

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.12 + phase: 0
(163 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
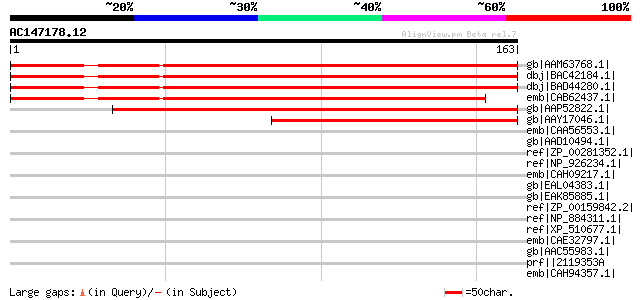
gb|AAM63768.1| unknown [Arabidopsis thaliana] 214 5e-55
dbj|BAC42184.1| unknown protein [Arabidopsis thaliana] gi|169743... 212 3e-54
dbj|BAD44280.1| unknown protein [Arabidopsis thaliana] 208 4e-53
emb|CAB62437.1| putative protein [Arabidopsis thaliana] gi|11280... 190 1e-47
gb|AAP52822.1| hypothetical protein [Oryza sativa (japonica cult... 180 1e-44
gb|AAY17046.1| p-166-4_1 [Pinus resinosa] 133 2e-30
emb|CAA56553.1| gamma-TIP-like protein [Hordeum vulgare subsp. v... 37 0.19
gb|AAD10494.1| gamma-type tonoplast intrinsic protein [Triticum ... 36 0.32
ref|ZP_00281352.1| COG0477: Permeases of the major facilitator s... 36 0.42
ref|NP_926234.1| hypothetical protein gll3288 [Gloeobacter viola... 36 0.42
emb|CAH09217.1| putative membrane protein [Bacteroides fragilis ... 35 0.72
gb|EAL04383.1| likely voltage-gated chloride channel fragment [C... 35 0.94
gb|EAK85885.1| hypothetical protein UM05025.1 [Ustilago maydis 5... 34 1.6
ref|ZP_00159842.2| COG0672: High-affinity Fe2+/Pb2+ permease [An... 34 1.6
ref|NP_884311.1| serine protease [Bordetella parapertussis 12822... 33 2.7
ref|XP_510677.1| PREDICTED: similar to hypothetical protein [Pan... 33 2.7
emb|CAE32797.1| serine protease [Bordetella bronchiseptica RB50]... 33 2.7
gb|AAC55983.1| polyprotein 33 2.7
prf||2119353A polyprotein gi|1096574|prf||2111488A polyprotein 33 2.7
emb|CAH94357.1| putative integral membrane protein [Streptomyces... 33 3.6
>gb|AAM63768.1| unknown [Arabidopsis thaliana]
Length = 158
Score = 214 bits (546), Expect = 5e-55
Identities = 108/163 (66%), Positives = 128/163 (78%), Gaps = 5/163 (3%)
Query: 1 MLLFSPISTPKTFTNHLNHPLRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAGGLI 60
MLL SPIS + H + +R+ ++ ++ AK+ +DT E AAIAGGL+
Sbjct: 1 MLLLSPISASLPPSFHRGNLIRRS----IKPLGRVVAKAKDNTDTGGF-LETAAIAGGLV 55
Query: 61 STPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGP 120
STPVIGWSLYTLKTTGCGLPPGP G IGALEGVSYLVVVGIV WS+YTKTKTGSGLPNGP
Sbjct: 56 STPVIGWSLYTLKTTGCGLPPGPAGLIGALEGVSYLVVVGIVGWSLYTKTKTGSGLPNGP 115
Query: 121 FGLLGAVEGLSYLALVAIVVVFGLQYFQQGYIPGPLPADQCFG 163
FGLLGAVEGLSYL+++AI+VVFG+Q+ G +PGPLP+DQCFG
Sbjct: 116 FGLLGAVEGLSYLSVLAILVVFGIQFLDNGSVPGPLPSDQCFG 158
>dbj|BAC42184.1| unknown protein [Arabidopsis thaliana] gi|16974355|gb|AAL31103.1|
