

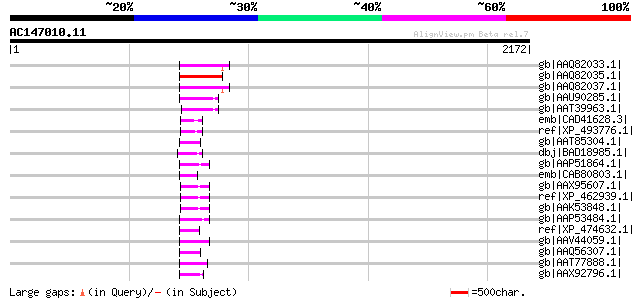
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.11 - phase: 0 /pseudo
(2172 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ82033.1| gag/pol polyprotein [Pisum sativum] 173 7e-41
gb|AAQ82035.1| gag/pol polyprotein [Pisum sativum] 168 2e-39
gb|AAQ82037.1| gag/pol polyprotein [Pisum sativum] 167 4e-39
gb|AAU90285.1| putative gag/pol polyprotein, 3'-partial [Solanum... 99 1e-18
gb|AAT39963.1| putative polyprotein [Solanum demissum] 91 3e-16
emb|CAD41628.3| OSJNBa0091D06.8 [Oryza sativa (japonica cultivar... 66 1e-08
ref|XP_493776.1| unnamed protein product [Oryza sativa (japonica... 64 8e-08
gb|AAT85304.1| reverse transcriptase (RNA-dependent DNA polymera... 57 5e-06
dbj|BAD18985.1| GAG-POL precursor [Vitis vinifera] 57 7e-06
gb|AAP51864.1| putative retroelement pol polyprotein [Oryza sati... 56 2e-05
emb|CAB80803.1| AT4g03800 [Arabidopsis thaliana] gi|3695395|gb|A... 55 3e-05
gb|AAX95607.1| RNase H, putative [Oryza sativa (japonica cultiva... 54 5e-05
ref|XP_462939.1| putative gag-pol protein [Oryza sativa (japonic... 54 5e-05
gb|AAK53848.1| Putative retroelement [Oryza sativa] 54 5e-05
gb|AAP53484.1| putative retroelement [Oryza sativa (japonica cul... 54 8e-05
ref|XP_474632.1| OSJNBa0039G19.2 [Oryza sativa (japonica cultiva... 54 8e-05
gb|AAV44059.1| hypothetical protein [Oryza sativa (japonica cult... 54 8e-05
gb|AAQ56307.1| putative gag-pol precursor [Oryza sativa (japonic... 53 1e-04
gb|AAT77888.1| putative retrotransposon gag protein [Oryza sativ... 53 1e-04
gb|AAX92796.1| retrotransposon protein, putative, unclassified [... 52 2e-04
>gb|AAQ82033.1| gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 173 bits (438), Expect = 7e-41
Identities = 101/231 (43%), Positives = 139/231 (59%), Gaps = 25/231 (10%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
V+AFDGS+++V GE+ LP+ IGPE F + F VMDI +YSCLLGRPWIH AGAV+STLHQ
Sbjct: 878 VRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQ 937
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEA-GSAEGT---AFQGLTIEG---AEPKKA 822
KLK++ NG++VTV GEE LVS LSSF +E G T AF+ + +E AE +K
Sbjct: 938 KLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVALEKVAYAEQRKP 997
Query: 823 GAAMASLKDAQKVIQDGQTAGWGKVIQLCENKRKEGLGFSPSSKVSSG-----VFHSAGF 877
GA++ S K A++V+ G+ GWGK++ L + K G+G+ P +G F SAG
Sbjct: 998 GASITSYKQAKEVVDSGKAEGWGKMVYLPVKEDKFGVGYEPLQAEQNGQAGPSTFTSAGL 1057
Query: 878 VNAISEEANGS-----------GLRPAFVTPGGIARDWDAIDIPSIMHVSE 917
+N A GS +RP PGG +W A ++ + ++E
Sbjct: 1058 MNHGDVSATGSEDCDSDCDLDNWVRPC--APGGSINNWTAEEVVQVTLLTE 1106
>gb|AAQ82035.1| gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 168 bits (425), Expect = 2e-39
Identities = 92/191 (48%), Positives = 124/191 (64%), Gaps = 12/191 (6%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
V+AFDGS+++V GE+ LP+ IGPE F + F VMDI +YSCLLGRPWIH AGAV+STLHQ
Sbjct: 878 VRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQ 937
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEA-GSAEGT---AFQGLTIEG---AEPKKA 822
KLK++ NG++VTV GEE LVS LSSF +E G T AF+ + +E AE +K
Sbjct: 938 KLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVALEKVAYAEQRKP 997
