

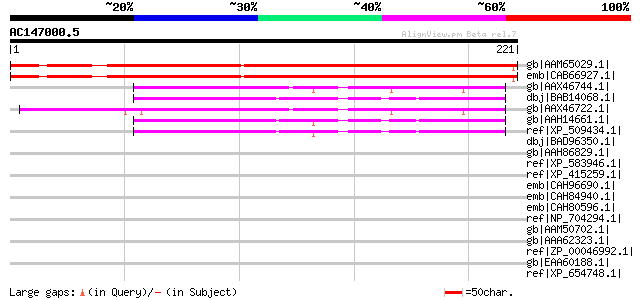
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.5 - phase: 0
(221 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM65029.1| unknown [Arabidopsis thaliana] 194 2e-48
emb|CAB66927.1| putative protein [Arabidopsis thaliana] gi|15229... 191 2e-47
gb|AAX46744.1| hypothetical protein FLJ12448 [Bos taurus] 48 2e-04
dbj|BAB14068.1| unnamed protein product [Homo sapiens] gi|125976... 47 4e-04
gb|AAX46722.1| hypothetical protein FLJ12448 [Bos taurus] 47 4e-04
gb|AAH14661.1| Hypothetical protein FLJ12448 [Homo sapiens] 46 9e-04
ref|XP_509434.1| PREDICTED: similar to Hypothetical protein FLJ1... 46 9e-04
dbj|BAD96350.1| hypothetical protein FLJ12448 variant [Homo sapi... 45 0.002
gb|AAH86829.1| Zgc:103536 [Danio rerio] gi|56693275|ref|NP_00100... 44 0.003
ref|XP_583946.1| PREDICTED: similar to hypothetical protein FLJ1... 44 0.003
ref|XP_415259.1| PREDICTED: similar to Hypothetical protein FLJ1... 44 0.003
emb|CAH96690.1| reticulocyte-binding protein, putative [Plasmodi... 40 0.062
emb|CAH84940.1| hypothetical protein PC301330.00.0 [Plasmodium c... 39 0.081
emb|CAH80596.1| conserved hypothetical protein [Plasmodium chaba... 39 0.081
ref|NP_704294.1| ubiquitin-protein ligase 1, putative [Plasmodiu... 38 0.18
gb|AAM50702.1| GM13065p [Drosophila melanogaster] 38 0.18
gb|AAA62323.1| histone H1-I gi|729654|sp|P40266|H11_GLYSA HISTON... 38 0.18
ref|ZP_00046992.1| COG0679: Predicted permeases [Lactobacillus g... 38 0.24
gb|EAA60188.1| hypothetical protein AN5093.2 [Aspergillus nidula... 37 0.40
ref|XP_654748.1| SMC4 protein, putative [Entamoeba histolytica H... 37 0.40
>gb|AAM65029.1| unknown [Arabidopsis thaliana]
Length = 220
Score = 194 bits (493), Expect = 2e-48
Identities = 102/222 (45%), Positives = 154/222 (68%), Gaps = 11/222 (4%)
Query: 1 MAEEELNNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQ 60
MA+ EL+ G SS ED D W+AAI+S+A T+ Y G SAT T+ +N +
Sbjct: 1 MAKRELSGGDSSS---EDEDPKWRAAINSIATTTVY------GASATKPAATQSHNNGDF 51
Query: 61 NPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFD 120
K K+ H Q+K LL++++E T++ V++P+ + ++ P +DCG+RLF+ GIVFD
Sbjct: 52 RLKPKKLTHGQIKVKNLLNEMVEKTLDFVEDPVNIPEDKPE-NDCGVRLFKRCATGIVFD 110
Query: 121 HADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKD 180
H DE + P K+P L P + ++ SK+F++R++SIAVDG+D++ AA +A KK+ ARL+AK+
Sbjct: 111 HVDEIRGPKKKPNLRPDKGVEGSSKEFKKRVKSIAVDGSDILTAAMEAAKKASARLDAKE 170
Query: 181 AAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK-IKH 221
AAK KAK+EEERI +LKK+RGE+WLPS+ + M+ ++K IKH
Sbjct: 171 VAAKDKAKKEEERIAELKKVRGEKWLPSIERAMKKEMKRIKH 212
>emb|CAB66927.1| putative protein [Arabidopsis thaliana]
gi|15229629|ref|NP_190558.1| expressed protein
[Arabidopsis thaliana] gi|11285662|pir||T46055
hypothetical protein T16K5.240 - Arabidopsis thaliana
Length = 220
Score = 191 bits (484), Expect = 2e-47