AT3g50680/T3A5_60 [Arabidopsis thaliana]
gi|15529176|gb|AAK97682.1| AT3g50680/T3A5_60
[Arabidopsis thaliana] gi|18409161|ref|NP_566937.1|
expressed protein [Arabidopsis thaliana]
gi|51971901|dbj|BAD44615.1| unknown protein [Arabidopsis
thaliana] gi|51971663|dbj|BAD44496.1| unknown protein
[Arabidopsis thaliana]
Length = 158
Score = 212 bits (540), Expect = 3e-54
Identities = 107/163 (65%), Positives = 127/163 (77%), Gaps = 5/163 (3%)
Query: 1 MLLFSPISTPKTFTNHLNHPLRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAGGLI 60
MLL SPIS + H + +R+ ++ ++ AK+ +DT E AAIAG L+
Sbjct: 1 MLLLSPISASLPPSFHRGNLIRRS----IKPLGRVVAKAKDNTDTGGF-LETAAIAGSLV 55
Query: 61 STPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGP 120
STPVIGWSLYTLKTTGCGLPPGP G IGALEGVSYLVVVGIV WS+YTKTKTGSGLPNGP
Sbjct: 56 STPVIGWSLYTLKTTGCGLPPGPAGLIGALEGVSYLVVVGIVGWSLYTKTKTGSGLPNGP 115
Query: 121 FGLLGAVEGLSYLALVAIVVVFGLQYFQQGYIPGPLPADQCFG 163
FGLLGAVEGLSYL+++AI+VVFG+Q+ G +PGPLP+DQCFG
Sbjct: 116 FGLLGAVEGLSYLSVLAILVVFGIQFLDNGSVPGPLPSDQCFG 158
>dbj|BAD44280.1| unknown protein [Arabidopsis thaliana]
Length = 158
Score = 208 bits (530), Expect = 4e-53
Identities = 106/163 (65%), Positives = 126/163 (77%), Gaps = 5/163 (3%)
Query: 1 MLLFSPISTPKTFTNHLNHPLRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAGGLI 60
MLL SPIS + H + +R+ ++ ++ AK+ +DT E AAIAG L+
Sbjct: 1 MLLLSPISASLPPSFHRGNLIRRS----IKPLGRVVAKAKDNTDTGGF-LETAAIAGSLV 55
Query: 61 STPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGP 120
STPVIGWSLYTLKTTGCGLPPGP G IGALEGVSYLVVVGIV WS+YTKTKTGSGLPNGP
Sbjct: 56 STPVIGWSLYTLKTTGCGLPPGPAGLIGALEGVSYLVVVGIVGWSLYTKTKTGSGLPNGP 115
Query: 121 FGLLGAVEGLSYLALVAIVVVFGLQYFQQGYIPGPLPADQCFG 163
FGLLGAVEGLSYL+++AI+VVFG+Q+ G +PGPLP+DQ FG
Sbjct: 116 FGLLGAVEGLSYLSVLAILVVFGIQFLDNGSVPGPLPSDQSFG 158
>emb|CAB62437.1| putative protein [Arabidopsis thaliana] gi|11280670|pir||T46145
hypothetical protein T3A5.60 - Arabidopsis thaliana
Length = 319
Score = 190 bits (483), Expect = 1e-47
Identities = 98/153 (64%), Positives = 117/153 (76%), Gaps = 5/153 (3%)
Query: 1 MLLFSPISTPKTFTNHLNHPLRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAGGLI 60
MLL SPIS + H + +R+ ++ ++ AK+ +DT E AAIAG L+
Sbjct: 1 MLLLSPISASLPPSFHRGNLIRRS----IKPLGRVVAKAKDNTDTGGF-LETAAIAGSLV 55
Query: 61 STPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGP 120
STPVIGWSLYTLKTTGCGLPPGP G IGALEGVSYLVVVGIV WS+YTKTKTGSGLPNGP
Sbjct: 56 STPVIGWSLYTLKTTGCGLPPGPAGLIGALEGVSYLVVVGIVGWSLYTKTKTGSGLPNGP 115
