

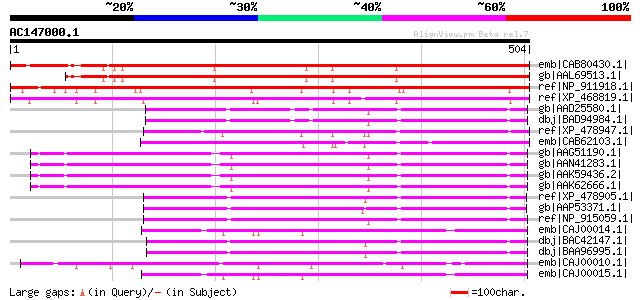
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.1 - phase: 0
(504 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB80430.1| putative protein [Arabidopsis thaliana] gi|44689... 568 e-160
gb|AAL69513.1| putative SHORT-ROOT (SHR) protein [Arabidopsis th... 527 e-148
ref|NP_911918.1| short-root protein-like [Oryza sativa (japonica... 471 e-131
ref|XP_468819.1| putative GRAS family transcription factor [Oryz... 454 e-126
gb|AAD25580.1| putative SCARECROW gene regulator [Arabidopsis th... 203 1e-50
dbj|BAD94984.1| putative SCARECROW gene regulator [Arabidopsis t... 202 2e-50
ref|XP_478947.1| putative short-root transcription factor [Oryza... 200 8e-50
emb|CAB62103.1| putative protein [Arabidopsis thaliana] gi|46518... 199 2e-49
gb|AAG51190.1| scarecrow-like protein [Arabidopsis thaliana] gi|... 196 2e-48
gb|AAN41283.1| putative scarecrow protein [Arabidopsis thaliana]... 196 2e-48
gb|AAK59436.2| putative scarecrow protein [Arabidopsis thaliana] 196 2e-48
gb|AAK62666.1| F17J6.12/F17J6.12 [Arabidopsis thaliana] 195 3e-48
ref|XP_478905.1| chitin-inducible gibberellin-responsive protein... 194 4e-48
gb|AAP53371.1| putative SCARECROW gene regulator-like [Oryza sat... 193 1e-47
ref|NP_915059.1| scarecrow-like protein [Oryza sativa (japonica ... 191 5e-47
emb|CAJ00014.1| Nodulation Signaling Pathway 1 protein homologue... 189 2e-46
dbj|BAC42147.1| putative SCARECROW gene regulator [Arabidopsis t... 188 3e-46
dbj|BAA96995.1| SCARECROW gene regulator-like [Arabidopsis thali... 188 3e-46
emb|CAJ00010.1| Nodulation Signaling Pathway 1 protein [Medicago... 182 2e-44
emb|CAJ00015.1| Nodulation Signaling Pathway 1 protein homologue... 181 6e-44
>emb|CAB80430.1| putative protein [Arabidopsis thaliana] gi|4468990|emb|CAB38304.1|
putative protein [Arabidopsis thaliana]
gi|8453100|gb|AAF75234.1| short-root protein
[Arabidopsis thaliana] gi|15235646|ref|NP_195480.1|
short-root transcription factor (SHR) [Arabidopsis
thaliana] gi|7485768|pir||T04722 hypothetical protein
F19F18.140 - Arabidopsis thaliana
Length = 531
Score = 568 bits (1463), Expect = e-160
Identities = 312/541 (57%), Positives = 390/541 (71%), Gaps = 48/541 (8%)
Query: 1 MDTLFRLVNFQQQQQQYQPDPSLNSTTTLT-TSSSSRSSRQTTYHY-YNQQEEDEECFNN 58
MDTLFRLV+ QQQQQ D + + ++L+ TS+++ S QT YHY + Q + EECFN
Sbjct: 1 MDTLFRLVSLQQQQQS---DSIITNQSSLSRTSTTTTGSPQTAYHYNFPQNDVVEECFN- 56
Query: 59 FYYMDHNNNNDEDLSSSSSKQHYYTYPYAST--TTITTPNTTYN--TINTPTTT------ 108
++MD +EDLSSSSS +++ + +T + TTP T Y+ T +TP++T
Sbjct: 57 -FFMD-----EEDLSSSSSHHNHHNHNNPNTYYSPFTTP-TQYHPATSSTPSSTAAAAAL 109
Query: 109 -----------DNYSFSPSHDYFNFEFSGHS-WSQNILLETARAFSDNNTNRIQQLMWML 156
D +FS +F+FS ++ W+ ++LLE ARAFSD +T R QQ++W L
Sbjct: 110 ASPYSSSGHHNDPSAFSIPQTPPSFDFSANAKWADSVLLEAARAFSDKDTARAQQILWTL 169
Query: 157 NELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTAS--EKTCSFDSTRKMLLKFQ 214
NELS+PYGDT+QKL+SYFLQALF+RM +G+R Y+T+ TA+ EKTCSF+STRK +LKFQ
Sbjct: 170 NELSSPYGDTEQKLASYFLQALFNRMTGSGERCYRTMVTAAATEKTCSFESTRKTVLKFQ 229
Query: 215 EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWPTLLEALATRSDDTPHLRLT 274
EVSPW TFGHVAANGAILEA++G K+HI+DIS+T+CTQWPTLLEALATRSDDTPHLRLT
Sbjct: 230 EVSPWATFGHVAANGAILEAVDGEAKIHIVDISSTFCTQWPTLLEALATRSDDTPHLRLT 289
Query: 275 TVVTAISGGSVQ----KVMKEIGSRMEKFARLMGVPFKFKII--FSDLRELNLCDLDIKE 328
TVV A + Q ++MKEIG+RMEKFARLMGVPFKF II DL E +L +LD+K
Sbjct: 290 TVVVANKFVNDQTASHRMMKEIGNRMEKFARLMGVPFKFNIIHHVGDLSEFDLNELDVKP 349
Query: 329 DEALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTIVEEEADL----EVCFGSDFVE 384
DE LAINCV ++H I+ G+ RD IS R L PR++T+VEEEADL E F +F+
Sbjct: 350 DEVLAINCVGAMHGIASRGSPRDAVISSFRRLRPRIVTVVEEEADLVGEEEGGFDDEFLR 409
Query: 385 GFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETAARWR 444
GF ECLRWFRV FE+ +ESF RTS+ERLMLER AGR IVDLVAC+P +S ERRETA +W
Sbjct: 410 GFGECLRWFRVCFESWEESFPRTSNERLMLERAAGRAIVDLVACEPSDSTERRETARKWS 469
Query: 445 RRLHGGGFNTVSFSDEVCDDVRALLRRYKEG-WSMTSSDGDTGIFLSWKDKPVVWASVWR 503
RR+ GF V +SDEV DDVRALLRRYKEG WSM GIFL W+D+PVVWAS WR
Sbjct: 470 RRMRNSGFGAVGYSDEVADDVRALLRRYKEGVWSMVQCPDAAGIFLCWRDQPVVWASAWR 529
Query: 504 P 504
P
Sbjct: 530 P 530
>gb|AAL69513.1| putative SHORT-ROOT (SHR) protein [Arabidopsis thaliana]
Length = 478
Score = 527 bits (1358), Expect = e-148
Identities = 285/485 (58%), Positives = 352/485 (71%), Gaps = 43/485 (8%)
Query: 55 CFNNFYYMDHNNNNDEDLSSSSSKQHYYTYPYAST--TTITTPNTTYN--TINTPTTT-- 108
CFN ++MD +EDLSSSSS +++ + +T + TTP T Y+ T +TP++T
Sbjct: 1 CFN--FFMD-----EEDLSSSSSHHNHHNHNNPNTYYSPFTTP-TQYHPATSSTPSSTAA 52
Query: 109 ---------------DNYSFSPSHDYFNFEFSGHS-WSQNILLETARAFSDNNTNRIQQL 152
D +FS +F+FS ++ W+ ++LLE ARAFSD +T R QQ+
Sbjct: 53 AAALASPYSSSGHHNDPSAFSIPQTPPSFDFSANAKWADSVLLEAARAFSDKDTARAQQI 112
Query: 153 MWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTAS--EKTCSFDSTRKML 210
+W LNELS+PYGDT+QKL+SYFLQALF+RM +G+R Y+T+ TA+ EKTCSF+STRK +
Sbjct: 113 LWTLNELSSPYGDTEQKLASYFLQALFNRMTGSGERCYRTMVTAAATEKTCSFESTRKTV 172
