

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.18 + phase: 0 /pseudo/partial
(211 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
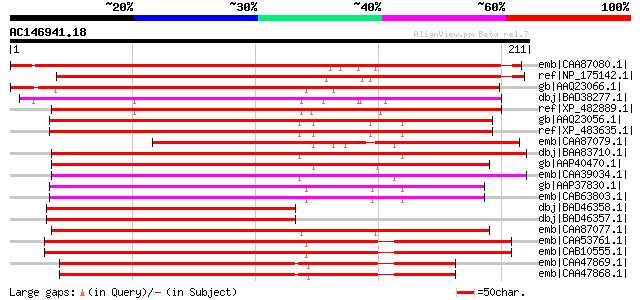
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA87080.1| heat shock transcription factor 5 [Glycine max] ... 267 2e-70
ref|NP_175142.1| heat shock transcription factor family protein ... 264 1e-69
gb|AAQ23066.1| heat shock factor RHSF12 [Oryza sativa (japonica ... 253 2e-66
dbj|BAD38277.1| putative heat shock transcription factor HSF5 [O... 226 3e-58
ref|XP_482889.1| putative heat shock transcription factor [Oryza... 210 2e-53
gb|AAQ23056.1| heat shock factor RHSF2 [Oryza sativa (japonica c... 187 2e-46
ref|XP_483635.1| putative heat shock factor RHSF2 [Oryza sativa ... 187 2e-46
emb|CAA87079.1| heat shock transcription factor 31 [Glycine max]... 181 1e-44
dbj|BAA83710.1| heat shock factor [Nicotiana tabacum] 173 3e-42
gb|AAP40470.1| putative heat shock factor 6 [Arabidopsis thalian... 171 1e-41
emb|CAA39034.1| heat stress transcription factor [Lycopersicon p... 171 2e-41
gb|AAP37830.1| At4g11660 [Arabidopsis thaliana] gi|7267866|emb|C... 171 2e-41
emb|CAB63803.1| heat shock factor 7 [Arabidopsis thaliana] 171 2e-41
dbj|BAD46358.1| putative heat shock factor [Oryza sativa (japoni... 170 3e-41
dbj|BAD46357.1| putative heat shock factor [Oryza sativa (japoni... 170 3e-41
emb|CAA87077.1| heat shock transcription factor 34 [Glycine max]... 164 1e-39
emb|CAA53761.1| heat shock factor [Arabidopsis thaliana] 162 8e-39
emb|CAB10555.1| heat shock transcription factor HSF1 [Arabidopsi... 161 1e-38
emb|CAA47869.1| heat shock transcription factor 8 [Lycopersicon ... 157 1e-37
emb|CAA47868.1| heat stress transcription factor 8 [Lycopersicon... 157 1e-37
>emb|CAA87080.1| heat shock transcription factor 5 [Glycine max]
gi|2129832|pir||S59539 heat shock transcription factor
HSF5 - soybean
Length = 370
Score = 267 bits (682), Expect = 2e-70
Identities = 141/244 (57%), Positives = 168/244 (68%), Gaps = 41/244 (16%)
Query: 1 MAFTLERCEDNMVFTMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEF 60
MA L+ CE +++ ++++ KSVPAPFLTKTYQLVDDP TDHIVSW +D+TTFVVWRPPEF
Sbjct: 1 MALLLDNCE-SILLSLDTHKSVPAPFLTKTYQLVDDPSTDHIVSWGEDDTTFVVWRPPEF 59
Query: 61 ARDLLPNFFKHNNFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQH 120
ARDLLPN+FKHNNFSSFVRQLNTYGF+K+V DRWEFAN++FKKG K+LLCEIHRRKT
Sbjct: 60 ARDLLPNYFKHNNFSSFVRQLNTYGFRKIVPDRWEFANEFFKKGEKNLLCEIHRRKTHHQ 119
Query: 121 YQQQYYEQS------------PQIF-------------QPDESIC--WIDSPLP------ 147
+ QQ + IF DE I W DSP
Sbjct: 120 HHQQVQAMNNHHHHHKFGLNVSSIFPFHNNRLSVSPSHDSDEVIIPNWCDSPPRGVAGVN 179
Query: 148 ---SPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPASPFEQR 204
