

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.5 - phase: 0
(145 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
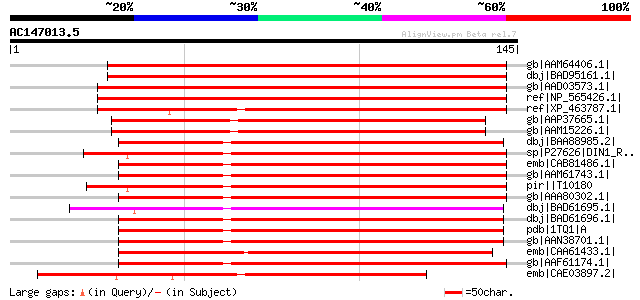
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM64406.1| senescence-associated protein [Arabidopsis thaliana] 144 3e-34
dbj|BAD95161.1| senescence-associated protein sen1-like protein ... 144 3e-34
gb|AAD03573.1| putative senescence-associated rhodanese protein ... 136 9e-32
ref|NP_565426.1| senescence-associated family protein [Arabidops... 136 9e-32
ref|XP_463787.1| putative senescence-associated protein [Oryza s... 120 9e-27
gb|AAP37665.1| At2g21045 [Arabidopsis thaliana] gi|42569206|ref|... 111 4e-24
gb|AAM15226.1| senescence-associated protein [Arabidopsis thalia... 111 4e-24
dbj|BAA88985.2| Ntdin [Nicotiana tabacum] 110 9e-24
sp|P27626|DIN1_RAPSA Senescence-associated protein DIN1 gi|74886... 106 1e-22
emb|CAB81486.1| senescence-associated protein sen1 [Arabidopsis ... 105 2e-22
gb|AAM61743.1| senescence-associated protein sen1 [Arabidopsis t... 105 2e-22
pir||T10180 senescence-associated protein din1, dark-inducible -... 105 2e-22
gb|AAA80302.1| senescence-associated protein [Arabidopsis thaliana] 105 3e-22
dbj|BAD61695.1| putative Ntdin [Oryza sativa (japonica cultivar-... 102 3e-21
dbj|BAD61696.1| putative Ntdin [Oryza sativa (japonica cultivar-... 101 3e-21
pdb|1TQ1|A Chain A, Solution Structure Of At5g66040, A Putative ... 100 6e-21
gb|AAN38701.1| At5g66040/K2A18_11 [Arabidopsis thaliana] gi|2153... 100 1e-20
emb|CAA61433.1| ketoconazole resistent protein [Arabidopsis thal... 97 6e-20
gb|AAF61174.1| OP1 [Cucumis sativus] 93 2e-18
emb|CAE03897.2| OSJNBb0026I12.5 [Oryza sativa (japonica cultivar... 91 4e-18
>gb|AAM64406.1| senescence-associated protein [Arabidopsis thaliana]
Length = 136
Score = 144 bits (364), Expect = 3e-34
Identities = 65/114 (57%), Positives = 89/114 (78%)
Query: 29 KVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVK 88
+VV++DV AK L+Q+GH YLDVRT +EF +GH +A KI+NIPY+L+TP+GRVKN F++
Sbjct: 13 EVVSVDVSQAKTLLQSGHQYLDVRTQDEFRRGHCEAAKIVNIPYMLNTPQGRVKNREFLE 72
Query: 89 QVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
QVSS + D ++VGCQSG RS AT+EL+A G+K V N+GGGY+ WV + P+
Sbjct: 73 QVSSLLNPADDILVGCQSGARSLKATTELVAAGYKKVRNVGGGYLAWVDHSFPI 126
>dbj|BAD95161.1| senescence-associated protein sen1-like protein [Arabidopsis
thaliana] gi|10177132|dbj|BAB10422.1|
senescence-associated protein sen1-like protein
[Arabidopsis thaliana] gi|20147103|gb|AAM10268.1|
AT5g66170/K2A18_25 [Arabidopsis thaliana]
gi|18086478|gb|AAL57692.1| AT5g66170/K2A18_25
[Arabidopsis thaliana] gi|18425042|ref|NP_569030.1|
senescence-associated family protein [Arabidopsis
thaliana]
Length = 136
Score = 144 bits (364), Expect = 3e-34
Identities = 65/114 (57%), Positives = 89/114 (78%)
Query: 29 KVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVK 88
+VV++DV AK L+Q+GH YLDVRT +EF +GH +A KI+NIPY+L+TP+GRVKN F++
Sbjct: 13 EVVSVDVSQAKTLLQSGHQYLDVRTQDEFRRGHCEAAKIVNIPYMLNTPQGRVKNQEFLE 72
Query: 89 QVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
QVSS + D ++VGCQSG RS AT+EL+A G+K V N+GGGY+ WV + P+
Sbjct: 73 QVSSLLNPADDILVGCQSGARSLKATTELVAAGYKKVRNVGGGYLAWVDHSFPI 126