Query: 211 LKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWPTLLEALATRSDDTPH 270
LKFQEVS W TFGHVAANGAILEA++G K+HI+DIS+T+CTQWPTLLEALATRSDDTPH
Sbjct: 173 LKFQEVSSWATFGHVAANGAILEAVDGEAKIHIVDISSTFCTQWPTLLEALATRSDDTPH 232
Query: 271 LRLTTVVTAISGGSVQ----KVMKEIGSRMEKFARLMGVPFKFKII--FSDLRELNLCDL 324
LRLTTVV A + Q ++MKEIG+RMEKFARLMGVPFKF II DL E +L +L
Sbjct: 233 LRLTTVVVANKFVNDQTASHRMMKEIGNRMEKFARLMGVPFKFNIIHHVGDLSEFDLNEL 292
Query: 325 DIKEDEALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTIVEEEADL----EVCFGS 380
D+K DE LAINCV ++H I+ G+ RD IS R L PR++T+VEEEADL E F
Sbjct: 293 DVKPDEVLAINCVGAMHGIASRGSPRDAVISSFRRLRPRIVTVVEEEADLVGEEEGGFDD 352
Query: 381 DFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETA 440
+F+ GF ECLRWFRV FE+ +ESF RTS+ERLMLER AGR IVDLVAC+P +S ERRETA
Sbjct: 353 EFLRGFGECLRWFRVCFESWEESFPRTSNERLMLERAAGRAIVDLVACEPSDSTERRETA 412
Query: 441 ARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEG-WSMTSSDGDTGIFLSWKDKPVVWA 499
+W RR+ GF V +SDEV DDVRALLRRYKEG WSM GIFL W+D+PVVWA
Sbjct: 413 RKWSRRMRNSGFGAVGYSDEVADDVRALLRRYKEGVWSMVQCPDAAGIFLCWRDQPVVWA 472
Query: 500 SVWRP 504
S WRP
Sbjct: 473 SAWRP 477
>ref|NP_911918.1| short-root protein-like [Oryza sativa (japonica cultivar-group)]
gi|50509213|dbj|BAD30442.1| short-root protein-like
[Oryza sativa (japonica cultivar-group)]
gi|23617232|dbj|BAC20900.1| short-root protein-like
[Oryza sativa (japonica cultivar-group)]
Length = 629
Score = 471 bits (1212), Expect = e-131
Identities = 283/604 (46%), Positives = 368/604 (60%), Gaps = 102/604 (16%)
Query: 1 MDTLFRLVNFQ---QQQQQYQPDPSLNSTTTLTTSSSSRSSRQTT---YHYYNQQEED-- 52
MDTLFRLV+ Q +QQQQ Q S NS +T TSS SRSS T Y YY+
Sbjct: 28 MDTLFRLVSLQAASEQQQQQQQSASYNSRST--TSSGSRSSSHQTNASYSYYHHSSNSGG 85
Query: 53 -------------EECFNNFYYMD-----------HNNNNDEDLSSSSSKQHYY------ 82
+ + +YY++ H DED SSSSS +H++
Sbjct: 86 GGGGGGGYYYGGQQPPPSQYYYLEPYQEECGNAPHHQLYMDEDFSSSSSSRHFHHGARVQ 145
Query: 83 --TYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYF----NFEFS----------- 125
P +ST T T P +T +T + F + F N +FS
Sbjct: 146 QQQPPASSTPTGTAPTPPLSTSSTAAGAGHGLFEAADLSFPPDLNLDFSSPASSSGGGTA 205
Query: 126 ---------GHSWSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQ 176
G W+ +LLE AR+ + ++ R+QQLMWMLNEL++PYGD +QKL+SYFLQ
Sbjct: 206 SSGAVGGGGGGRWASQLLLECARSVAARDSQRVQQLMWMLNELASPYGDVEQKLASYFLQ 265
Query: 177 ALFSRMNDAGDRTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILE--- 233
LF+R+ +G RT +TL AS++ SFDSTR+ L+FQE+SPW++FGHVAANGAILE
Sbjct: 266 GLFARLTASGPRTLRTLAAASDRNTSFDSTRRTALRFQELSPWSSFGHVAANGAILESFL 325
Query: 234 -----ALEGNPKLHIIDISNTYCTQWPTLLEALATRS-DDTPHLRLTTVVTAISG---GS 284
A + HI+D+SNT+CTQWPTLLEALATRS D+TPHL +TTVV+A +
Sbjct: 326 EVAAAASSETQRFHILDLSNTFCTQWPTLLEALATRSADETPHLSITTVVSAAPSAPTAA 385
Query: 285 VQKVMKEIGSRMEKFARLMGVPFKFKIIF--SDLRELNLCDLDIKE---DEALAINCVNS 339
VQ+VM+EIG RMEKFARLMGVPF+F+ + DL EL+L LD++E ALA+NCVNS
Sbjct: 386 VQRVMREIGQRMEKFARLMGVPFRFRAVHHSGDLAELDLDALDLREGGATTALAVNCVNS 445
Query: 340 LHS-ISGAGNHRDLFISLLRGLEPRVLTIVEEEADLEVC---------FGSD----FVEG 385
L + G RD F + LR L+PRV+T+VEEEADL G D F++
Sbjct: 446 LRGVVPGRARRRDAFAASLRRLDPRVVTVVEEEADLVASDPDASSATEEGGDTEAAFLKV 505
Query: 386 FKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETAARWRR 445
F E LR+F Y ++L+ESF +TS+ERL LER AGR IVDLV+C ES+ERRETAA W R
Sbjct: 506 FGEGLRFFSAYMDSLEESFPKTSNERLALERGAGRAIVDLVSCPASESMERRETAASWAR 565
Query: 446 RLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGD-----TGIFLSWKDKPVVWAS 500
R+ GF+ V+FS++V DDVR+LLRRY+EGWSM + D G+FL+WK++P+VWAS
Sbjct: 566 RMRSAGFSPVAFSEDVADDVRSLLRRYREGWSMREAGTDDSAAGAGVFLAWKEQPLVWAS 625
Query: 501 VWRP 504
WRP
Sbjct: 626 AWRP 629
>ref|XP_468819.1| putative GRAS family transcription factor [Oryza sativa (japonica
cultivar-group)] gi|41469537|gb|AAS07303.1| putative
GRAS family transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 603
Score = 454 bits (1169), Expect = e-126
Identities = 269/606 (44%), Positives = 350/606 (57%), Gaps = 105/606 (17%)
Query: 1 MDTLFRLVNFQQQQQQY---------QPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEE 51
MDTLFRLV+ QP S S+ T+S SS + TY++++
Sbjct: 1 MDTLFRLVSLHHHHHHQHAASPSPPDQPHKSYPSSRGSTSSPSSHHTHNHTYYHHSHSHY 60
Query: 52 DEECFNNFYYMD--------------------------HNNNNDEDLSSSSSKQHYY--T 83
+ N+YY H DED SSSSS + ++ T
Sbjct: 61 NNNSNTNYYYQGGGGGGGGYYYAEEQQPAAYLEECGNGHQFYMDEDFSSSSSSRQFHSGT 120
Query: 84 YPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHS--------------- 128
+S P+ T ++ + FS + +F G
Sbjct: 121 GAPSSAPVPPPPSATTSSAGGHGLFEAADFSFPQVDISLDFGGSPAVPSSSGAGAGAGAA 180
Query: 129 ------WSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRM 182
W+ +L+E ARA + ++ R+QQLMWMLNEL++PYGD DQKL+SYFLQ LF+R+
Sbjct: 181 PSSSGRWAAQLLMECARAVAGRDSQRVQQLMWMLNELASPYGDVDQKLASYFLQGLFARL 240
Query: 183 NDAGDRTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEAL------- 235
+G RT +TL TAS++ SFDSTR+ LKFQE+SPWT FGHVAANGAILE+
Sbjct: 241 TTSGPRTLRTLATASDRNASFDSTRRTALKFQELSPWTPFGHVAANGAILESFLEAAAAG 300
Query: 236 ----------EGNP--KLHIIDISNTYCTQWPTLLEALATR-SDDTPHLRLTTVV-TAIS 281
