

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.5 - phase: 0
(191 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
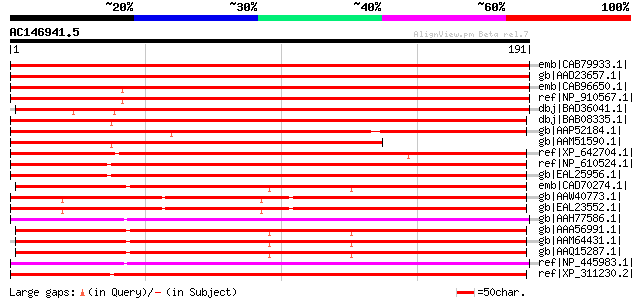
emb|CAB79933.1| synaptobrevin-like protein [Arabidopsis thaliana... 328 5e-89
gb|AAD23657.1| putative synaptobrevin [Arabidopsis thaliana] gi|... 323 2e-87
emb|CAB96650.1| putative protein [Arabidopsis thaliana] gi|21689... 316 2e-85
ref|NP_910567.1| ESTs AU082579(S2069),D40238(S2069) correspond t... 303 2e-81
dbj|BAD36041.1| putative synaptobrevin 1 [Oryza sativa (japonica... 280 2e-74
dbj|BAB08335.1| synaptobrevin-like protein [Arabidopsis thaliana... 250 2e-65
gb|AAP52184.1| putative synaptobrevin-like protein [Oryza sativa... 236 3e-61
gb|AAM51590.1| AT5g22360/MWD9_16 [Arabidopsis thaliana] gi|16323... 181 1e-44
ref|XP_642704.1| hypothetical protein DDB0169086 [Dictyostelium ... 176 2e-43
ref|NP_610524.1| CG1599-PA [Drosophila melanogaster] gi|7303846|... 154 1e-36
gb|EAL25956.1| GA14039-PA [Drosophila pseudoobscura] 154 2e-36
emb|CAD70274.1| synaptobrevin 1 [Oryza sativa (japonica cultivar... 149 3e-35
gb|AAW40773.1| vesicle-associated membrane protein 712, putative... 149 4e-35
gb|EAL23552.1| hypothetical protein CNBA1990 [Cryptococcus neofo... 149 4e-35
gb|AAH77586.1| Sybl1-prov protein [Xenopus laevis] 147 1e-34
gb|AAA56991.1| formerly called HAT24; synaptobrevin-related protein 147 2e-34
gb|AAM64431.1| putative synaptobrevin [Arabidopsis thaliana] gi|... 147 2e-34
gb|AAQ15287.1| synptobrevin-related protein [Pyrus pyrifolia] 146 3e-34
ref|NP_445983.1| synaptobrevin-like 1 [Rattus norvegicus] gi|113... 145 4e-34
ref|XP_311230.2| ENSANGP00000011948 [Anopheles gambiae str. PEST... 145 8e-34
>emb|CAB79933.1| synaptobrevin-like protein [Arabidopsis thaliana]
gi|3858935|emb|CAA16574.1| synaptobrevin-like protein
[Arabidopsis thaliana] gi|21593706|gb|AAM65673.1|
synaptobrevin-like protein [Arabidopsis thaliana]
gi|21928069|gb|AAM78063.1| AT4g32150/F10N7_40
[Arabidopsis thaliana] gi|15236687|ref|NP_194942.1|
synaptobrevin family protein [Arabidopsis thaliana]
gi|16612292|gb|AAL27509.1| AT4g32150/F10N7_40
[Arabidopsis thaliana] gi|4103357|gb|AAD01748.1|
vesicle-associated membrane protein 7C; synaptobrevin 7C
[Arabidopsis thaliana] gi|7488359|pir||T04630
synaptobrevin homolog F10N7.40 - Arabidopsis thaliana
gi|27805754|sp|O49377|V711_ARATH Vesicle-associated
membrane protein 711 (AtVAMP711) (v-SNARE synaptobrevin
7C) (AtVAMP7C)
Length = 219
Score = 328 bits (841), Expect = 5e-89
Identities = 158/191 (82%), Positives = 181/191 (94%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILYALVARGTVVL+EFT T+TNA+ IA+QILEK+PG+ND++VSYSQDRY+FHVKRTDG
Sbjct: 1 MAILYALVARGTVVLSEFTATSTNASTIAKQILEKVPGDNDSNVSYSQDRYVFHVKRTDG 60
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