Query: 121 FGLLGAVEGLSYLALVAIVVVFGLQYFQQGYIP 153
FGLLGAVEGLSYL+++AI+VVFG+Q+ G +P
Sbjct: 116 FGLLGAVEGLSYLSVLAILVVFGIQFLDNGSVP 148
>gb|AAP52822.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|37532466|ref|NP_920535.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|20043062|gb|AAM08870.1| Hypothetical protein [Oryza
sativa]
Length = 163
Score = 180 bits (456), Expect = 1e-44
Identities = 83/130 (63%), Positives = 100/130 (76%)
Query: 34 KITNMAKEGSDTNSSPTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPPGPGGSIGALEGV 93
++T K +S E+AA A GL S+ V+ WSLYTLKTTGCGLPPGPGG++GA EGV
Sbjct: 34 RLTTTCKAEPSGGNSTLELAAGAAGLASSSVVAWSLYTLKTTGCGLPPGPGGALGAAEGV 93
Query: 94 SYLVVVGIVAWSIYTKTKTGSGLPNGPFGLLGAVEGLSYLALVAIVVVFGLQYFQQGYIP 153
SYLVV ++ WS+ TK +TGSGLP GPFGLLGA EG+SYLA AI VVFG Q+F+ G +P
Sbjct: 94 SYLVVAALIGWSLTTKVRTGSGLPAGPFGLLGAAEGVSYLAAAAIAVVFGFQFFEVGSLP 153
Query: 154 GPLPADQCFG 163
GPLP+DQCFG
Sbjct: 154 GPLPSDQCFG 163
>gb|AAY17046.1| p-166-4_1 [Pinus resinosa]
Length = 79
Score = 133 bits (334), Expect = 2e-30
Identities = 59/79 (74%), Positives = 67/79 (84%)
Query: 85 GSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGPFGLLGAVEGLSYLALVAIVVVFGL 144
G++GALEG+S+LVV G++ WS+YTK KTGSGLP+GPFGLLGA EGLSYL LV IV VFGL
Sbjct: 1 GALGALEGISFLVVFGLIGWSVYTKVKTGSGLPSGPFGLLGAAEGLSYLTLVGIVAVFGL 60
Query: 145 QYFQQGYIPGPLPADQCFG 163
QY GYIPGP P DQCFG
Sbjct: 61 QYLDYGYIPGPTPGDQCFG 79
>emb|CAA56553.1| gamma-TIP-like protein [Hordeum vulgare subsp. vulgare]
gi|629784|pir||S47037 tonoplast intrinsic protein gamma
- barley
Length = 250
Score = 37.0 bits (84), Expect = 0.19
Identities = 33/112 (29%), Positives = 49/112 (43%), Gaps = 16/112 (14%)
Query: 48 SPTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPPGP-----------GGSIGALEGVSYL 96
SP +A AG + + ++L+ + G + G GG+I G+ Y
Sbjct: 48 SPDGVATPAGLISAAIAHAFALFVAVSVGANISGGHVNPAVTFGAFVGGNITLFRGLLYW 107
Query: 97 V--VVGIVAWSIYTKTKTGSGLPNGPFGLLGAVEGLSYLALVAIVVVFGLQY 146
V ++G A + TG GLP G FGL G G ++ IV+ FGL Y
Sbjct: 108 VAQLLGSTAACFLLRFSTG-GLPTGTFGLTGI--GAWEAVVLEIVMTFGLVY 156
>gb|AAD10494.1| gamma-type tonoplast intrinsic protein [Triticum aestivum]
Length = 250
Score = 36.2 bits (82), Expect = 0.32
Identities = 33/108 (30%), Positives = 48/108 (43%), Gaps = 17/108 (15%)
Query: 53 AAIAGGLISTPVI-GWSLYTLKTTGCGLPPGP-----------GGSIGALEGVSYLV--V 98
AA GLIS + ++L+ + G + G GG+I G+ Y + +
Sbjct: 52 AATPAGLISAAIAHAFALFVAVSVGANISGGHVNPAVTFGAFVGGNITLFRGLLYWIAQL 111