P +LHI+D+SNT+CTQWPTLLEALATR SDDTPHL +TTVV TA
Sbjct: 301 AAASSSSSSSSSTPPTRLHILDLSNTFCTQWPTLLEALATRSSDDTPHLSITTVVPTAAP 360
Query: 282 GGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF--SDLRELNLCDLDIKE---DEALAINC 336
+ Q+VM+EIG R+EKFARLMGVPF F+ + DL +L+L LD++E ALA+NC
Sbjct: 361 SAAAQRVMREIGQRLEKFARLMGVPFSFRAVHHSGDLADLDLAALDLREGGATAALAVNC 420
Query: 337 VNSLHSISGAGNHRDLFISLLRGLEPRVLTIVEEEADL---------EVCFGSDFVEGFK 387
VN+L G RD F++ LR LEPRV+T+VEEEADL E + FV+ F
Sbjct: 421 VNALR---GVARGRDAFVASLRRLEPRVVTVVEEEADLAAPEADASSEADTDAAFVKVFG 477
Query: 388 ECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETAARWRRRL 447
E LR+F Y ++L+ESF +TS+ERL LER GR IVDLV+C +S ERRETAA W RR+
Sbjct: 478 EGLRFFSAYMDSLEESFPKTSNERLSLERAVGRAIVDLVSCPASQSAERRETAASWARRM 537
Query: 448 HGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDT---------GIFLSWKDKPVVW 498
GF+ +FS++V DDVR+LLRRYKEGWSM + G T G FL+WK++PVVW
Sbjct: 538 RSAGFSPAAFSEDVADDVRSLLRRYKEGWSMRDAGGATDDAAGAAAAGAFLAWKEQPVVW 597
Query: 499 ASVWRP 504
AS W+P
Sbjct: 598 ASAWKP 603
>gb|AAD25580.1| putative SCARECROW gene regulator [Arabidopsis thaliana]
gi|20197984|gb|AAM15339.1| putative SCARECROW gene
regulator [Arabidopsis thaliana]
gi|6644392|gb|AAF21044.1| scarecrow-like 21 [Arabidopsis
thaliana] gi|15224425|ref|NP_178566.1| scarecrow-like
transcription factor 21 (SCL21) [Arabidopsis thaliana]
gi|25366592|pir||G84462 probable SCARECROW gene
regulator [imported] - Arabidopsis thaliana
gi|51968562|dbj|BAD42973.1| putative SCARECROW gene
regulator [Arabidopsis thaliana]
Length = 413
Score = 203 bits (516), Expect = 1e-50
Identities = 118/376 (31%), Positives = 202/376 (53%), Gaps = 20/376 (5%)
Query: 133 ILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKT 192
+L+ A+A S+NN + M L + + G+ Q+L +Y L+ L +R+ +G YK+
Sbjct: 53 VLVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKS 112
Query: 193 LTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCT 252
L + ++ F S +L EV P+ FG+++ANGAI EA++ ++HIID +
Sbjct: 113 LQSREPESYEFLSYVYVL---HEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGS 169
Query: 253 QWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKII 312
QW L++A A R P++R +T + GSV +K+ R+EK A+ VPF+F +
Sbjct: 170 QWIALIQAFAARPGGAPNIR----ITGVGDGSVLVTVKK---RLEKLAKKFDVPFRFNAV 222
Query: 313 FSDLRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIV 368
E+ + +LD+++ EAL +N LH + NHRD + +++ L P+V+T+V
Sbjct: 223 SRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLV 282
Query: 369 EEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVA 427
E+E + S F+ F E L ++ FE++D R ER+ +E+ R +V+++A
Sbjct: 283 EQECNTNT---SPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNIIA 339
Query: 428 CDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGI 487
C+ E +ER E +W+ R GF S + +RALLR Y G+++ DG +
Sbjct: 340 CEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYAIEERDG--AL 397
Query: 488 FLSWKDKPVVWASVWR 503
+L W D+ +V + W+
Sbjct: 398 YLGWMDRILVSSCAWK 413
>dbj|BAD94984.1| putative SCARECROW gene regulator [Arabidopsis thaliana]
gi|51970122|dbj|BAD43753.1| putative SCARECROW gene
regulator [Arabidopsis thaliana]
Length = 413
Score = 202 bits (514), Expect = 2e-50
Identities = 118/376 (31%), Positives = 202/376 (53%), Gaps = 20/376 (5%)
Query: 133 ILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKT 192
+L+ A+A S+NN + M L + + G+ Q+L +Y L+ L +R+ +G YK+
Sbjct: 53 VLVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKS 112
Query: 193 LTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCT 252
L + ++ F S +L EV P+ FG+++ANGAI EA++ ++HIID +
Sbjct: 113 LQSREPESYEFLSYVYVL---HEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGS 169
Query: 253 QWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKII 312
QW L++A A R P++R +T + GSV +K+ R+EK A+ VPF+F +
Sbjct: 170 QWIALIQAFAARPGGAPNIR----ITGVGDGSVLVTVKK---RLEKLAKKFDVPFRFNAV 222
Query: 313 FSDLRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIV 368
E+ + +LD+++ EAL +N LH + NHRD + +++ L P+V+T+V
Sbjct: 223 SRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLV 282
Query: 369 EEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVA 427
E+E + S F+ F E L ++ FE++D R ER+ +E+ R +V+++A
Sbjct: 283 EQECNTNT---SPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNIMA 339
Query: 428 CDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGI 487
C+ E +ER E +W+ R GF S + +RALLR Y G+++ DG +
Sbjct: 340 CEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYAIEERDG--AL 397
Query: 488 FLSWKDKPVVWASVWR 503
+L W D+ +V + W+
Sbjct: 398 YLGWMDRILVSSCAWK 413
>ref|XP_478947.1| putative short-root transcription factor [Oryza sativa (japonica
cultivar-group)] gi|28564823|dbj|BAC57752.1| putative
short-root transcription factor [Oryza sativa (japonica
cultivar-group)] gi|34393441|dbj|BAC82980.1| putative
short-root transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 472
Score = 200 bits (509), Expect = 8e-50
Identities = 126/403 (31%), Positives = 208/403 (51%), Gaps = 36/403 (8%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+ +L+ A A N+ QQ++W+LN ++ GD++Q+L++ FL AL SR + G