LTVLCMA+++ GRRIPFAFLE+IHQRFVR+YGRAV TA AYAMN+EFSRVL+QQ++Y+S+
Sbjct: 61 LTVLCMAEETAGRRIPFAFLEDIHQRFVRTYGRAVHTALAYAMNEEFSRVLSQQIDYYSN 120
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
DPNADRINR+KGEM+QVR VMIENIDKVLDRG+RLELLVDK ANMQGNTFRFRKQARRFR
Sbjct: 121 DPNADRINRIKGEMNQVRGVMIENIDKVLDRGERLELLVDKTANMQGNTFRFRKQARRFR 180
Query: 181 STVWWRNVKLT 191
S VWWRN KLT
Sbjct: 181 SNVWWRNCKLT 191
>gb|AAD23657.1| putative synaptobrevin [Arabidopsis thaliana]
gi|25412302|pir||C84647 probable synaptobrevin
[imported] - Arabidopsis thaliana
gi|15224711|ref|NP_180106.1| synaptobrevin family
protein [Arabidopsis thaliana]
gi|27805766|sp|Q9SIQ9|V712_ARATH Vesicle-associated
membrane protein 712 (AtVAMP712)
Length = 219
Score = 323 bits (828), Expect = 2e-87
Identities = 154/191 (80%), Positives = 177/191 (92%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILYALVARGTVVLAE + T+TNA+ IA+QILEKIPGN D+HVSYSQDRY+FHVKRTDG
Sbjct: 1 MSILYALVARGTVVLAELSTTSTNASTIAKQILEKIPGNGDSHVSYSQDRYVFHVKRTDG 60
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
LTVLCMAD+ GRRIPF+FLE+IHQRFVR+YGRA+ +A+AYAMNDEFSRVLNQQ+EY+S+
Sbjct: 61 LTVLCMADEDAGRRIPFSFLEDIHQRFVRTYGRAIHSAQAYAMNDEFSRVLNQQIEYYSN 120
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
DPNAD I+R+KGEM+QVR+VMIENID +LDRG+RLELLVDK ANMQGNTFRFRKQ RRF
Sbjct: 121 DPNADTISRIKGEMNQVRDVMIENIDNILDRGERLELLVDKTANMQGNTFRFRKQTRRFN 180
Query: 181 STVWWRNVKLT 191
+TVWWRN KLT
Sbjct: 181 NTVWWRNCKLT 191
>emb|CAB96650.1| putative protein [Arabidopsis thaliana] gi|21689691|gb|AAM67467.1|
unknown protein [Arabidopsis thaliana]
gi|20259407|gb|AAM14024.1| unknown protein [Arabidopsis
thaliana] gi|15238971|ref|NP_196676.1| synaptobrevin /
vesicle-associated membrane protein 713 (VAMP713)
[Arabidopsis thaliana] gi|27805763|sp|Q9LFP1|V713_ARATH
Vesicle-associated membrane protein 713 (AtVAMP713)
Length = 221
Score = 316 bits (809), Expect = 2e-85
Identities = 149/192 (77%), Positives = 183/192 (94%), Gaps = 1/192 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNN-DTHVSYSQDRYIFHVKRTD 59
M I++ALVARGTVVL+EF+ T+TNA++I++QILEK+PGN+ D+H+SYSQDRYIFHVKRTD
Sbjct: 1 MAIIFALVARGTVVLSEFSATSTNASSISKQILEKLPGNDSDSHMSYSQDRYIFHVKRTD 60
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFS 119
GLTVLCMAD++ GR IPFAFL++IHQRFV++YGRA+ +A+AY+MNDEFSRVL+QQME++S
Sbjct: 61 GLTVLCMADETAGRNIPFAFLDDIHQRFVKTYGRAIHSAQAYSMNDEFSRVLSQQMEFYS 120
Query: 120 SDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRF 179
+DPNADR++R+KGEMSQVRNVMIENIDKVLDRG+RLELLVDK NMQGNTFRFRKQARR+
Sbjct: 121 NDPNADRMSRIKGEMSQVRNVMIENIDKVLDRGERLELLVDKTENMQGNTFRFRKQARRY 180
Query: 180 RSTVWWRNVKLT 191
R+ +WWRNVKLT
Sbjct: 181 RTIMWWRNVKLT 192
>ref|NP_910567.1| ESTs AU082579(S2069),D40238(S2069) correspond to a region of the
predicted gene.~Similar to Arabidopsis thaliana
