dbj|BAD05342.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 132
Score = 117 bits (292), Expect = 1e-24
Identities = 55/82 (67%), Positives = 68/82 (82%), Gaps = 1/82 (1%)
Query: 49 ETKGFLKAMVDFLWESGKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFV 108
E G+++ +V+FLW SG++SY VWP M+F WKI++G +IGF GAA GSVGGVGGGGIFV
Sbjct: 38 EEVGYMRKVVNFLW-SGEASYHHVWPPMEFGWKIVLGILIGFFGAAFGSVGGVGGGGIFV 96
Query: 109 PMLALIIGFDPKSSTAISKCMI 130
PML LIIGFD KSSTAISK ++
Sbjct: 97 PMLTLIIGFDAKSSTAISKFIV 118
>emb|CAB79125.1| hypothetical protein [Arabidopsis thaliana]
gi|2911082|emb|CAA17544.1| hypothetical protein
[Arabidopsis thaliana] gi|7486594|pir||T04956
hypothetical protein F7J7.190 - Arabidopsis thaliana
Length = 431
Score = 116 bits (290), Expect = 2e-24
Identities = 57/161 (35%), Positives = 95/161 (58%), Gaps = 13/161 (8%)
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAA 134
E+K II+ ++ FL A + S GG+GGGG+F+P++ ++ G D K++++ S M+ G +
Sbjct: 51 ELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASSFSAFMVTGGS 110
Query: 135 LSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFI 194
++ V N+ L+DYDLALL +P ++LG+SIGVICN + +W++TVL +
Sbjct: 111 IANVISNL-----FGGKALLDYDLALLLEPCMLLGVSIGVICNRVLPEWLITVLFAVFLA 165
Query: 195 GTSTKALIKGINTWKKETMLKKETAK--------QLEEEPK 227
+ K G+ WK E+ + +E+ Q+EEE K
Sbjct: 166 WSILKTCRSGVKFWKLESEIARESGHGRPERGQGQIEEETK 206
>ref|NP_193857.2| expressed protein [Arabidopsis thaliana]
Length = 449
Score = 116 bits (290), Expect = 2e-24
Identities = 57/161 (35%), Positives = 95/161 (58%), Gaps = 13/161 (8%)
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAA 134
E+K II+ ++ FL A + S GG+GGGG+F+P++ ++ G D K++++ S M+ G +
Sbjct: 51 ELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASSFSAFMVTGGS 110
Query: 135 LSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFI 194
++ V N+ L+DYDLALL +P ++LG+SIGVICN + +W++TVL +
Sbjct: 111 IANVISNL-----FGGKALLDYDLALLLEPCMLLGVSIGVICNRVLPEWLITVLFAVFLA 165
Query: 195 GTSTKALIKGINTWKKETMLKKETAK--------QLEEEPK 227
+ K G+ WK E+ + +E+ Q+EEE K
Sbjct: 166 WSILKTCRSGVKFWKLESEIARESGHGRPERGQGQIEEETK 206
Score = 34.7 bits (78), Expect = 6.5
Identities = 28/110 (25%), Positives = 49/110 (44%), Gaps = 10/110 (9%)
Query: 88 IGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMI-MGAALSTVYYNMRLRN 146
+ FL LG + G+GGG + P+L L G P+ + A + M+ A +S V Y
Sbjct: 316 MSFLAGLLGGIFGIGGGMLISPLL-LQSGIPPQITAATTSFMVFFSATMSAVQY------ 368
Query: 147 PTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGT 196
L + + + D A +F + L +G++ +I+ +GT
Sbjct: 369 --LLLGMQNTDTAYVFSFICFLASLLGLVLVQKAVAQFGRASIIVFSVGT 416
>gb|AAF16643.1| T23J18.20 [Arabidopsis thaliana]
Length = 491
Score = 116 bits (290), Expect = 2e-24
Identities = 53/131 (40%), Positives = 86/131 (65%), Gaps = 1/131 (0%)
Query: 83 IVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNM 142
I+ +++ F A++ S GG+GGGG+F+ ++ +I G + K++++ S M+ G + + V N+
Sbjct: 97 IIAAVLSFFAASISSAGGIGGGGLFLSIMTIIAGLEMKTASSFSAFMVTGVSFANVGCNL 156
Query: 143 RLRNP-TLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGTSTKAL 201
LRNP + D LID+DLAL QP L+LG+SIGVICN MF +W+V L + ++ K
Sbjct: 157 FLRNPKSRDKTLIDFDLALTIQPCLLLGVSIGVICNRMFPNWLVLFLFAVFLAWSTMKTC 216
Query: 202 IKGINTWKKET 212
KG++ W E+
Sbjct: 217 KKGVSYWNLES 227
>ref|XP_646141.1| hypothetical protein DDB0190426 [Dictyostelium discoideum]
gi|60474235|gb|EAL72172.1| hypothetical protein
DDB0190426 [Dictyostelium discoideum]
Length = 549
Score = 112 bits (279), Expect = 3e-23
Identities = 56/142 (39%), Positives = 84/142 (58%)
Query: 83 IVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNM 142
IVG I+ F+G AL S GGVGGGGI++P+L L+ +DPK++ +S C++ G AL+ N
Sbjct: 137 IVGFILLFIGCALSSGGGVGGGGIYIPILILVSKWDPKTAIPLSNCLVAGCALANFIQNF 196
Query: 143 RLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGTSTKALI 202
R+P + LIDY +ALL +P+ + G GV + F ++ +LL+I TS K +
Sbjct: 197 PRRHPFSNKHLIDYSVALLIEPLTLAGTIFGVYLHTYFPPLVILLLLVITLGFTSYKTIS 256
Query: 203 KGINTWKKETMLKKETAKQLEE 224
KGI+ WK E K ++
Sbjct: 257 KGIDIWKSEKKKKNSLLSNTDD 278
Score = 34.3 bits (77), Expect = 8.5
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 2/77 (2%)
Query: 62 WESGKSSYEPVWPEMKFDWK-IIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPK 120
+E K + E+K+ +K I++ I+ + L S+ G+GGG I P+L L +G P
Sbjct: 385 YEKKKEEGREIEGEIKYTYKNILLLGILSVIAGCLASLLGIGGGMIKGPVL-LQMGLVPD 443
Query: 121 SSTAISKCMIMGAALST 137
+ A S MI+ + S+
Sbjct: 444 VTAATSSYMILFTSASS 460
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.351 0.156 0.553
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 695,940,148
Number of Sequences: 2540612
Number of extensions: 26444346
Number of successful extensions: 151940
Number of sequences better than 10.0: 208
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 144
Number of HSP's that attempted gapping in prelim test: 151603
Number of HSP's gapped (non-prelim): 382
length of query: 465
length of database: 863,360,394
effective HSP length: 132
effective length of query: 333
effective length of database: 527,999,610
effective search space: 175823870130
effective search space used: 175823870130
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC147178.14