Sbjct: 76 EQLLVHCANAIEANDATLTQQILWVLNNIAPADGDSNQRLTAAFLCALVSRASRTG--AC 133
Query: 191 KTLTTASEKTCSFDS------TRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHII 244
K +T A + T L F +++PW FG+ AAN AI+EA+EG P +HI+
Sbjct: 134 KAVTAAVADAVESAALHVHRFTAVELASFIDLTPWHRFGYTAANAAIVEAVEGFPVVHIV 193
Query: 245 DISNTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISG--GSVQKVMKEIGSRMEKFARL 302
D+S T+C Q PTL++ LA R++ P LRLT A S ++ +E+G+++ FAR
Sbjct: 194 DLSTTHCMQIPTLIDMLAGRAEGPPILRLTVADVAPSAPPPALDMPYEELGAKLVNFARS 253
Query: 303 MGVPFKFKII--------FSDLRELNLCDLDIKEDEALAINCVNSLHSI--SGAG----- 347
+ F+++ S + +L + L EAL +NC LH++ AG
Sbjct: 254 RNMSMDFRVVPTSPADALTSLVDQLRVQQLVSDGGEALVVNCHMLLHTVPDETAGSVSLT 313
Query: 348 ------NHRDLFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALD 401
+ R + + LR L+P ++ +V+E+AD D V + + + ++A+D
Sbjct: 314 TAQPPVSLRTMLLKSLRALDPTLVVVVDEDADFT---AGDVVGRLRAAFNFLWIPYDAVD 370
Query: 402 ESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEV 461
+ S +R E E G + +++A + E VER+E RW +R+ GF +F +E
Sbjct: 371 TFLPKGSEQRRWYEAEVGWKVENVLAQEGVERVERQEDRTRWGQRMRAAGFRAAAFGEEA 430
Query: 462 CDDVRALLRRYKEGWSMTSSDGDTGIFLSWKDKPVVWASVWRP 504
+V+A+L + GW M D D + L+WK VV+AS W P
Sbjct: 431 AGEVKAMLNDHAAGWGMKREDDD--LVLTWKGHNVVFASAWAP 471
>emb|CAB62103.1| putative protein [Arabidopsis thaliana] gi|46518431|gb|AAS99697.1|
At3g49950 [Arabidopsis thaliana]
gi|15229645|ref|NP_190564.1| scarecrow transcription
factor family protein [Arabidopsis thaliana]
gi|11357756|pir||T45848 hypothetical protein F3A4.30 -
Arabidopsis thaliana
Length = 410
Score = 199 bits (506), Expect = 2e-49
Identities = 127/393 (32%), Positives = 202/393 (51%), Gaps = 24/393 (6%)
Query: 128 SWSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGD 187
++ + +LL A A N+ Q++W+LN ++ P GD+ Q+L+S FL+AL SR
Sbjct: 25 NFMEQLLLHCATAIDSNDAALTHQILWVLNNIAPPDGDSTQRLTSAFLRALLSRAVSKTP 84
Query: 188 RTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDIS 247
T++ + + L F +++PW FG +AAN AIL A+EG +HI+D+S
Sbjct: 85 TLSSTISFLPQADELHRFSVVELAAFVDLTPWHRFGFIAANAAILTAVEGYSTVHIVDLS 144
Query: 248 NTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGGS--VQKVMKEIGSRMEKFARLMGV 305
T+C Q PTL++A+A+R + P L TVV++ + +E+GS++ FA +
Sbjct: 145 LTHCMQIPTLIDAMASRLNKPPPLLKLTVVSSSDHFPPFINISYEELGSKLVNFATTRNI 204
Query: 306 PFKFKII---FSD-----LRELNLCDLDIKEDEALAINCVNSLHSI------SGAGNHRD 351
+F I+ +SD L++L + +EAL +NC L I S + + R
Sbjct: 205 TMEFTIVPSTYSDGFSSLLQQLRIYPSSF--NEALVVNCHMMLRYIPEEPLTSSSSSLRT 262
Query: 352 LFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSER 411
+F+ LR L PR++T++EE+ DL + V K +F + F+ D S +R
Sbjct: 263 VFLKQLRSLNPRIVTLIEEDVDLT---SENLVNRLKSAFNYFWIPFDTTDTFMSE---QR 316
Query: 412 LMLEREAGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRR 471
E E I ++VA + E VER ET RW R+ F V ++ DV+A+L
Sbjct: 317 RWYEAEISWKIENVVAKEGAERVERTETKRRWIERMREAEFGGVRVKEDAVADVKAMLEE 376
Query: 472 YKEGWSMTSSDGDTGIFLSWKDKPVVWASVWRP 504
+ GW M D D + L+WK VV+A+VW P
Sbjct: 377 HAVGWGMKKEDDDESLVLTWKGHSVVFATVWVP 409
>gb|AAG51190.1| scarecrow-like protein [Arabidopsis thaliana]
gi|25405382|pir||E96542 scarecrow-like protein
[imported] - Arabidopsis thaliana
gi|9454552|gb|AAF87875.1| Putative transcription factor
[Arabidopsis thaliana]
Length = 526
Score = 196 bits (497), Expect = 2e-48
Identities = 137/493 (27%), Positives = 235/493 (46%), Gaps = 25/493 (5%)
Query: 21 PSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQH 80
P LN+ ++S++S SS ++ N FNN ++NN+ S++++ +
Sbjct: 49 PCLNNKNN-SSSTTSFSSNESPISQANNNNLSR--FNNHSPEENNNSPLSGSSATNTNET 105
Query: 81 YYTYPYAST-TTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETAR 139
+ T + P+ + N + S Y + E + +L E A+
Sbjct: 106 ELSLMLKDLETAMMEPDVDNSYNNQGGFGQQHGVVSSAMYRSMEMISRGDLKGVLYECAK 165
Query: 140 AFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEK 199
A + + L+ L ++ + G+ Q+L +Y L+ L +R+ +G YK L
Sbjct: 166 AVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASSGSSIYKALRCK--- 222
Query: 200 TCSFDSTRKMLLKFQ----EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWP 255
D T LL + E P+ FG+ +ANGAI EA++ +HIID + QW
Sbjct: 223 ----DPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWV 278
Query: 256 TLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSD 315
+L+ AL R P++R+T + S + Q ++ +G R+ K A + GVPF+F
Sbjct: 279 SLIRALGARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALC 338
Query: 316 LRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIVEEE 371
E+ + L ++ EALA+N LH + NHRD + L++ L P V+T+VE+E
Sbjct: 339 CTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQE 398
Query: 372 ADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVACDP 430
A+ + F+ F E + + FE++D +R ER+ +E+ R +V+L+AC+
Sbjct: 399 ANTNT---APFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEG 455
Query: 431 YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLS 490
E ER E +WR R H GF S V ++ LL Y E +++ DG ++L
Sbjct: 456 VEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLEERDG--ALYLG 513