vesicle-associated membrane protein 7C; synaptobrevin
7C. (AF025332) [Oryza sativa (japonica cultivar-group)]
gi|8096311|dbj|BAA95814.1| putative synaptobrevin 1
[Oryza sativa (japonica cultivar-group)]
Length = 221
Score = 303 bits (776), Expect = 2e-81
Identities = 142/192 (73%), Positives = 172/192 (88%), Gaps = 1/192 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNN-DTHVSYSQDRYIFHVKRTD 59
M ILYA+VARGTVVLAE + TNA A+ARQ+LE++PG D+HVSY+QDRY+FH KRTD
Sbjct: 1 MAILYAVVARGTVVLAEHSAAATNAGAVARQVLERLPGGGADSHVSYTQDRYVFHAKRTD 60
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFS 119
G+T LCMAD++ GRRIPFAFLE+IH RFV++YGRA LT+ AY MNDEFSRVL+QQM+Y+S
Sbjct: 61 GITALCMADEAAGRRIPFAFLEDIHGRFVKTYGRAALTSLAYGMNDEFSRVLSQQMDYYS 120
Query: 120 SDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRF 179
+DPNADRINR++GE+SQVR VMI+NIDKVL+RGDRL++LVDK ANMQGNT RF++QARRF
Sbjct: 121 NDPNADRINRMRGEISQVRTVMIDNIDKVLERGDRLDMLVDKTANMQGNTIRFKRQARRF 180
Query: 180 RSTVWWRNVKLT 191
R+T WWRNVKLT
Sbjct: 181 RNTTWWRNVKLT 192
>dbj|BAD36041.1| putative synaptobrevin 1 [Oryza sativa (japonica cultivar-group)]
Length = 226
Score = 280 bits (715), Expect = 2e-74
Identities = 138/195 (70%), Positives = 164/195 (83%), Gaps = 6/195 (3%)
Query: 3 ILYALVARGTVVLAEFTGTT-TNAAAIARQILEKIP-----GNNDTHVSYSQDRYIFHVK 56
I YA VARG VV+AE NA A+ARQIL+++ G D ++SY+QD ++FHVK
Sbjct: 4 IQYAAVARGAVVMAEHGDAAFPNAGAVARQILDRLSAGDGGGGGDCNISYTQDLHVFHVK 63
Query: 57 RTDGLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQME 116
RTDG+T LCMADD+ GRRIPFAFLE+IH RFV++YGRA LTA AYAMNDEFSRVL QQM+
Sbjct: 64 RTDGVTALCMADDAAGRRIPFAFLEDIHGRFVKTYGRAALTALAYAMNDEFSRVLGQQMD 123
Query: 117 YFSSDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQA 176
Y+S+DPNADRI+R++GEM QVRNVMI+NIDKVL+RGDRLELLVDK A MQGNT RF++QA
Sbjct: 124 YYSNDPNADRISRMRGEMDQVRNVMIDNIDKVLERGDRLELLVDKTATMQGNTMRFKRQA 183
Query: 177 RRFRSTVWWRNVKLT 191
RRFR+TVWWRNVKLT
Sbjct: 184 RRFRNTVWWRNVKLT 198
>dbj|BAB08335.1| synaptobrevin-like protein [Arabidopsis thaliana]
gi|15242224|ref|NP_197628.1| synaptobrevin family
protein [Arabidopsis thaliana]
gi|27805762|sp|Q9FMR5|V714_ARATH Vesicle-associated
membrane protein 714 (AtVAMP714)
Length = 221
Score = 250 bits (638), Expect = 2e-65
Identities = 115/191 (60%), Positives = 158/191 (82%), Gaps = 1/191 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKI-PGNNDTHVSYSQDRYIFHVKRTD 59
M I+YA+VARGTVVLAEF+ T N A+ R+ILEK+ P +D + +SQDRYIFH+ R+D
Sbjct: 1 MAIVYAVVARGTVVLAEFSAVTGNTGAVVRRILEKLSPEISDERLCFSQDRYIFHILRSD 60
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFS 119
GLT LCMA+D+ GRR+PF++LEEIH RF+++YG+ A AYAMNDEFSRVL+QQME+FS
Sbjct: 61 GLTFLCMANDTFGRRVPFSYLEEIHMRFMKNYGKVAHNAPAYAMNDEFSRVLHQQMEFFS 120
Query: 120 SDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRF 179
S+P+ D +NR++GE+S++R+VM+ENI+K+++RGDR+ELLVDK A MQ ++F FRKQ++R