Query: 823 GAAMASLKDAQKVIQDGQTAGWGKVIQLCENKRKEGLGFSP-----SSKVSSGVFHSAGF 877
GA++ S K A++V+ G+ GWGK++ L + K G+G+ P + + F SAG
Sbjct: 998 GASITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEPLRAEQNGQAGPSTFTSAGL 1057
Query: 878 VNAISEEANGS 888
+N A GS
Sbjct: 1058 MNHGDVSATGS 1068
>gb|AAQ82037.1| gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 167 bits (423), Expect = 4e-39
Identities = 98/231 (42%), Positives = 136/231 (58%), Gaps = 25/231 (10%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
V+AFD S+++V GE+ LP+ IGPE F + F VMDI +YSCLLGRPWIH AGAV+STLHQ
Sbjct: 876 VRAFDRSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQ 935
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEA-GSAEGT---AFQGLTIEG---AEPKKA 822
KLK++ NG++VTV GEE LVS LSSF +E G T F+ + +E AE +K
Sbjct: 936 KLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQVFETVALEKVAYAEQRKP 995
Query: 823 GAAMASLKDAQKVIQDGQTAGWGKVIQLCENKRKEGLGFSPSSKVSSG-----VFHSAGF 877
G ++ S K A++V+ G+ GWGK++ L + K G+G+ P +G F SAG
Sbjct: 996 GVSITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEPLQAEQNGQAGPSTFTSAGL 1055
Query: 878 VNAISEEANGS-----------GLRPAFVTPGGIARDWDAIDIPSIMHVSE 917
+N A GS +RP PGG +W A ++ + ++E
Sbjct: 1056 MNHGDVSATGSEDCDSDCDLDNWVRPC--APGGSINNWTAEEVVQVTLLTE 1104
>gb|AAU90285.1| putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 99.4 bits (246), Expect = 1e-18
Identities = 66/171 (38%), Positives = 94/171 (54%), Gaps = 14/171 (8%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
V+AFDG++ + +GEI+L + IG +F V FQV++INASY+ LLGRPW+H AGAV STLHQ
Sbjct: 1265 VRAFDGTKTDAIGEIELILKIGHVDFTVNFQVLNINASYNLLLGRPWVHRAGAVPSTLHQ 1324
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSA-EGTAFQGLTI-------EGAEPKK 821
+KF + + V VH E V + SS I+A + E +Q + EG K
Sbjct: 1325 MVKFEYDRQEVIVHSEGDLSVYKDSSLPFIKANNENEALVYQAFEVVVVEHILEGNLISK 1384
Query: 822 AGAAMASLKDAQKVIQDGQTAGWGKVIQLCENKRKEGLGFSPSSKVSSGVF 872
MAS+ ++++ G G G I L +G + S + S G F
Sbjct: 1385 PQLPMASVMMVNEMLKHGFEPGKGLGIFL------QGRAYPVSPRKSLGTF 1429
>gb|AAT39963.1| putative polyprotein [Solanum demissum]
Length = 1483
Score = 91.3 bits (225), Expect = 3e-16
Identities = 62/163 (38%), Positives = 88/163 (53%), Gaps = 14/163 (8%)
Query: 718 KNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFIRNG 777
++ +GEI+L + IGP +F V FQV++INASY+ LLGRPW+H AGAV STLHQ +KF +
Sbjct: 213 EDAIGEIELILKIGPVDFTVNFQVLNINASYNLLLGRPWVHRAGAVPSTLHQMVKFEYDR 272
Query: 778 KLVTVHGEEAYLVSQLSSFSCIEAGSA-EGTAFQGL-------TIEGAEPKKAGAAMASL 829
+ V VH E V + SS I+A + E +Q +EG K MAS+
Sbjct: 273 QEVIVHCERDLSVYKDSSLPFIKANNENEALVYQAFEVVVVEHNLEGNLISKPQLPMASV 332
Query: 830 KDAQKVIQDGQTAGWGKVIQLCENKRKEGLGFSPSSKVSSGVF 872
++++ G G G I L +G + S + S G F
Sbjct: 333 MMVNEMLKHGFKLGKGLGIFL------QGRAYPVSPRKSLGTF 369
>emb|CAD41628.3| OSJNBa0091D06.8 [Oryza sativa (japonica cultivar-group)]
gi|50926818|ref|XP_473330.1| OSJNBa0091D06.8 [Oryza
sativa (japonica cultivar-group)]
Length = 2657
Score = 66.2 bits (160), Expect = 1e-08
Identities = 40/93 (43%), Positives = 54/93 (58%), Gaps = 6/93 (6%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
FD K+ LG I + I + +F V F V++ N SYS LLGRPWIH V STLHQ LK
Sbjct: 1514 FDNQGKSTLGAITVKIQMSTFSFKVHFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLK 1573