Identities = 101/222 (45%), Positives = 153/222 (68%), Gaps = 11/222 (4%)
Query: 1 MAEEELNNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQ 60
MA+ EL+ G SS ED D W+AAI+S+A T+ Y G SAT T+ + +
Sbjct: 1 MAKRELSGGDSSS---EDEDPKWRAAINSIATTTVY------GASATKPAATQSHNYGDF 51
Query: 61 NPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFD 120
K K+ H Q+K LL++++E T++ V++P+ + ++ P +DCG+RLF+ GIVFD
Sbjct: 52 RLKPKKLTHGQIKVKNLLNEMVEKTLDFVEDPVNIPEDKPE-NDCGVRLFKRCATGIVFD 110
Query: 121 HADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKD 180
H DE + P K+P L P + ++ SK+F++R++SIAVDG+D++ AA +A KK+ ARL+AK+
Sbjct: 111 HVDEIRGPKKKPNLRPDKGVEGSSKEFKKRVKSIAVDGSDILTAAVEAAKKASARLDAKE 170
Query: 181 AAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK-IKH 221
AAK KAK+EEERI +LKK+RGE+WLPS+ + M+ ++K IKH
Sbjct: 171 VAAKDKAKKEEERIAELKKVRGEKWLPSIERAMKKEMKRIKH 212
>gb|AAX46744.1| hypothetical protein FLJ12448 [Bos taurus]
Length = 257
Score = 48.1 bits (113), Expect = 2e-04
Identities = 51/176 (28%), Positives = 77/176 (42%), Gaps = 19/176 (10%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D EIVKEP + ++D G RLF S
Sbjct: 64 EQDGNELQTTPEFRAHVAKKLGALLDSSITISEIVKEPRKSEVQQGALEDDGFRLFFTSI 123
Query: 115 PGIVFDHADEPQPPMKR-PKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAA-------- 165
PG A PQP KR P +D DE+ RR R AV +D++ +
Sbjct: 124 PGGPEKEA-APQPCRKRLPSSSSSDDGDEE----LRRCREAAVSASDILQESAIHGHVSV 178
Query: 166 ----NDAYKKSLARLEAKDAAAKAKAKREEERIEK-LKKIRGERWLPSMAKEMQAK 216
KK + ++ D AA A +K E R EK ++ G++ P K+ + K
Sbjct: 179 EKKKKRKLKKKAKKEDSADVAATATSKAEVGRQEKESAQLNGDQAPPGTKKKKRKK 234
>dbj|BAB14068.1| unnamed protein product [Homo sapiens] gi|12597631|ref|NP_075046.1|
hypothetical protein LOC64897 [Homo sapiens]
Length = 262
Score = 47.0 bits (110), Expect = 4e-04
Identities = 44/162 (27%), Positives = 71/162 (43%), Gaps = 9/162 (5%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D E KEP + ++D G RLF S
Sbjct: 70 EQDGNELQTTPEFRAHVAKKLGALLDSFITISEAAKEPAKAKVQKVALEDDGFRLFFTSV 129
Query: 115 PGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLA 174
PG + + PQP KR ED DE+ RR R AV +D++ ++ S
Sbjct: 130 PG-GREKEESPQPRRKRQPSSSSEDSDEE----WRRCREAAVSASDIL---QESAIHSPG 181
Query: 175 RLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAK 216
+E K+A K K K++ +++ + SMA + K
Sbjct: 182 TVE-KEAKKKRKLKKKAKKVASVDSAVAATTPTSMATVQKQK 222
>gb|AAX46722.1| hypothetical protein FLJ12448 [Bos taurus]
Length = 256
Score = 47.0 bits (110), Expect = 4e-04
Identities = 62/236 (26%), Positives = 96/236 (40%), Gaps = 29/236 (12%)
Query: 5 ELNNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNN----NDTKKKH---- 56
+L + SSS +E+ + +AA+ + G ATN N +H
Sbjct: 3 DLESSSSSSSDEEELERCREAALPAWGLEQRPRGPEKPGVDATNAKLPANQPSLRHKVDE 62
Query: 57 ---NDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHS 113
+ N+ +P+ + + K L L D EIVKEP + ++D G RLF S
Sbjct: 63 HEQDGNELQTTPEFRAHVAKKLGALLDSSITISEIVKEPRKSEVQQGALEDDGFRLFFTS 122