Sbjct: 121 SNPSVDTLNRVRGEVSEIRSVMVENIEKIMERGDRIELLVDKTATMQDSSFHFRKQSKRL 180
Query: 180 RSTVWWRNVKL 190
R +W +N KL
Sbjct: 181 RRALWMKNAKL 191
>gb|AAP52184.1| putative synaptobrevin-like protein [Oryza sativa (japonica
cultivar-group)] gi|37531190|ref|NP_919897.1| putative
synaptobrevin-like protein [Oryza sativa (japonica
cultivar-group)] gi|20177639|gb|AAM14694.1| Putative
synaptobrevin-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 223
Score = 236 bits (601), Expect = 3e-61
Identities = 115/196 (58%), Positives = 153/196 (77%), Gaps = 9/196 (4%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRT-- 58
M I+YA+VARGTVVLAEF+ + NA A+AR+ILEK+P + ++ + ++QDRYIFHV R+
Sbjct: 1 MAIVYAVVARGTVVLAEFSAVSGNAGAVARRILEKLPPDAESRLCFAQDRYIFHVLRSPP 60
Query: 59 ----DGLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQ 114
DGLT LCMA+D+ GRRIPF +LE+I RF+++YGR A AYAMNDEFSRVL+QQ
Sbjct: 61 PAAADGLTFLCMANDTFGRRIPFLYLEDIQMRFIKNYGRIAHNALAYAMNDEFSRVLHQQ 120
Query: 115 MEYFSSDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRK 174
MEYFSS+P+AD +NRL+GE + VM++NI+K+LDRG+R+ LLVDK + MQ + F FRK
Sbjct: 121 MEYFSSNPSADTLNRLRGE---IHTVMVDNIEKILDRGERISLLVDKTSTMQDSAFHFRK 177
Query: 175 QARRFRSTVWWRNVKL 190
Q+RR R +W +N KL
Sbjct: 178 QSRRLRRALWMKNAKL 193
>gb|AAM51590.1| AT5g22360/MWD9_16 [Arabidopsis thaliana] gi|16323190|gb|AAL15329.1|
AT5g22360/MWD9_16 [Arabidopsis thaliana]
Length = 138
Score = 181 bits (458), Expect = 1e-44
Identities = 85/138 (61%), Positives = 114/138 (82%), Gaps = 1/138 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKI-PGNNDTHVSYSQDRYIFHVKRTD 59
M I+YA+VARGTVVLAEF+ T N A+ R+ILEK+ P +D + +SQDRYIFH+ R+D
Sbjct: 1 MAIVYAVVARGTVVLAEFSAVTGNTGAVVRRILEKLSPEISDERLCFSQDRYIFHILRSD 60
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFS 119
GLT LCMA+D+ GRR+PF++LEEIH RF+++YG+ A AYAMNDEFSRVL+QQME+FS
Sbjct: 61 GLTFLCMANDTFGRRVPFSYLEEIHMRFMKNYGKVAHNAPAYAMNDEFSRVLHQQMEFFS 120
Query: 120 SDPNADRINRLKGEMSQV 137
S+P+ D +NR++GE+S+V
Sbjct: 121 SNPSVDTLNRVRGEVSEV 138
>ref|XP_642704.1| hypothetical protein DDB0169086 [Dictyostelium discoideum]
gi|28828593|gb|AAO51196.1| similar to Arabidopsis
thaliana (Mouse-ear cress). Synaptobrevin-like protein
[Dictyostelium discoideum] gi|60470800|gb|EAL68772.1|
hypothetical protein DDB0169086 [Dictyostelium
discoideum]
Length = 260
Score = 176 bits (447), Expect = 2e-43
Identities = 85/191 (44%), Positives = 130/191 (67%), Gaps = 2/191 (1%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M I+Y+LVARG+ VLAEFT T N I R+IL+ IP N DT +SY ++YIFH +D
Sbjct: 1 MPIIYSLVARGSSVLAEFTSTNGNFVTITRRILDLIPPN-DTKMSYVYEKYIFHYLVSDT 59
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
LT LCMAD+ GRRIPF FL+++ RF YG TA AY MN +FSR L M+++S+
Sbjct: 60 LTYLCMADEEFGRRIPFTFLDDVKNRFKSMYGDKGKTAIAYGMNSDFSRTLENLMDHYSN 119