>gb|AAD03573.1| putative senescence-associated rhodanese protein [Arabidopsis
thaliana] gi|7486986|pir||T00839 hypothetical protein
At2g17850 [imported] - Arabidopsis thaliana
Length = 150
Score = 136 bits (343), Expect = 9e-32
Identities = 63/117 (53%), Positives = 87/117 (73%)
Query: 26 SGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLN 85
S KV+TIDV+ A+ L+ +G+ +LDVRTVEEF+KGHVD+ + N+PY L TP+G+ N N
Sbjct: 2 SEPKVITIDVNQAQKLLDSGYTFLDVRTVEEFKKGHVDSENVFNVPYWLYTPQGQEINPN 61
Query: 86 FVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
F+K VSS C++ D L++GC+SG RS AT L++ GFK V NM GGY+ WV+ + PV
Sbjct: 62 FLKHVSSLCNQTDHLILGCKSGVRSLHATKFLVSSGFKTVRNMDGGYIAWVNKRFPV 118
>ref|NP_565426.1| senescence-associated family protein [Arabidopsis thaliana]
Length = 130
Score = 136 bits (343), Expect = 9e-32
Identities = 63/117 (53%), Positives = 87/117 (73%)
Query: 26 SGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLN 85
S KV+TIDV+ A+ L+ +G+ +LDVRTVEEF+KGHVD+ + N+PY L TP+G+ N N
Sbjct: 2 SEPKVITIDVNQAQKLLDSGYTFLDVRTVEEFKKGHVDSENVFNVPYWLYTPQGQEINPN 61
Query: 86 FVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
F+K VSS C++ D L++GC+SG RS AT L++ GFK V NM GGY+ WV+ + PV
Sbjct: 62 FLKHVSSLCNQTDHLILGCKSGVRSLHATKFLVSSGFKTVRNMDGGYIAWVNKRFPV 118
>ref|XP_463787.1| putative senescence-associated protein [Oryza sativa (japonica
cultivar-group)] gi|41053235|dbj|BAD08196.1| putative
senescence-associated protein [Oryza sativa (japonica
cultivar-group)] gi|41052901|dbj|BAD07813.1| putative
senescence-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 137
Score = 120 bits (300), Expect = 9e-27
Identities = 64/118 (54%), Positives = 81/118 (68%), Gaps = 3/118 (2%)
Query: 26 SGAKVVTIDVHAAKNLIQT-GHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNL 84
S VVT+DV AA +LI + GH Y+DVRT EE KGH+ + +N+P++ TP+GR KN
Sbjct: 14 SPVPVVTVDVAAASDLITSAGHRYVDVRTEEEMNKGHLHNS--LNVPFMFVTPQGREKNP 71
Query: 85 NFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
FV+Q SS KE+ +VVGCQSGKRS A +LL GFKNV NMGGGY W+ N P+
Sbjct: 72 LFVEQFSSLVSKEEHVVVGCQSGKRSELACVDLLEAGFKNVKNMGGGYAAWLDNGFPI 129
>gb|AAP37665.1| At2g21045 [Arabidopsis thaliana] gi|42569206|ref|NP_565497.2|
senescence-associated family protein [Arabidopsis
thaliana]
Length = 140
Score = 111 bits (277), Expect = 4e-24
Identities = 54/107 (50%), Positives = 73/107 (67%), Gaps = 2/107 (1%)
Query: 30 VVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQ 89
V T+DV+ AK + TGH YLDVRT EEF K HV+ + +NIPY+ T +GRV N +F+ Q
Sbjct: 12 VETVDVYTAKGFLSTGHRYLDVRTNEEFAKSHVE--EALNIPYMFKTDEGRVINPDFLSQ 69
Query: 90 VSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWV 136
V+S C K++ L+V C +G R A +LL +G+ +V NMGGGY WV
Sbjct: 70 VASVCKKDEHLIVACNAGGRGSRACVDLLNEGYDHVANMGGGYSAWV 116
>gb|AAM15226.1| senescence-associated protein [Arabidopsis thaliana]
gi|20197696|gb|AAM15209.1| senescence-associated protein
[Arabidopsis thaliana]
Length = 169
Score = 111 bits (277), Expect = 4e-24
Identities = 54/107 (50%), Positives = 73/107 (67%), Gaps = 2/107 (1%)
Query: 30 VVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQ 89
V T+DV+ AK + TGH YLDVRT EEF K HV+ + +NIPY+ T +GRV N +F+ Q