Query: 99 VGIVAWSIYTKTKTGSGLPNGPFGLLGAVEGLSYLALVAIVVVFGLQY 146
+G A + TG GLP G FGL G G ++ IV+ FGL Y
Sbjct: 112 LGSTAACFLLRFSTG-GLPTGTFGLSGI--GAWEAVVLEIVMTFGLVY 156
>ref|ZP_00281352.1| COG0477: Permeases of the major facilitator superfamily
[Burkholderia fungorum LB400]
Length = 552
Score = 35.8 bits (81), Expect = 0.42
Identities = 26/75 (34%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query: 85 GSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGPFGL-----LGAVEGLSYLALVAIV 139
GS+ A S+ V A I+T +G PFG LG + G Y LV IV
Sbjct: 39 GSLAAFISKSFFSGVNPTAGFIFTLLSFAAGFAVRPFGAIVFGRLGDLVGRKYTFLVTIV 98
Query: 140 VVFGLQYFQQGYIPG 154
++ GL F G++PG
Sbjct: 99 IM-GLSTFLVGFLPG 112
>ref|NP_926234.1| hypothetical protein gll3288 [Gloeobacter violaceus PCC 7421]
gi|35213859|dbj|BAC91229.1| gll3288 [Gloeobacter
violaceus PCC 7421]
Length = 319
Score = 35.8 bits (81), Expect = 0.42
Identities = 30/132 (22%), Positives = 56/132 (41%), Gaps = 21/132 (15%)
Query: 40 KEGSDTNSSPTEIAAIAGGLISTPVIG----WSLYTLKTTGCGLPPGPGGSIGALEGVSY 95
K+ T+ + +AGGL ++ +G W L L T P LEG+
Sbjct: 32 KKAGHTHLNRWVWGGVAGGLAASTAVGLLFGWLLGNLSTANQKYAPAVEP---LLEGIFG 88
Query: 96 LVVVGIVAWSIYTKTKTGSGLPNGPFGLLGAVEG--------LSYLALVAIV------VV 141
++ +G+++W + T+ L +G G +++L L ++ V+
Sbjct: 89 VLAIGMLSWMLIWMTRQARTLKGQVEASVGQALGQERAAGWAIAWLILFTVLREGFETVL 148
Query: 142 FGLQYFQQGYIP 153
F YF+QG++P
Sbjct: 149 FVTSYFRQGFVP 160
>emb|CAH09217.1| putative membrane protein [Bacteroides fragilis NCTC 9343]
gi|60682987|ref|YP_213131.1| putative membrane protein
[Bacteroides fragilis NCTC 9343]
gi|53715025|ref|YP_101017.1| hypothetical protein BF3741
[Bacteroides fragilis YCH46] gi|52217890|dbj|BAD50483.1|
conserved hypothetical protein [Bacteroides fragilis
YCH46]
Length = 156
Score = 35.0 bits (79), Expect = 0.72
Identities = 29/93 (31%), Positives = 44/93 (47%), Gaps = 7/93 (7%)
Query: 58 GLISTPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIVAWSI---YTKTKTG- 113
GL V+ + + G L P G+I +L G+ LV+V +VAWS+ T T G
Sbjct: 56 GLFGIAVVATVVAAIFQFGSALKDNPKGAIRSLLGLILLVLVLVVAWSMGSGETLTIQGY 115
Query: 114 SGLPNGPFGLLGA---VEGLSYLALVAIVVVFG 143
G N PF L + + +L LV ++ + G
Sbjct: 116 EGTDNVPFWLKLTDMFLYSIYFLMLVTVLAIIG 148
>gb|EAL04383.1| likely voltage-gated chloride channel fragment [Candida albicans
SC5314] gi|46444957|gb|EAL04228.1| likely voltage-gated
chloride channel fragment [Candida albicans SC5314]
Length = 477
Score = 34.7 bits (78), Expect = 0.94