Query: 114 KPGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAA-------- 165
PG A PQP KR +D DE+ RR R AV +D++ +
Sbjct: 123 IPGGPEKEA-APQPCRKRLPSSSSDDGDEE----LRRCREAAVSASDILQESAIHGHVSV 177
Query: 166 ----NDAYKKSLARLEAKDAAAKAKAKREEERIEK-LKKIRGERWLPSMAKEMQAK 216
KK + ++ D AA A +K R EK ++ G++ P K+ + K
Sbjct: 178 EKKKKRKLKKKAKKEDSADVAAAATSKAAVGRQEKESAQLNGDQAPPGTKKKKRKK 233
>gb|AAH14661.1| Hypothetical protein FLJ12448 [Homo sapiens]
Length = 263
Score = 45.8 bits (107), Expect = 9e-04
Identities = 45/163 (27%), Positives = 72/163 (43%), Gaps = 10/163 (6%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D E KEP + ++D G RLF S
Sbjct: 70 EQDGNELQTTPEFRAHVAKKLGALLDSFITISEAAKEPAKAKVQKVALEDDGFRLFFTSV 129
Query: 115 PGIVFDHADEPQPPMKR-PKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSL 173
PG + + PQP KR P ED DE+ RR R AV +D++ ++ S
Sbjct: 130 PG-GREKEESPQPRRKRQPSSSSSEDSDEE----WRRCREAAVSASDIL---QESAIHSP 181
Query: 174 ARLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAK 216
+E K+A K K K++ +++ + SMA + K
Sbjct: 182 GTVE-KEAKKKRKLKKKAKKVASVDSAVAATTPTSMATVQKQK 223
>ref|XP_509434.1| PREDICTED: similar to Hypothetical protein FLJ12448 [Pan
troglodytes]
Length = 430
Score = 45.8 bits (107), Expect = 9e-04
Identities = 45/163 (27%), Positives = 72/163 (43%), Gaps = 10/163 (6%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D E KEP + ++D G RLF S
Sbjct: 70 EQDGNELQTTPEFRAHVAKKLGALLDSFITISEAAKEPAKAKVQKVALEDDGFRLFFTSV 129
Query: 115 PGIVFDHADEPQPPMKR-PKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSL 173
PG + + PQP KR P ED DE+ RR R AV +D++ ++ S
Sbjct: 130 PG-GREKEESPQPRRKRQPSSSSSEDSDEE----WRRCREAAVSASDIL---QESAIHSP 181
Query: 174 ARLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAK 216
+E K+A K K K++ +++ + SMA + K
Sbjct: 182 GTVE-KEAKKKRKLKKKAKKVASVDSAVAATTPTSMATVQKQK 223
>dbj|BAD96350.1| hypothetical protein FLJ12448 variant [Homo sapiens]
Length = 262
Score = 44.7 bits (104), Expect = 0.002
Identities = 43/162 (26%), Positives = 70/162 (42%), Gaps = 9/162 (5%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D E K P + ++D G RLF S
Sbjct: 70 EQDGNELQTTPEFRAHVAKKLGALLDSFITISEAAKAPAKAKVQKVALEDDGFRLFFTSV 129
Query: 115 PGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLA 174
PG + + PQP KR ED DE+ RR R AV +D++ ++ S
Sbjct: 130 PG-GREKEESPQPRRKRQPSSSSEDSDEE----WRRCREAAVSASDIL---QESAIHSPG 181
Query: 175 RLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAK 216
+E K+A K K K++ +++ + SMA + K
Sbjct: 182 TVE-KEAKKKRKLKKKAKKVASVDSAVAATTPTSMATVQKQK 222
>gb|AAH86829.1| Zgc:103536 [Danio rerio] gi|56693275|ref|NP_001008602.1|
hypothetical protein LOC494059 [Danio rerio]
Length = 234
Score = 44.3 bits (103), Expect = 0.003
Identities = 48/223 (21%), Positives = 92/223 (40%), Gaps = 23/223 (10%)
Query: 13 SSGDEDGDAAWKAAIDSVAGTSSYVTSF-MNGFSATNNNDTKKKHNDNQNPKSPKIKHYQ 71