Query: 121 DPNADRINRLKGEMSQVRNVMIENI-DKVLDRGDRLELLVDKAANMQGNTFRFRKQARRF 179
D ++R E+ +V+N+++ +I ++L RG+++E+LV++ + +F+F+KQ+++
Sbjct: 120 TTRVDTMSRTMAEIDEVKNILVSDIAPQLLKRGEKIEMLVERTDTLNQQSFKFKKQSKQL 179
Query: 180 RSTVWWRNVKL 190
+ +WW+NVKL
Sbjct: 180 KCAMWWKNVKL 190
>ref|NP_610524.1| CG1599-PA [Drosophila melanogaster] gi|7303846|gb|AAF58892.1|
CG1599-PA [Drosophila melanogaster]
gi|17946573|gb|AAL49317.1| RH15778p [Drosophila
melanogaster]
Length = 218
Score = 154 bits (390), Expect = 1e-36
Identities = 73/190 (38%), Positives = 123/190 (64%), Gaps = 1/190 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILY++++RGT VLA+F N A + I+ +I G ++ ++Y+ Y+ H +
Sbjct: 1 MPILYSVISRGTTVLAKFAECVGNFAEVTEHIIGRI-GVHNHKMTYTHGDYLIHYTCENK 59
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
L +C+ D+ R F FL +I Q+F+++YG V TA AY+MN EFS++L QQM YFS
Sbjct: 60 LVYMCITDNEFERSRAFLFLADIKQKFIQTYGLQVATAIAYSMNTEFSKILAQQMVYFSQ 119
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
D I+R+ G++ +++++M++NID + DRG++LELLV+K N+ N+ FRK +R
Sbjct: 120 SREVDTISRVHGQIDELKDIMVKNIDSLRDRGEKLELLVNKTENLSNNSVAFRKASRNLA 179
Query: 181 STVWWRNVKL 190
++W+N+++
Sbjct: 180 RQMFWKNIRV 189
>gb|EAL25956.1| GA14039-PA [Drosophila pseudoobscura]
Length = 218
Score = 154 bits (388), Expect = 2e-36
Identities = 73/190 (38%), Positives = 123/190 (64%), Gaps = 1/190 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILY++++RGT VLA+F N A + I+ +I G ++ ++Y+ Y+ H +
Sbjct: 1 MPILYSVISRGTTVLAKFAECVGNFAEVTEHIIGRI-GVHNHKMTYTHGDYLIHYSCDNK 59
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
L +C+ D+ + F FL +I Q+F+++YG V TA AYAMN EFS+VL QQM YFS
Sbjct: 60 LVYMCITDNEFEKSRAFFFLADIKQKFIQTYGLQVATAIAYAMNTEFSKVLAQQMVYFSQ 119
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
+ D I+R+ G++ +++++M++NID + DRG++LELLV+K N+ N+ FRK +R
Sbjct: 120 SSDVDNISRVHGQIDELKDIMVKNIDSLRDRGEKLELLVNKTENLSNNSVAFRKASRNLA 179
Query: 181 STVWWRNVKL 190
++W+ +++
Sbjct: 180 RQMFWKKIRV 189
>emb|CAD70274.1| synaptobrevin 1 [Oryza sativa (japonica cultivar-group)]
gi|50509098|dbj|BAD30158.1| synaptobrevin-like protein
[Oryza sativa (japonica cultivar-group)]
Length = 220
Score = 149 bits (377), Expect = 3e-35
Identities = 79/190 (41%), Positives = 122/190 (63%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++YA VARGTVVLAE+T T N IA Q L+K+P +N+ +Y+ D + F+ DG T
Sbjct: 6 LIYAFVARGTVVLAEYTEFTGNFTTIAAQCLQKLPASNNKF-TYNCDGHTFNYLVEDGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A +SVGR+IP AFL+ + + F + YG TA A ++N EF L + M+Y
Sbjct: 65 YCVVAVESVGRQIPIAFLDRVKEDFTKRYGGGKAATAAANSLNREFGSKLKEHMQYCVDH 124
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P ++ ++K ++S+V+ VM+ENI+KVLDRG+++ELLVDK N++ FR+Q + R
Sbjct: 125 PEEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRQQGTKVR 184
Query: 181 STVWWRNVKL 190