Identities = 31/103 (30%), Positives = 42/103 (40%), Gaps = 12/103 (11%)
Query: 43 SDTNSSPTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPPGPGGSIGALEG--VSYLVVVG 100
S S +EI I G + +GW +K+ G L G G S+G EG V Y V VG
Sbjct: 179 SAAGSGISEIKCIVSGFVMDGFLGWPTLFIKSLGLPLAIGSGLSVGK-EGPSVHYAVCVG 237
Query: 101 IVAWSIYTKTKTGSGLPNGPFGLLGAVEGLSYLALVAIVVVFG 143
+ TK + + A E L+ A + V FG
Sbjct: 238 NSIAKLITKYRKSAS---------RAREFLTATAAAGVAVAFG 271
>gb|EAK85885.1| hypothetical protein UM05025.1 [Ustilago maydis 521]
gi|49077600|ref|XP_402640.1| hypothetical protein
UM05025.1 [Ustilago maydis 521]
Length = 591
Score = 33.9 bits (76), Expect = 1.6
Identities = 27/87 (31%), Positives = 36/87 (41%), Gaps = 12/87 (13%)
Query: 47 SSPTEIAAIAGGLISTPVIGWSLYT------------LKTTGCGLPPGPGGSIGALEGVS 94
SSP ++IAG + I + YT L+ G GLPPG GG G G S
Sbjct: 484 SSPAPTSSIAGSIGIGAEIPFLAYTTPNQDMVNNVSWLRAGGGGLPPGVGGFGGGGGGGS 543
Query: 95 YLVVVGIVAWSIYTKTKTGSGLPNGPF 121
+ W+ +T TG G + F
Sbjct: 544 SSHKSVVQEWNEFTAGSTGGGSSSSGF 570
>ref|ZP_00159842.2| COG0672: High-affinity Fe2+/Pb2+ permease [Anabaena variabilis ATCC
29413]
Length = 315
Score = 33.9 bits (76), Expect = 1.6
Identities = 33/131 (25%), Positives = 56/131 (42%), Gaps = 15/131 (11%)
Query: 38 MAKEGSDTNSSPTEIAAIAGGLISTPVIGWSL-YTLKTTGCGLPPGPGGSIGALEGVSYL 96
+ K+ + +P A +A G++ + +IG + +K G P LEGV +
Sbjct: 30 LLKKAKQSRLNPWVYAGVAVGIVISALIGVLFTWVIKAVGAANPQYTVVVEPMLEGVFSV 89
Query: 97 VVVGIVAWSIYTKTKTGSGLPNGPFGLL--------GAVEGLSYLALVAIV------VVF 142
+ + +++W + TK + G + A G+ L LVA+V V+F
Sbjct: 90 LAIAMLSWMLIWMTKQARFMKAQVEGAVTDALTQNSNAGWGVFSLILVAVVREGFETVLF 149
Query: 143 GLQYFQQGYIP 153
FQQG IP
Sbjct: 150 IAANFQQGLIP 160
>ref|NP_884311.1| serine protease [Bordetella parapertussis 12822]
gi|33573369|emb|CAE37353.1| serine protease [Bordetella
parapertussis]
Length = 1076
Score = 33.1 bits (74), Expect = 2.7
Identities = 27/89 (30%), Positives = 40/89 (44%), Gaps = 11/89 (12%)
Query: 37 NMAKEGSDTNSSPTEIAAIAGGL--ISTPVIGWSLY---TLKTTGCGLPPGPG------G 85
++A S SPT + GG +S ++G +L L+ TG L PG G
Sbjct: 529 SLAGGSSLGGGSPTTPIVVTGGADAVSGAILGGNLDIGGALRMTGASLAPGNSIGTVTVG 588
Query: 86 SIGALEGVSYLVVVGIVAWSIYTKTKTGS 114
S+GAL G +YL V S ++G+
Sbjct: 589 SVGALSGSTYLAEVNAKGQSDLLVVRSGN 617
>ref|XP_510677.1| PREDICTED: similar to hypothetical protein [Pan troglodytes]
Length = 861
Score = 33.1 bits (74), Expect = 2.7
Identities = 19/49 (38%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Query: 69 LYTLKTTGCGLPPGPGGSIGALEGVSYLV-VVGIVAWSIYTKTKTGSGL 116