SS +++ A K A+ S + NG + + +K +H+ N+ +P+ + +
Sbjct: 4 SSSEDENTARLKEAVWSFKPEDVKINGKEKNGRQSHRADVSKHEHDGNELGTTPEFRSHV 63
Query: 72 LKAL-KLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMK 130
K L LD + E+ + + + D+ RLF S PG + + P PP +
Sbjct: 64 AKKLGAYLDGCIS---EVCSDTVEPAQSENREDEEDFRLFSSSTPGKWMEQS-LPPPPKR 119
Query: 131 RPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAA----NDAYKKSLARLEAKDAAAKAK 186
RP VP + + RIR AV +D++ A ++ ++ + E +D K K
Sbjct: 120 RP--VPSS--SDSDSEMEMRIREAAVSLSDILGPAAQNLSEKTEEKSTKEETEDTVTKKK 175
Query: 187 AKRE---------EERIEKLKKIRGERWLPSMAKEMQAKIKIK 220
++ + EK + G + + + ++ K K K
Sbjct: 176 KRKTSSEESQDKVNHQTEKQSNVEGNQEQTTAGERLKKKKKKK 218
>ref|XP_583946.1| PREDICTED: similar to hypothetical protein FLJ12448 [Bos taurus]
Length = 251
Score = 43.9 bits (102), Expect = 0.003
Identities = 42/137 (30%), Positives = 62/137 (44%), Gaps = 7/137 (5%)
Query: 55 KHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSK 114
+ + N+ +P+ + + K L L D EIVKEP + ++D G RLF S
Sbjct: 22 EQDGNELQTTPEFRAHVAKKLGALLDSSITISEIVKEPRKSEVQQGALEDDGFRLFFTSI 81
Query: 115 PGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLA 174
PG A PQP KR +D DE+ RR R AV +D++ A ++
Sbjct: 82 PGGPEKEA-APQPCRKRLPSSSSDDGDEE----LRRCREAAVSASDIL--QESAIHGHVS 134
Query: 175 RLEAKDAAAKAKAKREE 191
+ K K KAK+E+
Sbjct: 135 VEKKKKRKLKKKAKKED 151
>ref|XP_415259.1| PREDICTED: similar to Hypothetical protein FLJ12448 [Gallus gallus]
Length = 302
Score = 43.9 bits (102), Expect = 0.003
Identities = 43/191 (22%), Positives = 74/191 (38%), Gaps = 18/191 (9%)
Query: 20 DAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHND-----NQNPKSPKIKHYQLKA 74
+AAW A + T + F +++ ND N+ +P+ + + K
Sbjct: 49 EAAWDCAKQAAVRTEARGGGFTKDQRQAARPSLRREVNDHDEDGNELQTTPEFRAHVAKK 108
Query: 75 LKLLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMKRPKL 134
L + D ++ P P + + D G RLF S PG P P +RP
Sbjct: 109 LGAMLDSFITVLKDSSGPSPACVQQSDCGDDGFRLFSSSVPGDCGQSEQSPAPRRRRPAS 168
Query: 135 VPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAAKAKAKREEER- 193
D D++ +K++ + +A D K+S ++D++ E R
Sbjct: 169 SSDSDSDQEWQKYQ-----------EAAVSAADILKQSAFPELSQDSSQNMCQGYVEHRQ 217
Query: 194 -IEKLKKIRGE 203
+K KKI+GE
Sbjct: 218 KKKKKKKIKGE 228
>emb|CAH96690.1| reticulocyte-binding protein, putative [Plasmodium berghei]
Length = 1757
Score = 39.7 bits (91), Expect = 0.062
Identities = 50/235 (21%), Positives = 95/235 (40%), Gaps = 52/235 (22%)
Query: 27 IDSVAGTSSYVTSFMNGFSATNNNDTKK---------------------------KHNDN 59
I+S+ T+S S+ + F T N TKK K N
Sbjct: 3 IESIKNTTSIGKSYTDKFLNTLTNKTKKLENIFKDASLNEHEQTNNELKKYFNNLKENLG 62
Query: 60 QNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDE----------------DPNVD 103
++PKS + + K KL +DI++ ++I K + E + N+
Sbjct: 63 KDPKSTLSQQFNEKE-KLFNDIIQKNVDINKNISNIEIEIYASIHNISDETENEIEENIK 121
Query: 104 DCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIA 163
++F K + + +E + +K+ E EK+KK+ I I D I