Query: 491 WKDKPVVWASVWR 503
WK++P++ + WR
Sbjct: 514 WKNQPLITSCAWR 526
>gb|AAN41283.1| putative scarecrow protein [Arabidopsis thaliana]
gi|30694805|ref|NP_175475.2| scarecrow-like
transcription factor 5 (SCL5) [Arabidopsis thaliana]
Length = 597
Score = 196 bits (497), Expect = 2e-48
Identities = 137/493 (27%), Positives = 235/493 (46%), Gaps = 25/493 (5%)
Query: 21 PSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQH 80
P LN+ ++S++S SS ++ N FNN ++NN+ S++++ +
Sbjct: 120 PCLNNKNN-SSSTTSFSSNESPISQANNNNLSR--FNNHSPEENNNSPLSGSSATNTNET 176
Query: 81 YYTYPYAST-TTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETAR 139
+ T + P+ + N + S Y + E + +L E A+
Sbjct: 177 ELSLMLKDLETAMMEPDVDNSYNNQGGFGQQHGVVSSAMYRSMEMISRGDLKGVLYECAK 236
Query: 140 AFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEK 199
A + + L+ L ++ + G+ Q+L +Y L+ L +R+ +G YK L
Sbjct: 237 AVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASSGSSIYKALRCK--- 293
Query: 200 TCSFDSTRKMLLKFQ----EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWP 255
D T LL + E P+ FG+ +ANGAI EA++ +HIID + QW
Sbjct: 294 ----DPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWV 349
Query: 256 TLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSD 315
+L+ AL R P++R+T + S + Q ++ +G R+ K A + GVPF+F
Sbjct: 350 SLIRALGARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALC 409
Query: 316 LRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIVEEE 371
E+ + L ++ EALA+N LH + NHRD + L++ L P V+T+VE+E
Sbjct: 410 CTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQE 469
Query: 372 ADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVACDP 430
A+ + F+ F E + + FE++D +R ER+ +E+ R +V+L+AC+
Sbjct: 470 ANTNT---APFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEG 526
Query: 431 YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLS 490
E ER E +WR R H GF S V ++ LL Y E +++ DG ++L
Sbjct: 527 VEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLEERDG--ALYLG 584
Query: 491 WKDKPVVWASVWR 503
WK++P++ + WR
Sbjct: 585 WKNQPLITSCAWR 597
>gb|AAK59436.2| putative scarecrow protein [Arabidopsis thaliana]
Length = 587
Score = 196 bits (497), Expect = 2e-48
Identities = 137/493 (27%), Positives = 235/493 (46%), Gaps = 25/493 (5%)
Query: 21 PSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQH 80
P LN+ ++S++S SS ++ N FNN ++NN+ S++++ +
Sbjct: 110 PCLNNKNN-SSSTTSFSSNESPISQANNNNLSR--FNNHSPEENNNSPLSGSSATNTNET 166
Query: 81 YYTYPYAST-TTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETAR 139
+ T + P+ + N + S Y + E + +L E A+
Sbjct: 167 ELSLMLKDLETAMMEPDVDNSYNNQGGFGQQHGVVSSAMYRSMEMISRGDLKGVLYECAK 226
Query: 140 AFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEK 199
A + + L+ L ++ + G+ Q+L +Y L+ L +R+ +G YK L
Sbjct: 227 AVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASSGSSIYKALRCK--- 283
Query: 200 TCSFDSTRKMLLKFQ----EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWP 255
D T LL + E P+ FG+ +ANGAI EA++ +HIID + QW
Sbjct: 284 ----DPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWV 339
Query: 256 TLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSD 315
+L+ AL R P++R+T + S + Q ++ +G R+ K A + GVPF+F
Sbjct: 340 SLIRALGARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALC 399
Query: 316 LRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIVEEE 371
E+ + L ++ EALA+N LH + NHRD + L++ L P V+T+VE+E
Sbjct: 400 CTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQE 459
Query: 372 ADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVACDP 430
A+ + F+ F E + + FE++D +R ER+ +E+ R +V+L+AC+
Sbjct: 460 ANTNT---APFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEG 516
Query: 431 YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLS 490
E ER E +WR R H GF S V ++ LL Y E +++ DG ++L
Sbjct: 517 VEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLEERDG--ALYLG 574
Query: 491 WKDKPVVWASVWR 503
WK++P++ + WR
Sbjct: 575 WKNQPLITSCAWR 587
>gb|AAK62666.1| F17J6.12/F17J6.12 [Arabidopsis thaliana]
Length = 526
Score = 195 bits (495), Expect = 3e-48
Identities = 137/493 (27%), Positives = 234/493 (46%), Gaps = 25/493 (5%)
Query: 21 PSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQH 80
P LN+ ++S++S SS ++ N FNN ++NN+ S++++ +
Sbjct: 49 PCLNNKNN-SSSTTSFSSNESPISQANNNNLSR--FNNHSPEENNNSPLSGSSATNTNET 105
Query: 81 YYTYPYAST-TTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETAR 139
+ T + P+ + N + S Y + E + +L E A+
Sbjct: 106 ELSLMLKDLETAMMEPDVDNSYNNQGGFGQQHGVVSSAMYRSMEMISRGDLKGVLYECAK 165
Query: 140 AFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEK 199
A + + L+ L ++ + G+ Q+L +Y L+ L +R+ +G YK L
Sbjct: 166 AVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASSGSSIYKALRCK--- 222
Query: 200 TCSFDSTRKMLLKFQ----EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWP 255
D T LL + E P+ FG+ +ANGAI EA++ +HIID + QW
Sbjct: 223 ----DPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWV 278
Query: 256 TLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSD 315
+L+ AL R P++R+T + S + Q ++ +G R+ K A + GVPF+F