Sbjct: 41 VETVDVYTAKGFLSTGHRYLDVRTNEEFAKSHVE--EALNIPYMFKTDEGRVINPDFLSQ 98
Query: 90 VSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWV 136
V+S C K++ L+V C +G R A +LL +G+ +V NMGGGY WV
Sbjct: 99 VASVCKKDEHLIVACNAGGRGSRACVDLLNEGYDHVANMGGGYSAWV 145
>dbj|BAA88985.2| Ntdin [Nicotiana tabacum]
Length = 185
Score = 110 bits (274), Expect = 9e-24
Identities = 56/110 (50%), Positives = 69/110 (61%), Gaps = 2/110 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A L+Q GH YLDVRT EEF GH A INIPY+ G KN NF+++V
Sbjct: 75 SVPVRVALELLQAGHRYLDVRTAEEFSDGH--APGAINIPYMFRIGSGMTKNPNFLEEVL 132
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
K+D ++VGCQ GKRSF ATS+LLA GF V ++ GGY W N LP
Sbjct: 133 ERFGKDDEIIVGCQLGKRSFMATSDLLAAGFTGVTDIAGGYAAWTENGLP 182
>sp|P27626|DIN1_RAPSA Senescence-associated protein DIN1 gi|7488607|pir||T10192
senescence-associated protein din1, dark-inducible -
radish gi|169690|gb|AAA33867.1| din1
Length = 183
Score = 106 bits (264), Expect = 1e-22
Identities = 54/122 (44%), Positives = 79/122 (64%), Gaps = 3/122 (2%)
Query: 22 VLCSSGAKVVT-IDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGR 80
V + A+V T + V A+ L Q G+ +LDVRT +EF GH ++ IN+PY+ G
Sbjct: 62 VAAEAAARVPTSVPVRVARELAQAGYKHLDVRTPDEFSIGH--PSRAINVPYMYRVGSGM 119
Query: 81 VKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKL 140
VKN +F++QVSS K D +++GC+SG+RS A++ELL GF V ++ GGY+ W N+L
Sbjct: 120 VKNPSFLRQVSSHFRKHDEIIIGCESGERSLMASTELLTAGFTGVTDIAGGYVPWTENEL 179
Query: 141 PV 142
PV
Sbjct: 180 PV 181
>emb|CAB81486.1| senescence-associated protein sen1 [Arabidopsis thaliana]
gi|3367595|emb|CAA20047.1| senescence-associated protein
sen1 [Arabidopsis thaliana] gi|20147107|gb|AAM10270.1|
AT4g35770/F8D20_280 [Arabidopsis thaliana]
gi|18958011|gb|AAL79579.1| AT4g35770/F8D20_280
[Arabidopsis thaliana] gi|15233328|ref|NP_195302.1|
senescence-associated protein (SEN1) [Arabidopsis
thaliana] gi|1046270|gb|AAA80303.1|
senescence-associated protein [Arabidopsis thaliana]
gi|7488328|pir||T04682 senescence-associated protein
sen1 - Arabidopsis thaliana
Length = 182
Score = 105 bits (263), Expect = 2e-22
Identities = 50/111 (45%), Positives = 74/111 (66%), Gaps = 2/111 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A+ L Q G+ YLDVRT +EF GH T+ IN+PY+ G VKN +F++QVS
Sbjct: 72 SVPVRVARELAQAGYRYLDVRTPDEFSIGH--PTRAINVPYMYRVGSGMVKNPSFLRQVS 129
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
S K D +++GC+SG+ SF A+++LL GF + ++ GGY+ W N+LPV
Sbjct: 130 SHFRKHDEIIIGCESGQMSFMASTDLLTAGFTAITDIAGGYVAWTENELPV 180
>gb|AAM61743.1| senescence-associated protein sen1 [Arabidopsis thaliana]
Length = 182
Score = 105 bits (263), Expect = 2e-22
Identities = 50/111 (45%), Positives = 74/111 (66%), Gaps = 2/111 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A+ L Q G+ YLDVRT +EF GH T+ IN+PY+ G VKN +F++QVS
Sbjct: 72 SVPVRVARELAQAGYRYLDVRTPDEFSIGH--PTRAINVPYMYRVGSGMVKNPSFLRQVS 129
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
S K D +++GC+SG+ SF A+++LL GF + ++ GGY+ W N+LPV
Sbjct: 130 SHFRKHDEIIIGCESGQMSFMASTDLLTAGFTAITDIAGGYVAWTENELPV 180
>pir||T10180 senescence-associated protein din1, dark-inducible - radish
gi|2190012|dbj|BAA20356.1| din1 [Raphanus sativus]