Sbjct: 122 SLNTQIFEKVKTNVA--NLNEIKEKLKQYNFDDFEK--EKNKKYNDEINKIKGD----IT 173
Query: 164 AANDAYKKSLARLEAKDAAAKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK 218
++ K++A+L + ++++ A + +I+K +K+ + KE++ KIK
Sbjct: 174 TLDNKIDKNIAKLTEIEKSSESYASEIKIQIDKSEKVTDKTIYKEEPKEIEKKIK 228
>emb|CAH84940.1| hypothetical protein PC301330.00.0 [Plasmodium chabaudi]
Length = 614
Score = 39.3 bits (90), Expect = 0.081
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 8/76 (10%)
Query: 48 NNNDTKKKHNDNQNPKSPKIKHYQLKALKLLDDILE---NTIEIVKEPIPVLDEDPN--- 101
NN +T K+H DN+ KS + + LK K +DI + N I+ + + + +L PN
Sbjct: 504 NNPETMKQHEDNEQNKSNNVMYKHLKPPKNSNDISQLHFNEIKNMLKDLTILQSGPNWKT 563
Query: 102 --VDDCGIRLFRHSKP 115
+DD LF++ +P
Sbjct: 564 YKMDDLPKELFKNIRP 579
>emb|CAH80596.1| conserved hypothetical protein [Plasmodium chabaudi]
Length = 836
Score = 39.3 bits (90), Expect = 0.081
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 8/76 (10%)
Query: 48 NNNDTKKKHNDNQNPKSPKIKHYQLKALKLLDDILE---NTIEIVKEPIPVLDEDPN--- 101
NN +T K+H DN+ KS + + LK K +DI + N I+ + + + +L PN
Sbjct: 504 NNPETMKQHEDNEQNKSNNVMYKHLKPPKNSNDISQLHFNEIKNMLKDLTILQSGPNWKT 563
Query: 102 --VDDCGIRLFRHSKP 115
+DD LF++ +P
Sbjct: 564 YKMDDLPKELFKNIRP 579
>ref|NP_704294.1| ubiquitin-protein ligase 1, putative [Plasmodium falciparum 3D7]
gi|23499033|emb|CAD51113.1| ubiquitin-protein ligase 1,
putative [Plasmodium falciparum 3D7]
Length = 8591
Score = 38.1 bits (87), Expect = 0.18
Identities = 18/63 (28%), Positives = 30/63 (47%)
Query: 7 NNGYCSSSGDEDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQNPKSPK 66
NNG+ + GD D D+ + +S + + Y + N + NNND +NDN N
Sbjct: 6250 NNGFNPNEGDNDSDSDSNSDSNSDSDSDDYNDNSDNNNNNDNNNDNNDNNNDNNNENQNN 6309
Query: 67 IKH 69
+ +
Sbjct: 6310 VNN 6312
>gb|AAM50702.1| GM13065p [Drosophila melanogaster]
Length = 435
Score = 38.1 bits (87), Expect = 0.18
Identities = 28/97 (28%), Positives = 55/97 (55%), Gaps = 6/97 (6%)
Query: 124 EPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAA 183
E +P K P +V + +KSK ++ +++ V +++ AA ++ KK L AKD A
Sbjct: 13 ELKPIKKEPGVVEDGKVTKKSKTKPKK-KTVKVPKDEVKAAVQESPKK----LSAKDLA- 66
Query: 184 KAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIKIK 220
K K K+ +++ +KLKK + + + E +A++K++
Sbjct: 67 KIKKKKAKKQAQKLKKQQNKLAPKEIKTEPEAQVKVE 103
>gb|AAA62323.1| histone H1-I gi|729654|sp|P40266|H11_GLYSA HISTONE H1-I
Length = 233
Score = 38.1 bits (87), Expect = 0.18
Identities = 30/96 (31%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 123 DEPQPPMKRPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAA 182
D+P+ K+ P +I++K KK + AV+ AA A KK++A+ AK AA
Sbjct: 131 DKPEAAPKKKAPRPKREIEKKEKKVVAKKPKPAVEKK----AAKPAAKKAVAKPAAKKAA 186
Query: 183 AKAKAKREEERIEKLKKIRGERWLPSMAKEMQAKIK 218
AK AK + K + P+ K AK K
Sbjct: 187 AKPAAKEPAAKASPKKAAAKPKAKPTPKKSTAAKPK 222
>ref|ZP_00046992.1| COG0679: Predicted permeases [Lactobacillus gasseri]