S +N + +TALSEDN+RLRR N ML+SEL+HMK LYNDIIYF+QNHV P +P
Sbjct: 180 NNNSSSNNYNTVTALSEDNERLRRSNNMLMSELAHMKKLYNDIIYFVQNHVKPVAP---- 235
Query: 205 SNNS 208
SNN+
Sbjct: 236 SNNN 239
>ref|NP_175142.1| heat shock transcription factor family protein [Arabidopsis
thaliana] gi|12321016|gb|AAG50634.1| heat shock
transcription factor, putative [Arabidopsis thaliana]
gi|25296103|pir||C96511 probable heat shock
transcription factor [imported] - Arabidopsis thaliana
Length = 348
Score = 264 bits (675), Expect = 1e-69
Identities = 133/218 (61%), Positives = 155/218 (71%), Gaps = 32/218 (14%)
Query: 20 KSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVR 79
K+VPAPFLTKTYQLVDDP TDH+VSW DD+TTFVVWRPPEFARDLLPN+FKHNNFSSFVR
Sbjct: 29 KAVPAPFLTKTYQLVDDPATDHVVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVR 88
Query: 80 QLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYE------------ 127
QLNTYGF+K+V DRWEFAN++FK+G KHLLCEIHRRKT Q QQ+
Sbjct: 89 QLNTYGFRKIVPDRWEFANEFFKRGEKHLLCEIHRRKTSQMIPQQHSPFMSHHHAPPQIP 148
Query: 128 ---------QSPQIFQPDESICWI-DSP------LPSPKSNTDILTALSEDNQRLRRKNF 171
P++ P+E W DSP +P +TALSEDN+RLRR N
Sbjct: 149 FSGGSFFPLPPPRVTTPEEDHYWCDDSPPSRPRVIPQQIDTAAQVTALSEDNERLRRSNT 208
Query: 172 MLLSELSHMKNLYNDIIYFIQNHVSPASPFEQRSNNSA 209
+L+SEL+HMK LYNDIIYF+QNHV P +P SNNS+
Sbjct: 209 VLMSELAHMKKLYNDIIYFVQNHVKPVAP----SNNSS 242
>gb|AAQ23066.1| heat shock factor RHSF12 [Oryza sativa (japonica cultivar-group)]
gi|50939531|ref|XP_479293.1| putative heat shock
transcription factor HSF5 [Oryza sativa (japonica
cultivar-group)] gi|33146640|dbj|BAC79970.1| putative
heat shock transcription factor HSF5 [Oryza sativa
(japonica cultivar-group)] gi|50510174|dbj|BAD31269.1|
putative heat shock transcription factor HSF5 [Oryza
sativa (japonica cultivar-group)]
Length = 310
Score = 253 bits (647), Expect = 2e-66
Identities = 132/230 (57%), Positives = 159/230 (68%), Gaps = 32/230 (13%)
Query: 1 MAFTLERCEDNMVFTME----------SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDET 50
MAF +ERC + MV +ME + K VPAPFLTKTYQLVDDP TDHIVSW +D+T
Sbjct: 1 MAFLVERCGE-MVVSMEMGPHGGGGAAAGKPVPAPFLTKTYQLVDDPCTDHIVSWGEDDT 59
Query: 51 TFVVWRPPEFARDLLPNFFKHNNFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLC 110
TFVVWRPPEFARDLLPN+FKHNNFSSFVRQLNTYGF+K+VADRWEFAN++F+KGAKHLL
Sbjct: 60 TFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTYGFRKIVADRWEFANEFFRKGAKHLLA 119
Query: 111 EIHRRKTPQ-------------HYQQQYYEQSP--------QIFQPDESICWIDSPLPSP 149
EIHRRK+ Q H+ + P + Q + + +
Sbjct: 120 EIHRRKSSQPPPPPMPHQPYHHHHHLNPFSLPPPPPAYHHHHLIQEEPATTAHCTVAGDG 179
Query: 150 KSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPAS 199
D L ALSEDN++LRR+N +LLSEL+HMK LYNDIIYF+QNHV+P +