+W +N+K+
Sbjct: 185 RKMWLQNMKI 194
>gb|AAW40773.1| vesicle-associated membrane protein 712, putative [Cryptococcus
neoformans var. neoformans JEC21]
gi|58258359|ref|XP_566592.1| vesicle-associated membrane
protein 712, putative [Cryptococcus neoformans var.
neoformans JEC21]
Length = 306
Score = 149 bits (376), Expect = 4e-35
Identities = 80/192 (41%), Positives = 123/192 (63%), Gaps = 4/192 (2%)
Query: 1 MGILYALVARGTVVLAEF-TGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTD 59
M +++AL+ARGT VLAE TGT A IL KIP NN QDR I +V ++
Sbjct: 88 MSLIHALIARGTTVLAEHATGTAELKPAAQITILSKIPPNNSKLTYVWQDRLIHYVS-SN 146
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSY-GRAVLTAEAYAMNDEFSRVLNQQMEYF 118
G+ L MADDSVGRR+PFAFL ++ +RF Y +++A A+++ +EF L + M +
Sbjct: 147 GVIYLVMADDSVGRRMPFAFLADLERRFTAQYESDDIVSAGAHSL-EEFEPELAKLMHQY 205
Query: 119 SSDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARR 178
+S P AD + + + +++ V+++M++NID +L RG+RL+LLVDK + G + FR+ AR
Sbjct: 206 TSSPPADPLRQAQSDLNNVKDIMVQNIDSILQRGERLDLLVDKTDTLAGQAYAFRRGARS 265
Query: 179 FRSTVWWRNVKL 190
R WW+N+++
Sbjct: 266 VRRQQWWKNMRI 277
>gb|EAL23552.1| hypothetical protein CNBA1990 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 306
Score = 149 bits (376), Expect = 4e-35
Identities = 80/192 (41%), Positives = 123/192 (63%), Gaps = 4/192 (2%)
Query: 1 MGILYALVARGTVVLAEF-TGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTD 59
M +++AL+ARGT VLAE TGT A IL KIP NN QDR I +V ++
Sbjct: 88 MSLIHALIARGTTVLAEHATGTAELKPAAQITILSKIPPNNSKLTYVWQDRLIHYVS-SN 146
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSY-GRAVLTAEAYAMNDEFSRVLNQQMEYF 118
G+ L MADDSVGRR+PFAFL ++ +RF Y +++A A+++ +EF L + M +
Sbjct: 147 GVIYLVMADDSVGRRMPFAFLADLERRFTAQYESDDIVSAGAHSL-EEFEPELAKLMHQY 205
Query: 119 SSDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARR 178
+S P AD + + + +++ V+++M++NID +L RG+RL+LLVDK + G + FR+ AR
Sbjct: 206 TSSPPADPLRQAQSDLNNVKDIMVQNIDSILQRGERLDLLVDKTDTLAGQAYAFRRGARS 265
Query: 179 FRSTVWWRNVKL 190
R WW+N+++
Sbjct: 266 VRRQQWWKNMRI 277
>gb|AAH77586.1| Sybl1-prov protein [Xenopus laevis]
Length = 220
Score = 147 bits (372), Expect = 1e-34
Identities = 76/191 (39%), Positives = 113/191 (58%), Gaps = 1/191 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M IL+A+VARGT +LA+ N + QIL KIP N+ ++YS Y+FH D
Sbjct: 1 MAILFAVVARGTTILAKHAWCGGNFLEVTEQILAKIPSENNK-LTYSHGSYLFHYMCQDR 59
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
+ LC+ DD R F FL EI +RF +YG TA YAMN EFS VL+ Q++Y S
Sbjct: 60 IIYLCITDDDFERSRAFNFLNEIKKRFQTTYGSRAQTALPYAMNSEFSSVLSAQLKYHSE 119
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
+ + DR+ + ++ +++ +M+ NID V RG+RLELL+DK N+ ++ F+ +R
Sbjct: 120 NKSVDRVAETQAQVDELKGIMVRNIDLVAQRGERLELLIDKTENLVDSSVTFKTTSRNLA 179
Query: 181 STVWWRNVKLT 191
+ +N+KLT
Sbjct: 180 RAMCMKNLKLT 190