Sbjct: 279 SLIRALGARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALC 338
Query: 316 LRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIVEEE 371
E+ + L ++ EALA+N LH + NHRD + L++ L P V+T+VE+E
Sbjct: 339 CTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQE 398
Query: 372 ADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVACDP 430
A+ + F+ F E + + FE++D +R ER+ +E+ R +V+L+AC+
Sbjct: 399 ANTNT---APFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEG 455
Query: 431 YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLS 490
E ER E +WR R H GF S V + LL Y E +++ DG ++L
Sbjct: 456 VEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIEGLLESYSEKYTLEERDG--ALYLG 513
Query: 491 WKDKPVVWASVWR 503
WK++P++ + WR
Sbjct: 514 WKNQPLITSCAWR 526
>ref|XP_478905.1| chitin-inducible gibberellin-responsive protein [Oryza sativa
(japonica cultivar-group)] gi|51963506|ref|XP_506430.1|
PREDICTED OJ1127_E01.113 gene product [Oryza sativa
(japonica cultivar-group)] gi|25989334|gb|AAL61821.1|
chitin-inducible gibberellin-responsive protein [Oryza
sativa (japonica cultivar-group)]
gi|27817840|dbj|BAC55608.1| chitin-inducible
gibberellin-responsive protein [Oryza sativa (japonica
cultivar-group)]
Length = 544
Score = 194 bits (494), Expect = 4e-48
Identities = 117/377 (31%), Positives = 197/377 (52%), Gaps = 13/377 (3%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+ +L+ ARA + N+ I ++ L ++ + G+ ++L +Y ++ L +R+ +G Y
Sbjct: 175 KELLIACARAVEEKNSFAIDMMIPELRKIVSVSGEPLERLGAYMVEGLVARLASSGISIY 234
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
K L K+ S L E P+ FG+++ANGAI EA++G ++HIID +
Sbjct: 235 KALKCKEPKSSDLLSYMHFLY---EACPYFKFGYMSANGAIAEAVKGEDRIHIIDFHISQ 291
Query: 251 CTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK 310
QW +LL+ALA R P +R+T + ++S + ++ +G R+ A L VPF+F
Sbjct: 292 GAQWISLLQALAARPGGPPTVRITGIDDSVSAYARGGGLELVGRRLSHIASLCKVPFEFH 351
Query: 311 IIFSDLRELNLCDLDIKEDEALAINCVNSLHSIS----GAGNHRDLFISLLRGLEPRVLT 366
+ ++ L + EALA+N LH I NHRD + +++ L P+VLT
Sbjct: 352 PLAISGSKVEAAHLGVIPGEALAVNFTLELHHIPDESVSTANHRDRLLRMVKSLSPKVLT 411
Query: 367 IVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDL 425
+VE E++ + F + F E L ++ FE++D + R ER+ +E+ R IV+L
Sbjct: 412 LVEMESNTNT---APFPQRFAETLDYYTAIFESIDLTLPRDDRERINMEQHCLAREIVNL 468
Query: 426 VACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDT 485
+AC+ E ER E +W+ RL GF S V +R LL+ Y + + + DG
Sbjct: 469 IACEGEERAERYEPFGKWKARLTMAGFRPSPLSSLVNATIRTLLQSYSDNYKLAERDG-- 526
Query: 486 GIFLSWKDKPVVWASVW 502
++L WK +P+V +S W
Sbjct: 527 ALYLGWKSRPLVVSSAW 543
>gb|AAP53371.1| putative SCARECROW gene regulator-like [Oryza sativa (japonica
cultivar-group)] gi|37533564|ref|NP_921084.1| putative
SCARECROW gene regulator-like [Oryza sativa (japonica
cultivar-group)] gi|20043021|gb|AAM08829.1| Putative
SCARECROW gene regulator-like [Oryza sativa (japonica
cultivar-group)]
Length = 524
Score = 193 bits (490), Expect = 1e-47
Identities = 115/378 (30%), Positives = 200/378 (52%), Gaps = 15/378 (3%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+ +++ +A ++N+ + L+ L ++ + GD Q+L +Y L+ L +R++ +G + Y
Sbjct: 155 KQVIIACGKAVAENDVRLTELLISELGQMVSVSGDPLQRLGAYMLEGLVARLSSSGSKIY 214
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
K+L + S +L E+ P+ FG+++ANGAI EA++G +HIID
Sbjct: 215 KSLKCKEPTSSELMSYMHLLY---EICPFFKFGYMSANGAIAEAIKGENFVHIIDFQIAQ 271
Query: 251 CTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK 310
+QW TL++ALA R P LR+T + + S + + +G R+ K A+ G+PF+F
Sbjct: 272 GSQWMTLIQALAARPGGPPFLRITGIDDSNSAYARGGGLDIVGMRLYKVAQSFGLPFEFN 331
Query: 311 IIFSDLRELNLCDLDIKEDEALAINCVNSLH-----SISGAGNHRDLFISLLRGLEPRVL 365
+ + E+ L LDI+ E + +N LH S+S NHRD + +++ L PR++
Sbjct: 332 AVPAASHEVYLEHLDIRVGEVIVVNFAYQLHHTPDESVS-TENHRDRILRMVKSLSPRLV 390
Query: 366 TIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLERE-AGRGIVD 424
T+VE+E++ F + E L ++ FE++D + R R+ E+ R IV+
Sbjct: 391 TLVEQESNTNT---RPFFPRYLETLDYYTAMFESIDVALPRDDKRRMSAEQHCVARDIVN 447
Query: 425 LVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGD 484
L+AC+ E VER E +W+ RL GF S V ++ LL Y + + DG
Sbjct: 448 LIACEGAERVERHEVFGKWKARLTMAGFRPYPLSSVVNSTIKTLLHTYNSFYRLEERDG- 506
Query: 485 TGIFLSWKDKPVVWASVW 502
++L WK++ +V +S W
Sbjct: 507 -VLYLGWKNRVLVVSSAW 523
>ref|NP_915059.1| scarecrow-like protein [Oryza sativa (japonica cultivar-group)]
gi|21952821|dbj|BAC06237.1| putative chitin-inducible
gibberellin-responsive protein [Oryza sativa (japonica
cultivar-group)] gi|20161431|dbj|BAB90355.1| putative
chitin-inducible gibberellin-responsive protein [Oryza
sativa (japonica cultivar-group)]
Length = 553
Score = 191 bits (485), Expect = 5e-47
Identities = 114/378 (30%), Positives = 202/378 (53%), Gaps = 13/378 (3%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+ +L + A A SD N + Q ++ L ++ + GD Q++++Y ++ L +R+ +G Y
Sbjct: 184 KQLLFDCAMALSDYNVDEAQAIITDLRQMVSIQGDPSQRIAAYLVEGLAARIVASGKGIY 243
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
K L+ T S ++L E+ P FG +AAN AILEA +G ++HIID