Query: 773 FIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAE 805
F+ +GK G + +V SS++ E+ A+
Sbjct: 1574 FL-DGK-----GAQQRIVGNSSSYTIQESYHAD 1600
>ref|XP_493776.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|9927274|dbj|BAB08213.2| Similar to Arabidopsis
thaliana chromosome II BAC F26H6; putative retroelement
pol polyprotein (AC006920) [Oryza sativa (japonica
cultivar-group)]
Length = 2876
Score = 63.5 bits (153), Expect = 8e-08
Identities = 37/93 (39%), Positives = 50/93 (52%), Gaps = 6/93 (6%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
FD K LG I + I + +F V F V++ N SYS LLGRPWIH V STLHQ LK
Sbjct: 1432 FDNQGKPTLGAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLK 1491
Query: 773 FIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAE 805
F+ +G + + S S ++ E+ A+
Sbjct: 1492 FLDG------NGVQQRITSNFSPYTIQESYHAD 1518
>gb|AAT85304.1| reverse transcriptase (RNA-dependent DNA polymerase) domain
containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 1372
Score = 57.4 bits (137), Expect = 5e-06
Identities = 31/87 (35%), Positives = 46/87 (52%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 484 LKDFGGNPSETKGVLNVELTVGNKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 543
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSF 796
L + K+ V + A L+ L F
Sbjct: 544 CLIQWQGDKIQIVPADRAKLIELLKEF 570
>dbj|BAD18985.1| GAG-POL precursor [Vitis vinifera]
Length = 549
Score = 57.0 bits (136), Expect = 7e-06
Identities = 33/103 (32%), Positives = 53/103 (51%), Gaps = 9/103 (8%)
Query: 704 GEVLSWVKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAV 763
G +LS F+GS LG+I LP+ GP V F V+ + ++ +LGR W+H A+
Sbjct: 439 GRILS---GFNGSSTTSLGDIILPVQAGPVTLNVQFSVVQELSPFNIILGRTWLHYMKAI 495
Query: 764 TSTLHQKLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEG 806
ST HQ + F+ + + ++G SQL++ C + G
Sbjct: 496 PSTYHQMVIFLTHDGQIDLYG------SQLAARQCYQIAREAG 532
>gb|AAP51864.1| putative retroelement pol polyprotein [Oryza sativa (japonica
cultivar-group)] gi|37530550|ref|NP_919577.1| putative
retroelement pol polyprotein [Oryza sativa (japonica
cultivar-group)] gi|18581535|gb|AAK52540.2| Putative
retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 55.8 bits (133), Expect = 2e-05
Identities = 35/126 (27%), Positives = 60/126 (46%), Gaps = 3/126 (2%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ +G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 411 LKDFGGNPSETMGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTIHQ 470
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGAEPKKAGAAMASL 829
L + K+ V + + S C E + T++ + K+ G + S
Sbjct: 471 CLIQWQGDKIEIVPADSQLKMENPS--YCFEGVMEGSNVYTKDTVDDLDDKQ-GQGLMSA 527
Query: 830 KDAQKV 835
D +++
Sbjct: 528 DDLEEI 533
>emb|CAB80803.1| AT4g03800 [Arabidopsis thaliana] gi|3695395|gb|AAC62796.1| contains
similarity to reverse transcriptase (Pfam: PF00078 rvt,
E=4.3e-08) [Arabidopsis thaliana] gi|7487690|pir||T01961
hypothetical protein T5H22.5 - Arabidopsis thaliana
Length = 637
Score = 55.1 bits (131), Expect = 3e-05
Identities = 30/74 (40%), Positives = 40/74 (53%)
Query: 712 AFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKL 771