>gb|AAA56991.1| formerly called HAT24; synaptobrevin-related protein
Length = 221
Score = 147 bits (371), Expect = 2e-34
Identities = 76/190 (40%), Positives = 123/190 (64%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++Y+ VARGTV+L EFT N +IA Q L+K+P +N+ +Y+ D + F+ +G T
Sbjct: 6 LIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKF-TYNCDGHTFNYLVENGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A DS GR+IP AFLE + + F + YG TA+A ++N EF L + M+Y +
Sbjct: 65 YCVVAVDSAGRQIPMAFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQYCMAH 124
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P+ ++ ++K ++S+V+ VM+ENI+KVLDRG+++ELLVDK N++ FR Q + R
Sbjct: 125 PDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQGTQMR 184
Query: 181 STVWWRNVKL 190
+W++N+K+
Sbjct: 185 RKMWFQNMKI 194
>gb|AAM64431.1| putative synaptobrevin [Arabidopsis thaliana]
gi|22135820|gb|AAM91096.1| At2g33120/F25I18.14
[Arabidopsis thaliana] gi|21389653|gb|AAM48025.1|
putative synaptobrevin [Arabidopsis thaliana]
gi|2924792|gb|AAC04921.1| putative synaptobrevin
[Arabidopsis thaliana] gi|18958028|gb|AAL79587.1|
At2g33120/F25I18.14 [Arabidopsis thaliana]
gi|18252975|gb|AAL62414.1| putative synaptobrevin
[Arabidopsis thaliana] gi|16930421|gb|AAL31896.1|
At2g33120/F25I18.14 [Arabidopsis thaliana]
gi|25408247|pir||F84741 probable synaptobrevin
[imported] - Arabidopsis thaliana
gi|15225796|ref|NP_180871.1| synaptobrevin-related
protein / vesicle-associated membrane protein 722
(VAMP722) (SAR1) [Arabidopsis thaliana]
gi|21542460|sp|P47192|V722_ARATH Vesicle-associated
membrane protein 722 (AtVAMP722) (Synaptobrevin-related
protein 1)
Length = 221
Score = 147 bits (370), Expect = 2e-34
Identities = 76/190 (40%), Positives = 122/190 (64%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++Y+ VARGTV+L EFT N +IA Q L+K+P +N+ +Y+ D + F+ +G T
Sbjct: 6 LIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKF-TYNCDGHTFNYLVENGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A DS GR+IP AFLE + + F + YG TA+A ++N EF L + M+Y
Sbjct: 65 YCVVAVDSAGRQIPMAFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQYCMDH 124
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P+ ++ ++K ++S+V+ VM+ENI+KVLDRG+++ELLVDK N++ FR Q + R
Sbjct: 125 PDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQGTQMR 184
Query: 181 STVWWRNVKL 190
+W++N+K+
Sbjct: 185 RKMWFQNMKI 194
>gb|AAQ15287.1| synptobrevin-related protein [Pyrus pyrifolia]
Length = 194
Score = 146 bits (369), Expect = 3e-34
Identities = 77/190 (40%), Positives = 122/190 (63%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++Y+ VARGTV+LAE+T T N +IA Q L+K+P N+ +Y+ D + F+ +G T
Sbjct: 6 LIYSFVARGTVILAEYTEFTGNFTSIASQCLQKLPATNNKF-TYNCDGHTFNYLVDNGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A ++VGR++P AFLE I + F YG TA A ++N EF L + M+Y