+Y +T CG GP G+I A+ V+ V +VG +AW I T GL
Sbjct: 142 VYQFRTKRCGQATGPAGNIMAVNSVTLAVNLVGCLAWLIGGGGATNFGL 190
>emb|CAE32797.1| serine protease [Bordetella bronchiseptica RB50]
gi|33601284|ref|NP_888844.1| serine protease [Bordetella
bronchiseptica RB50]
Length = 1076
Score = 33.1 bits (74), Expect = 2.7
Identities = 27/89 (30%), Positives = 40/89 (44%), Gaps = 11/89 (12%)
Query: 37 NMAKEGSDTNSSPTEIAAIAGGL--ISTPVIGWSLY---TLKTTGCGLPPGPG------G 85
++A S SPT + GG +S ++G +L L+ TG L PG G
Sbjct: 529 SLAGGSSLGGGSPTTPIVVTGGADAVSGAILGGNLDIGGALRMTGASLAPGNSIGTVTVG 588
Query: 86 SIGALEGVSYLVVVGIVAWSIYTKTKTGS 114
S+GAL G +YL V S ++G+
Sbjct: 589 SVGALSGSTYLAEVNAKGQSDLLVVRSGN 617
>gb|AAC55983.1| polyprotein
Length = 2954
Score = 33.1 bits (74), Expect = 2.7
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 9/54 (16%)
Query: 49 PTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIV 102
P AA+A L+ TPV+GW+ TGC +G + V+YL V+G V
Sbjct: 652 PVVEAALAPELVCTPVVGWAAQEWWFTGC---------LGVMCVVAYLNVLGSV 696
>prf||2119353A polyprotein gi|1096574|prf||2111488A polyprotein
Length = 2962
Score = 33.1 bits (74), Expect = 2.7
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 9/54 (16%)
Query: 49 PTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIV 102
P AA+A L+ TPV+GW+ TGC +G + V+YL V+G V
Sbjct: 660 PVVEAALAPELVCTPVVGWAAQEWWFTGC---------LGVMCVVAYLNVLGSV 704
>emb|CAH94357.1| putative integral membrane protein [Streptomyces griseus subsp.
griseus]
Length = 347
Score = 32.7 bits (73), Expect = 3.6
Identities = 29/88 (32%), Positives = 41/88 (45%), Gaps = 2/88 (2%)
Query: 38 MAKEGSDTNSSPTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLV 97
M + S T+ S +A AG L +TP I T G G GP A + L
Sbjct: 1 MRADNSATHPSSIAVAEPAGPLRTTPAIRPEAAEAATAGRGPGSGPARQPSAGRSGTVLA 60
Query: 98 VVGIVAWSIYTKTKTGSGLPN-GPFGLL 124
+G+VA+S+ T T GL + GP+ L+
Sbjct: 61 GLGVVAFSL-TFPATVWGLESFGPWSLV 87
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 334,626,623
Number of Sequences: 2540612
Number of extensions: 15949831
Number of successful extensions: 40514
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 40488
Number of HSP's gapped (non-prelim): 47
length of query: 163
length of database: 863,360,394
effective HSP length: 118
effective length of query: 45
effective length of database: 563,568,178
effective search space: 25360568010
effective search space used: 25360568010
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC147178.12