Length = 182
Score = 105 bits (263), Expect = 2e-22
Identities = 54/121 (44%), Positives = 77/121 (63%), Gaps = 3/121 (2%)
Query: 23 LCSSGAKVVT-IDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRV 81
+ + A+V T + V A+ L Q G+ YLDVRT +EF GH IN+PY+ G V
Sbjct: 62 VAAEAARVPTSVPVRVARELAQAGYKYLDVRTPDEFSIGH--PCSAINVPYMYRVGSGMV 119
Query: 82 KNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
KN +F++QVSS K D +++GC+SG+RS A++ELL GF V ++ GGY+ W N+LP
Sbjct: 120 KNPSFLRQVSSHFRKHDEIIIGCESGERSLMASTELLTAGFTGVTDIAGGYVAWTENELP 179
Query: 142 V 142
V
Sbjct: 180 V 180
>gb|AAA80302.1| senescence-associated protein [Arabidopsis thaliana]
Length = 182
Score = 105 bits (261), Expect = 3e-22
Identities = 49/111 (44%), Positives = 75/111 (67%), Gaps = 2/111 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A+++ Q G+ YLDVRT +EF GH T+ IN+PY+ G VKN +F++QVS
Sbjct: 72 SVPVRVARDVAQAGYRYLDVRTPDEFSIGH--PTRAINVPYMYRVGSGMVKNPSFLRQVS 129
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
S K D +++GC+SG+ SF A+++LL GF + ++ GGY+ W N+LPV
Sbjct: 130 SHFRKHDEIIIGCESGQMSFMASTDLLTAGFTAITDIAGGYVAWTENELPV 180
>dbj|BAD61695.1| putative Ntdin [Oryza sativa (japonica cultivar-group)]
Length = 170
Score = 102 bits (253), Expect = 3e-21
Identities = 58/132 (43%), Positives = 75/132 (55%), Gaps = 10/132 (7%)
Query: 18 LLVFVLCSSGAKVVTID--------VHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIIN 69
LL V C A+ + D V A L Q GH YLDVRT EF GH +N
Sbjct: 38 LLGSVRCGGAAEALGADMAVPRSVPVRVAHELQQAGHRYLDVRTEGEFAGGH--PVGAVN 95
Query: 70 IPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMG 129
IPY+ T G KN +F+++VS++ KED ++VGCQSGKRS A SEL + GF V ++
Sbjct: 96 IPYMYKTGSGLTKNTHFLEKVSTTFGKEDEIIVGCQSGKRSLMAASELCSAGFTAVTDIA 155
Query: 130 GGYMEWVSNKLP 141
GG+ W N+LP
Sbjct: 156 GGFSAWKENELP 167
>dbj|BAD61696.1| putative Ntdin [Oryza sativa (japonica cultivar-group)]
Length = 116
Score = 101 bits (252), Expect = 3e-21
Identities = 52/110 (47%), Positives = 69/110 (62%), Gaps = 2/110 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A L Q GH YLDVRT EF GH +NIPY+ T G KN +F+++VS
Sbjct: 6 SVPVRVAHELQQAGHRYLDVRTEGEFAGGH--PVGAVNIPYMYKTGSGLTKNTHFLEKVS 63
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
++ KED ++VGCQSGKRS A SEL + GF V ++ GG+ W N+LP
Sbjct: 64 TTFGKEDEIIVGCQSGKRSLMAASELCSAGFTAVTDIAGGFSAWKENELP 113
>pdb|1TQ1|A Chain A, Solution Structure Of At5g66040, A Putative Protein From
Arabidosis Thaliana
Length = 129
Score = 100 bits (250), Expect = 6e-21
Identities = 52/110 (47%), Positives = 69/110 (62%), Gaps = 2/110 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A +L+ GH YLDVRT EEF +GH A IN+PY+ G KN +F++QVS
Sbjct: 19 SVSVTVAHDLLLAGHRYLDVRTPEEFSQGH--ACGAINVPYMNRGASGMSKNTDFLEQVS 76
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
S + D ++VGCQSG RS AT++LL GF V ++ GGY W N LP
Sbjct: 77 SHFGQSDNIIVGCQSGGRSIKATTDLLHAGFTGVKDIVGGYSAWAKNGLP 126
>gb|AAN38701.1| At5g66040/K2A18_11 [Arabidopsis thaliana]
gi|21536991|gb|AAM61332.1| senescence-associated protein
[Arabidopsis thaliana] gi|10177119|dbj|BAB10409.1|
senescence-associated protein sen1-like protein;
ketoconazole resistance protein-like [Arabidopsis
thaliana] gi|30698182|ref|NP_851278.1|