AF LG I LP+ G + +V F V D A+Y+ +LG PWI AV ST HQ L
Sbjct: 238 AFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWL 297
Query: 772 KFIRNGKLVTVHGE 785
KF + + T+ G+
Sbjct: 298 KFPTSNGVETIWGD 311
>gb|AAX95607.1| RNase H, putative [Oryza sativa (japonica cultivar-group)]
Length = 2282
Score = 54.3 bits (129), Expect = 5e-05
Identities = 37/124 (29%), Positives = 62/124 (49%), Gaps = 5/124 (4%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
F G+ G ++ +T+G + TF V+D N SYS LLGR WIH + ST+HQ L
Sbjct: 1220 FGGNLSETKGVSNVELTVGNKTIPTTFFVIDGNGSYSLLLGRDWIHANCCIPSTMHQCLI 1279
Query: 773 FIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGAEPKKAGAAMASLKD 831
++ K+ V + ++ + SC G EG+ + T++ + K+ G S D
Sbjct: 1280 QWQDDKIEIVPADSQL---KMENPSCYFEGVVEGSNVYIKDTVDDLDDKQ-GQGFISADD 1335
Query: 832 AQKV 835
+++
Sbjct: 1336 LEEI 1339
>ref|XP_462939.1| putative gag-pol protein [Oryza sativa (japonica cultivar-group)]
Length = 2273
Score = 54.3 bits (129), Expect = 5e-05
Identities = 37/124 (29%), Positives = 62/124 (49%), Gaps = 5/124 (4%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
F G+ G ++ +T+G + TF V+D N SYS LLGR WIH + ST+HQ L
Sbjct: 1220 FGGNLSETKGVSNVELTVGNKTIPTTFFVIDGNGSYSLLLGRDWIHANCCIPSTMHQCLI 1279
Query: 773 FIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGAEPKKAGAAMASLKD 831
++ K+ V + ++ + SC G EG+ + T++ + K+ G S D
Sbjct: 1280 QWQDDKIEIVPADSQL---KMENPSCYFEGVVEGSNVYIKDTVDDLDDKQ-GQGFISADD 1335
Query: 832 AQKV 835
+++
Sbjct: 1336 LEEI 1339
>gb|AAK53848.1| Putative retroelement [Oryza sativa]
Length = 2014
Score = 54.3 bits (129), Expect = 5e-05
Identities = 37/124 (29%), Positives = 62/124 (49%), Gaps = 5/124 (4%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
F G+ G ++ +T+G + TF V+D N SYS LLGR WIH + ST+HQ L
Sbjct: 967 FGGNLSETKGVSNVELTVGNKTIPTTFFVIDGNGSYSLLLGRDWIHANCCIPSTMHQCLI 1026
Query: 773 FIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGAEPKKAGAAMASLKD 831
++ K+ V + ++ + SC G EG+ + T++ + K+ G S D
Sbjct: 1027 QWQDDKIEIVPADSQL---KMENPSCYFEGVVEGSNVYIKDTVDDLDDKQ-GQGFISADD 1082
Query: 832 AQKV 835
+++
Sbjct: 1083 LEEI 1086
>gb|AAP53484.1| putative retroelement [Oryza sativa (japonica cultivar-group)]
gi|37533790|ref|NP_921197.1| putative retroelement
[Oryza sativa (japonica cultivar-group)]
gi|21672097|gb|AAM74459.1| Putative retroelement [Oryza
sativa (japonica cultivar-group)]
Length = 858
Score = 53.5 bits (127), Expect = 8e-05
Identities = 37/134 (27%), Positives = 63/134 (46%), Gaps = 11/134 (8%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 413 LKDFGGNPSETKGVLNVELTVGSKTVPTTFFVIDRKGSYSLLLGRDWIHANCCIPSTMHQ 472
Query: 770 --------KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGAEPKK 821
K+K + + V V + L ++ C+ +G F T++ + K+
Sbjct: 473 CLIQWQGDKIKIVPADRSVNVASADLAL-WEMDGIDCLSGKVWDGD-FLKDTVDDLDDKQ 530
Query: 822 AGAAMASLKDAQKV 835
G S D +++
Sbjct: 531 -GQGFMSANDLEEI 543
>ref|XP_474632.1| OSJNBa0039G19.2 [Oryza sativa (japonica cultivar-group)]
gi|38347142|emb|CAD39495.2| OSJNBa0039G19.2 [Oryza
sativa (japonica cultivar-group)]
Length = 1136
Score = 53.5 bits (127), Expect = 8e-05
Identities = 29/82 (35%), Positives = 45/82 (54%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G ++L +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 424 LKDFGGNPSETKGVLNLELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 483