Length = 393
Score = 37.7 bits (86), Expect = 0.24
Identities = 25/79 (31%), Positives = 38/79 (47%), Gaps = 2/79 (2%)
Query: 52 TKKKHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPN-VDDCGIRLF 110
TKKK N N N K K++ +K K L+ + + + + +PV D + + + GI
Sbjct: 157 TKKKANANPNNKGTKVEVEDIKG-KGLNKTKKKVVVTIPDGVPVTDAEAKALKEKGIERE 215
Query: 111 RHSKPGIVFDHADEPQPPM 129
+ DHADEP PM
Sbjct: 216 IDLARAEIADHADEPHQPM 234
>gb|EAA60188.1| hypothetical protein AN5093.2 [Aspergillus nidulans FGSC A4]
gi|67537846|ref|XP_662697.1| hypothetical protein
AN5093_2 [Aspergillus nidulans FGSC A4]
gi|49095538|ref|XP_409230.1| hypothetical protein
AN5093.2 [Aspergillus nidulans FGSC A4]
Length = 384
Score = 37.0 bits (84), Expect = 0.40
Identities = 39/155 (25%), Positives = 64/155 (41%), Gaps = 19/155 (12%)
Query: 48 NNNDTKKKHNDNQNPKSPKIKHYQLKALKLLDDILENTIEIVKEPIPVLDEDPNVDDCGI 107
NNN+ +N+N N + + Q D N+ + ++PI +D +VDD I
Sbjct: 86 NNNNNSHNNNNNNNNTNTNSNNSQDADSNADSDHNSNSDQNQQKPIIETTDDHSVDDQHI 145
Query: 108 RLFRHSKPGIVFDHADEPQPPMK----RPKLVPGEDIDEKSKKFRRRIRSIAVDGNDLIA 163
S DEP P+ RP P + + +++ +RR RS+ +D +A
Sbjct: 146 DTVSDS---------DEPSSPIMGIGLRPISKPVSVVVDNTRR-KRRARSLVASEDDDVA 195
Query: 164 AANDAYK-----KSLARLEAKDAAAKAKAKREEER 193
A + K K L R K A A+ + E +
Sbjct: 196 APSTRQKRARANKYLTRKTQKAIQAAARVRDPESK 230
>ref|XP_654748.1| SMC4 protein, putative [Entamoeba histolytica HM-1:IMSS]
gi|56471819|gb|EAL49361.1| SMC4 protein, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 1226
Score = 37.0 bits (84), Expect = 0.40
Identities = 43/197 (21%), Positives = 81/197 (40%), Gaps = 20/197 (10%)
Query: 17 EDGDAAWKAAIDSVAGTSSYVTSFMNGFSATNNNDTKKKHNDNQNPKSPKIKHYQLKALK 76
++G+ ++ + GTS Y+ G S +K ND + K+ + K + +
Sbjct: 167 KNGEDGMLEFLEDIIGTSQYIEGIEKGTSEL------EKLNDEVHEKTNRFKIIERERNG 220
Query: 77 LLDDILENTIEIVKEPIPVLDEDPNVDDCGIRLFRHSKPGIVFDHADEPQPPMKRPKLVP 136
+L + EN I +KE + N++ ++ + + +E + ++ K++
Sbjct: 221 ILKE-KENAISYLKEEYKMCGLKNNLNKLELKAVEEKE----VEGKEEREKLEQKRKIIQ 275
Query: 137 ---GEDIDEKSKKFR------RRIRSIAVDGNDLIAAANDAYKKSLARLEAKDAAAKAKA 187
E + EK+ K + R I+ I + N A KK E K KA
Sbjct: 276 DAKNEKLKEKTTKDKIIEEKERDIKKIEKEYEKQKGLINSAKKKKARAEEEKKQNEKAVL 335
Query: 188 KREEERIEKLKKIRGER 204
+ E+E E KKI+ E+
Sbjct: 336 RNEKEIKEMEKKIKDEK 352
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 394,913,843
Number of Sequences: 2540612
Number of extensions: 17425647
Number of successful extensions: 90877
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 141
Number of HSP's that attempted gapping in prelim test: 90472
Number of HSP's gapped (non-prelim): 466
length of query: 221
length of database: 863,360,394
effective HSP length: 123
effective length of query: 98
effective length of database: 550,865,118
effective search space: 53984781564
effective search space used: 53984781564
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC147000.5