Sbjct: 65 YCVVAVEAVGRQVPIAFLERIKEDFTGRYGGGKAATAVANSLNKEFGSKLKEHMQYCVDH 124
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P +++++K ++S+V+ VM+ENI+KVLDRG+++ELLVDK N++ FR+Q + R
Sbjct: 125 PEEISKLSKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRQQGTQMR 184
Query: 181 STVWWRNVKL 190
+W +N+KL
Sbjct: 185 RKMWLQNMKL 194
>ref|NP_445983.1| synaptobrevin-like 1 [Rattus norvegicus] gi|11360410|pir||JC7258
vesicle-associated membrane protein-7 - rat
gi|9502258|gb|AAF88059.1| vesicle-associated membrane
protein 7 [Rattus norvegicus]
Length = 220
Score = 145 bits (367), Expect = 4e-34
Identities = 75/191 (39%), Positives = 112/191 (58%), Gaps = 1/191 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M IL+A+VARGT +LA+ N + QIL KIP N+ ++YS Y+FH D
Sbjct: 1 MAILFAVVARGTTILAKHAWCGGNFLEVTEQILAKIPSENNK-LTYSHGNYLFHYICQDR 59
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
+ LC+ DD R F FL E+ +RF +YG TA YAMN EFS VL Q+++ S
Sbjct: 60 IVYLCITDDDFERSRAFGFLNEVKKRFQTTYGSRAQTALPYAMNSEFSSVLAAQLKHHSE 119
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
+ + DR+ + ++ +++ +M+ NID V RG+RLELL+DK N+ ++ F+ +R
Sbjct: 120 NQSLDRVTETQAQVDELKGIMVRNIDLVAQRGERLELLIDKTENLVDSSVTFKTTSRNLA 179
Query: 181 STVWWRNVKLT 191
+ +NVKLT
Sbjct: 180 RAMCVKNVKLT 190
>ref|XP_311230.2| ENSANGP00000011948 [Anopheles gambiae str. PEST]
gi|55243028|gb|EAA06868.2| ENSANGP00000011948 [Anopheles
gambiae str. PEST]
Length = 218
Score = 145 bits (365), Expect = 8e-34
Identities = 71/190 (37%), Positives = 117/190 (61%), Gaps = 1/190 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILY++V+RG VLA + N + QI+ KI D ++Y Y+ H +
Sbjct: 1 MPILYSMVSRGQTVLARYADCIGNFTEVTEQIIPKIQ-LIDHKLTYLHGNYLIHYICDNR 59
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
+ +C+ DD R F FL++I++RF+ YG V TA +YAMN EF+R L QM F+
Sbjct: 60 VIYMCITDDRFDRSRAFLFLQDINERFICMYGLTVATAISYAMNTEFARTLAAQMRQFNE 119
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
D I+R+ GE+ +++++M++NI+ + +RG+RLELLV++ N++ N+ FR+ +R
Sbjct: 120 GQEQDAISRVNGEIDELKDIMVKNIENITNRGERLELLVNQTENLRNNSVTFRQTSRNLA 179
Query: 181 STVWWRNVKL 190
T++WRNV++
Sbjct: 180 RTMFWRNVRM 189
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 288,049,737
Number of Sequences: 2540612
Number of extensions: 10772828
Number of successful extensions: 38465
Number of sequences better than 10.0: 383
Number of HSP's better than 10.0 without gapping: 356
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 37917
Number of HSP's gapped (non-prelim): 393
length of query: 191
length of database: 863,360,394
effective HSP length: 121
effective length of query: 70
effective length of database: 555,946,342
effective search space: 38916243940
effective search space used: 38916243940
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146941.5