Query: 770 KLKFIRNGKLVTVHGEEAYLVS 791
L + K+ V +++ V+
Sbjct: 484 CLIQWQGNKIEIVLADKSVNVA 505
>gb|AAV44059.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|55168117|gb|AAV43984.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1681
Score = 53.5 bits (127), Expect = 8e-05
Identities = 37/127 (29%), Positives = 63/127 (49%), Gaps = 5/127 (3%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 803 LKHFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKRSYSLLLGRDWIHTNCCIPSTMHQ 862
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGAEPKKAGAAMAS 828
L + K+ V + S++ + S G EG+ + T++ + K+ G S
Sbjct: 863 CLIQWQGHKIEVVPADSQ---SKMENPSYYFEGVVEGSDVYAKDTVDDLDDKQ-GQGFMS 918
Query: 829 LKDAQKV 835
D +++
Sbjct: 919 ADDLEEI 925
>gb|AAQ56307.1| putative gag-pol precursor [Oryza sativa (japonica cultivar-group)]
Length = 1672
Score = 53.1 bits (126), Expect = 1e-04
Identities = 31/97 (31%), Positives = 49/97 (49%), Gaps = 7/97 (7%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 703 LKDFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 762
Query: 770 KLKFIRNGKLVTVHGEEAYLVS-------QLSSFSCI 799
L + KL V + + V+ ++ S C+
Sbjct: 763 CLIQWQGDKLEIVPADRSVNVASANLALWEMDSIDCL 799
>gb|AAT77888.1| putative retrotransposon gag protein [Oryza sativa (japonica
cultivar-group)]
Length = 1251
Score = 52.8 bits (125), Expect = 1e-04
Identities = 35/120 (29%), Positives = 60/120 (49%), Gaps = 4/120 (3%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 748 LKDFGGNPSETRGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 807
Query: 770 KLKFIRNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT-AFQGLTIEGAEPKKAGAAMAS 828
L + K+ V + S++ + S G EG+ + T++ + K+ M++
Sbjct: 808 CLIQWQGDKIEIVPADSQ---SKMENPSYYFEGVVEGSNVYAKDTVDDLDDKQGQGFMSA 864
>gb|AAX92796.1| retrotransposon protein, putative, unclassified [Oryza sativa
(japonica cultivar-group)]
Length = 1163
Score = 52.4 bits (124), Expect = 2e-04
Identities = 33/106 (31%), Positives = 53/106 (49%), Gaps = 8/106 (7%)
Query: 710 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 769
+K F G+ G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 913 LKDFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHTNCCIPSTMHQ 972
Query: 770 KLKFIRNGKLVTVHGEEAYLVS-------QLSSFSCIEAGSAEGTA 808
L + K+ V + + V+ ++ C+ +G A G A
Sbjct: 973 CLIQWQGDKIEIVPVDMSVNVASADLGLWEMDGLDCL-SGKANGQA 1017
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.354 0.154 0.541
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,951,709,574
Number of Sequences: 2540612
Number of extensions: 104300287
Number of successful extensions: 543200
Number of sequences better than 10.0: 300
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 542832
Number of HSP's gapped (non-prelim): 415
length of query: 2172
length of database: 863,360,394
effective HSP length: 144
effective length of query: 2028
effective length of database: 497,512,266
effective search space: 1008954875448
effective search space used: 1008954875448
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 83 (36.6 bits)
Medicago: description of AC147010.11