Sbjct: 244 KALSCKEPPTLYQLSAMQILF---EICPCFRFGFMAANYAILEACKGEDRVHIIDFDINQ 300
Query: 251 CTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK 310
+Q+ TL++ L ++ HLR+T V + +K IG R+EK A G+ F+F+
Sbjct: 301 GSQYITLIQFLKNNANKPRHLRITGVDDPETVQRTVGGLKVIGQRLEKLAEDCGISFEFR 360
Query: 311 IIFSDLRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLT 366
+ +++ ++ LD EAL +N LH + N RD + +++GL+P+++T
Sbjct: 361 AVGANIGDVTPAMLDCCPGEALVVNFAFQLHHLPDESVSIMNERDQLLRMVKGLQPKLVT 420
Query: 367 IVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDL 425
+VE++A+ + F F+E ++ F++LD + R S +R+ +ER+ R IV++
Sbjct: 421 LVEQDANTNT---APFQTRFREVYDYYAALFDSLDATLPRESPDRMNVERQCLAREIVNI 477
Query: 426 VACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDT 485
+AC+ + VER E A +WR R+ GF FS V +R+LL+ Y + + G
Sbjct: 478 LACEGPDRVERYEVAGKWRARMTMAGFTPCPFSSNVISGIRSLLKSYCDRYKFEEDHG-- 535
Query: 486 GIFLSWKDKPVVWASVWR 503
G+ W +K ++ +S W+
Sbjct: 536 GLHFGWGEKTLIVSSAWQ 553
>emb|CAJ00014.1| Nodulation Signaling Pathway 1 protein homologue 1 [Populus
trichocarpa]
Length = 556
Score = 189 bits (480), Expect = 2e-46
Identities = 130/395 (32%), Positives = 202/395 (50%), Gaps = 33/395 (8%)
Query: 129 WSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDR 188
W++ +L A A N +R+Q L+++L+EL++ GD + +L++Y L+AL ++ +
Sbjct: 165 WAEQLLNPCAAAIPIGNMSRVQHLLYVLHELASLTGDANHRLAAYGLRALTHHLSSSS-- 222
Query: 189 TYKTLTTASEKTCSFDST-----RKMLLKFQEVSPWTTFGHVAANGAILEAL--EGNPK- 240
TL++AS T +F ST +K LLKF EVSPW F H AN +IL+ L E +P+
Sbjct: 223 ---TLSSASTGTITFASTEPKFFQKSLLKFYEVSPWFAFPHNIANASILQVLAQEQDPRR 279
Query: 241 -LHIIDISNTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGG--------SVQKVMKE 291
LH++DI ++ QWPTLLEAL R P L TV+TA S S+
Sbjct: 280 NLHVLDIGVSHGVQWPTLLEALTRRPGGPPPLVRITVITAASENDQTTETPFSIGPPGDN 339
Query: 292 IGSRMEKFARLMGVPFKFKIIFS-DLRELNLCDLDIKEDEALAINCVNSLHSIS-GAGNH 349
SR+ FA+ M + + K + + L++L+ +D K DEAL + LH ++ +
Sbjct: 340 FSSRLLGFAKSMNINLQIKRLDNHSLQKLSGRIIDTKPDEALIVCAQFRLHHLNHNTPDE 399
Query: 350 RDLFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESF-SRTS 408
R F+ +LR LEP+ + + E D DF GF + + + ++ +F R S
Sbjct: 400 RTEFLRVLRRLEPKGVILTENNMDCSCNSCGDFATGFSRRVEYLWRFLDSTSSAFKGRES 459
Query: 409 SERLMLEREAGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRAL 468
ER M+E EA + + + E E +W R+ G GF F ++ D RAL
Sbjct: 460 VERRMMEGEAAKALTN--------RGEMNEGKEKWCERMRGVGFVGELFGEDAIDGARAL 511
Query: 469 LRRYKEGWSMTSSDGDTGIFLSWKDKPVVWASVWR 503
LR+Y W M D + L WK +PV + S+W+
Sbjct: 512 LRKYDSNWEMRGEGKDGCVGLWWKGQPVSFCSLWK 546
>dbj|BAC42147.1| putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 411
Score = 188 bits (478), Expect = 3e-46
Identities = 108/375 (28%), Positives = 196/375 (51%), Gaps = 12/375 (3%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
L+ A+A S+N+ +M L ++ + G+ Q+L +Y L+ L +++ +G YK L
Sbjct: 44 LVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASSGSSIYKAL 103
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
E + + +L EV P+ FG+++ANGAI EA++ ++HIID +Q
Sbjct: 104 NRCPEPASTELLSYMHIL--YEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQ 161
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF 313
W TL++A A R P +R+T + S + + +G+R+ K A+ VPF+F +
Sbjct: 162 WVTLIQAFAARPGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVS 221
Query: 314 SDLRELNLCDLDIKEDEALAINCVNSLHSIS----GAGNHRDLFISLLRGLEPRVLTIVE 369
+ E+ +L ++ EALA+N LH + NHRD + +++ L P+V+T+VE
Sbjct: 222 VSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVE 281
Query: 370 EEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVAC 428
+E++ + F F E + ++ FE++D + R +R+ +E+ R +V+++AC
Sbjct: 282 QESNTNT---AAFFPRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIAC 338
Query: 429 DPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIF 488
+ + VER E +WR R GF S V +++LLR Y + + + DG ++
Sbjct: 339 EGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDKYRLEERDG--ALY 396
Query: 489 LSWKDKPVVWASVWR 503
L W + +V + W+
Sbjct: 397 LGWMHRDLVASCAWK 411
>dbj|BAA96995.1| SCARECROW gene regulator-like [Arabidopsis thaliana]
gi|8132289|gb|AAF73237.1| phytochrome A signal
transduction 1 protein [Arabidopsis thaliana]
gi|42573614|ref|NP_974903.1| phytochrome A signal
transduction 1 (PAT1) [Arabidopsis thaliana]
gi|15238903|ref|NP_199626.1| phytochrome A signal
transduction 1 (PAT1) [Arabidopsis thaliana]
Length = 490
Score = 188 bits (478), Expect = 3e-46
Identities = 108/375 (28%), Positives = 196/375 (51%), Gaps = 12/375 (3%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
L+ A+A S+N+ +M L ++ + G+ Q+L +Y L+ L +++ +G YK L
Sbjct: 123 LVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASSGSSIYKAL 182
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
E + + +L EV P+ FG+++ANGAI EA++ ++HIID +Q
Sbjct: 183 NRCPEPASTELLSYMHIL--YEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQ 240
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF 313
W TL++A A R P +R+T + S + + +G+R+ K A+ VPF+F +
Sbjct: 241 WVTLIQAFAARPGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVS 300