senescence-associated family protein [Arabidopsis
thaliana] gi|15146322|gb|AAK83644.1| AT5g66040/K2A18_11
[Arabidopsis thaliana] gi|62903514|sp|Q39129|STR16_ARATH
Thiosulfate sulfurtransferase (Rhodanese)
(Senescence-associated protein) (Sulfurtransferase
protein 16) (AtStr16)
Length = 120
Score = 100 bits (248), Expect = 1e-20
Identities = 52/110 (47%), Positives = 69/110 (62%), Gaps = 2/110 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A +L+ GH YLDVRT EEF +GH A IN+PY+ G KN +F++QVS
Sbjct: 10 SVSVTVAHDLLLAGHRYLDVRTPEEFSQGH--ACGAINVPYMNRGASGMSKNPDFLEQVS 67
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
S + D ++VGCQSG RS AT++LL GF V ++ GGY W N LP
Sbjct: 68 SHFGQSDNIIVGCQSGGRSIKATTDLLHAGFTGVKDIVGGYSAWAKNGLP 117
>emb|CAA61433.1| ketoconazole resistent protein [Arabidopsis thaliana]
Length = 140
Score = 97.4 bits (241), Expect = 6e-20
Identities = 50/107 (46%), Positives = 67/107 (61%), Gaps = 1/107 (0%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A +L+ GH YLDVRT EEF +GHV I N+PY+ G KN +F++QVS
Sbjct: 10 SVSVTVAHDLLLAGHRYLDVRTPEEFSQGHVPVGSI-NVPYMNRGASGMSKNPDFLEQVS 68
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSN 138
S + D ++VGCQSG RS AT++LL GF V ++ GGY W N
Sbjct: 69 SHFGQSDNIIVGCQSGGRSIKATTDLLHAGFTGVKDIVGGYSAWAKN 115
>gb|AAF61174.1| OP1 [Cucumis sativus]
Length = 150
Score = 92.8 bits (229), Expect = 2e-18
Identities = 48/111 (43%), Positives = 67/111 (60%), Gaps = 2/111 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A L+Q G YLDVRT EE+ GH A INIPY+ G +N +F+ +V+
Sbjct: 40 SVPVRVALELLQAGQRYLDVRTPEEYSVGH--APGAINIPYMYRVGSGMTRNPHFLAEVA 97
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
K+D ++VGC SGKRS A ++LLA G+ V ++ GGY W N LP+
Sbjct: 98 IYFRKDDEIIVGCLSGKRSLMAAADLLASGYNYVTDIAGGYEAWSRNGLPM 148
>emb|CAE03897.2| OSJNBb0026I12.5 [Oryza sativa (japonica cultivar-group)]
gi|50921895|ref|XP_471308.1| OSJNBb0026I12.5 [Oryza
sativa (japonica cultivar-group)]
Length = 125
Score = 91.3 bits (225), Expect = 4e-18
Identities = 52/117 (44%), Positives = 78/117 (66%), Gaps = 8/117 (6%)
Query: 9 LPRCLAFFLLLVFVLCSSGAK-----VVTIDVHAAKNLIQTG-HIYLDVRTVEEFEKGHV 62
LP +L V +L + G++ V T+ V AA +L+ +G H YLDVRT EEF+KGHV
Sbjct: 4 LPAMFVICILAVPLLPALGSEPPSTPVPTVGVTAASHLVGSGGHSYLDVRTEEEFKKGHV 63
Query: 63 DATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLA 119
+ + +N+P+L TP+G+ KN F++QV+ DKED ++VGC SG RS A+++L+A
Sbjct: 64 ENS--LNVPFLFFTPQGKEKNTKFIEQVALHYDKEDNIIVGCLSGVRSELASADLIA 118
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 237,943,292
Number of Sequences: 2540612
Number of extensions: 8767629
Number of successful extensions: 22467
Number of sequences better than 10.0: 666
Number of HSP's better than 10.0 without gapping: 346
Number of HSP's successfully gapped in prelim test: 320
Number of HSP's that attempted gapping in prelim test: 21838
Number of HSP's gapped (non-prelim): 690
length of query: 145
length of database: 863,360,394
effective HSP length: 121
effective length of query: 24
effective length of database: 555,946,342
effective search space: 13342712208
effective search space used: 13342712208
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147013.5