Query: 314 SDLRELNLCDLDIKEDEALAINCVNSLHSIS----GAGNHRDLFISLLRGLEPRVLTIVE 369
+ E+ +L ++ EALA+N LH + NHRD + +++ L P+V+T+VE
Sbjct: 301 VSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVE 360
Query: 370 EEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVAC 428
+E++ + F F E + ++ FE++D + R +R+ +E+ R +V+++AC
Sbjct: 361 QESNTNT---AAFFPRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIAC 417
Query: 429 DPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIF 488
+ + VER E +WR R GF S V +++LLR Y + + + DG ++
Sbjct: 418 EGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDKYRLEERDG--ALY 475
Query: 489 LSWKDKPVVWASVWR 503
L W + +V + W+
Sbjct: 476 LGWMHRDLVASCAWK 490
>emb|CAJ00010.1| Nodulation Signaling Pathway 1 protein [Medicago truncatula]
gi|66947619|emb|CAJ00005.1| Nodulation Signaling Pathway
1 protein [Medicago truncatula]
Length = 554
Score = 182 bits (463), Expect = 2e-44
Identities = 147/520 (28%), Positives = 246/520 (47%), Gaps = 44/520 (8%)
Query: 11 QQQQQQYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMD--HNNNN 68
Q QYQ D + NS+ ++++ ++ TT + + FNN + D N
Sbjct: 46 QDISNQYQIDANTNSSNATNSTTNIVAASTTT----STTSLEPNSFNNIPFSDLPKKRNA 101
Query: 69 DEDLSSSSSKQHYYTYPYASTTTITTPN---TTYNTINTPTTTDNYSFSPSH--DYFNFE 123
+++LS Q+ S + N T+ + + + ++ + N
Sbjct: 102 EDELSLKKQPQNQKNKRLKSRPMNESDNGDAALEGTVVRKSGGNKKGAAKANGSNSNNGN 161
Query: 124 FSGHSWSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMN 183
W++ +L A A + N NR+Q L+++L+EL++ GD + +L+++ L+AL ++
Sbjct: 162 NKDGRWAEQLLNPCAVAITGGNLNRVQHLLYVLHELASTTGDANHRLAAHGLRALTHHLS 221
Query: 184 DAGDRTYK-TLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPK-- 240
+ T T+T AS + F +K LLKF E SPW +F + AN +IL+ L P
Sbjct: 222 SSSSSTPSGTITFASTEPRFF---QKSLLKFYEFSPWFSFPNNIANASILQVLAEEPNNL 278
Query: 241 --LHIIDISNTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKE------- 291
LHI+DI ++ QWPT LEAL+ R P L TVV A S + M+
Sbjct: 279 RTLHILDIGVSHGVQWPTFLEALSRRPGGPPPLVRLTVVNASSSTENDQNMETPFSIGPC 338
Query: 292 ---IGSRMEKFARLMGVPFKFKIIFSD-LRELNLCDLDIKEDEALAINCVNSLHSISGAG 347
S + +A+ + V + K + + L+ LN +D DE L + LH ++
Sbjct: 339 GDTFSSGLLGYAQSLNVNLQIKKLDNHPLQTLNAKSVDTSSDETLIVCAQFRLHHLNHNN 398
Query: 348 -NHRDLFISLLRGLEPRVLTIVEEEADLEVCFGS--DFVEGFKECLRWFRVYFEALDESF 404
+ R F+ +LRG+EP+ ++ E ++E C S DF GF + + + ++ +F
Sbjct: 399 PDERSEFLKVLRGMEPK--GVILSENNMECCCSSCGDFATGFSRRVEYLWRFLDSTSSAF 456
Query: 405 -SRTSSERLMLEREAGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCD 463
+R S ER M+E EA + + + E ERRE +W R+ GF F ++ D
Sbjct: 457 KNRDSDERKMMEGEAAKALTN-----QREMNERRE---KWCERMKEAGFAGEVFGEDAID 508
Query: 464 DVRALLRRYKEGWSMTSSDGDTGIFLSWKDKPVVWASVWR 503
RALLR+Y W M + T + L WK +PV + S+W+
Sbjct: 509 GGRALLRKYDNNWEMKVEENSTSVELWWKSQPVSFCSLWK 548
>emb|CAJ00015.1| Nodulation Signaling Pathway 1 protein homologue 2 [Populus
trichocarpa]
Length = 562
Score = 181 bits (458), Expect = 6e-44
Identities = 126/395 (31%), Positives = 202/395 (50%), Gaps = 33/395 (8%)
Query: 129 WSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDR 188
W++ +L A A + N +R+Q L+++L+EL++P GD + +L+++ L+AL ++ +
Sbjct: 173 WAEQLLNPCAAAITIGNLSRVQHLLYVLHELASPTGDANHRLAAHGLRALTHHLSSSS-- 230
Query: 189 TYKTLTTASEKTCSFDST-----RKMLLKFQEVSPWTTFGHVAANGAILEAL--EGNPK- 240
T+++AS T +F ST +K LLKF EVSPW F + AN +IL+ L E +P
Sbjct: 231 ---TVSSASIGTVTFASTEPKFFQKALLKFYEVSPWFAFPNNIANASILQVLGQEQDPTR 287
Query: 241 -LHIIDISNTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGG--------SVQKVMKE 291
LHI+DI ++ QWPTLLEAL R P L TV+TA + S+
Sbjct: 288 ILHILDIGVSHGVQWPTLLEALTRRPGGPPPLVRITVITATTENDQSTEPPFSIGPPGDN 347
Query: 292 IGSRMEKFARLMGVPFKFKIIFS-DLRELNLCDLDIKEDEALAINCVNSLHSIS-GAGNH 349
SR+ FA+ M + + K + L++L+ +D K +EAL + LH ++ +
Sbjct: 348 FPSRLLGFAKFMNINLEIKRLDGYPLQKLSGRIIDAKPEEALIVCAQFRLHHLNHNTPDE 407
Query: 350 RDLFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESF-SRTS 408
R F +LR LEP+ + + E D DF GF +++ + ++ +F R S
Sbjct: 408 RTEFFRVLRRLEPKGVILSENNMDCSCNSCGDFATGFSRRVQYLWRFLDSTSSAFKGRES 467
Query: 409 SERLMLEREAGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRAL 468
ER M+E E + + + E E +W R+ G GF F ++ D RAL
Sbjct: 468 EERRMMEGEGSKALTN--------RGEMNEGIDKWCERMQGVGFVGEMFGEDAIDGARAL 519
Query: 469 LRRYKEGWSMTSSDGDTGIFLSWKDKPVVWASVWR 503
LR+Y W M + D + L WK +PV + S+W+
Sbjct: 520 LRKYDGNWEMRTGGKDGCVGLWWKGQPVSFCSLWK 554
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 888,211,101
Number of Sequences: 2540612
Number of extensions: 37802695
Number of successful extensions: 216684
Number of sequences better than 10.0: 1060
Number of HSP's better than 10.0 without gapping: 357
Number of HSP's successfully gapped in prelim test: 761
Number of HSP's that attempted gapping in prelim test: 197893
Number of HSP's gapped (non-prelim): 9157
length of query: 504
length of database: 863,360,394
effective HSP length: 132
effective length of query: 372
effective length of database: 527,999,610
effective search space: 196415854920
effective search space used: 196415854920
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC147000.1