Sbjct: 180 GEGGDFLAALSEDNRQLRRRNSLLLSELAHMKKLYNDIIYFLQNHVAPVT 229
>dbj|BAD38277.1| putative heat shock transcription factor HSF5 [Oryza sativa
(japonica cultivar-group)]
Length = 394
Score = 226 bits (577), Expect = 3e-58
Identities = 126/253 (49%), Positives = 149/253 (58%), Gaps = 57/253 (22%)
Query: 5 LERC---EDNMVFTMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDE-TTFVVWRPPEF 60
+ERC D +QK+VPAPFLTKTYQLVDDP TDHIVSW DD +TFVVWRPPEF
Sbjct: 1 MERCGSWSDCEAAAAAAQKAVPAPFLTKTYQLVDDPATDHIVSWGDDRVSTFVVWRPPEF 60
Query: 61 ARDLLPNFFKHNNFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKT--- 117
ARD+LPN+FKHNNFSSFVRQLNTYGF+KVV +RWEFAN++F+KG K LL EIHRRKT
Sbjct: 61 ARDILPNYFKHNNFSSFVRQLNTYGFRKVVPERWEFANEFFRKGEKQLLTEIHRRKTSSA 120
Query: 118 ------------PQHYQQQYY-----EQSPQIFQPDESIC-------------W------ 141
P H+ ++ Q F D+ + W
Sbjct: 121 STASPSPPPFFAPPHFPLFHHPGVAAAQHHHAFVGDDGVVAAHGIGMPFPQPHWREPNLP 180
Query: 142 -------IDSPLPSPKS-------NTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDI 187
+ P PSP S L E+N+RLRR N LL EL+HM+ LYNDI
Sbjct: 181 VATRLLALGGPAPSPSSAEAGGAGRAATAAVLMEENERLRRSNTALLQELAHMRKLYNDI 240
Query: 188 IYFIQNHVSPASP 200
IYF+QNHV P +P
Sbjct: 241 IYFVQNHVRPVAP 253
>ref|XP_482889.1| putative heat shock transcription factor [Oryza sativa (japonica
cultivar-group)] gi|42408639|dbj|BAD09860.1| putative
heat shock transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 380
Score = 210 bits (534), Expect = 2e-53
Identities = 111/233 (47%), Positives = 143/233 (60%), Gaps = 50/233 (21%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDE-----TTFVVWRPPEFARDLLPNFFKHN 72
S VPAPFLTKTYQLVDDP TDH+VSW DD+ ++FVVWRPPEFARD+LPN+FKH+
Sbjct: 18 SASVVPAPFLTKTYQLVDDPATDHVVSWEDDDGGESASSFVVWRPPEFARDILPNYFKHS 77
Query: 73 NFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKT--------------- 117
NFSSFVRQLNTYGF+KVV +RWEFAN++F+KG K LLCEIHRRK+
Sbjct: 78 NFSSFVRQLNTYGFRKVVPERWEFANEFFRKGEKQLLCEIHRRKSAAATWPPFPPPPPPF 137
Query: 118 --PQHY-------------------------QQQYYEQSPQIFQPDESICWIDSPLPSP- 149
P+H+ ++ ++E + P + P+ +P
Sbjct: 138 FAPRHFAAGAFFRHGDGMLHGRLGALVTTTERRHWFESAALPVAPSSRLLSQLGPVIAPA 197
Query: 150 --KSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPASP 200
+ T AL ++N RL R N L+ EL+HM+ LY+DIIYF+QNHV P +P
Sbjct: 198 RRAAATPEEEALMQENHRLLRGNAALVQELAHMRKLYSDIIYFVQNHVRPVAP 250
>gb|AAQ23056.1| heat shock factor RHSF2 [Oryza sativa (japonica cultivar-group)]
Length = 616
Score = 187 bits (474), Expect = 2e-46
Identities = 99/206 (48%), Positives = 129/206 (62%), Gaps = 26/206 (12%)
Query: 17 ESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSS 76
+ Q++VP PFLTKTYQLVDDP D ++SW+DD +TFVVWRP EFARDLLP +FKHNNFSS
Sbjct: 183 QQQRTVPTPFLTKTYQLVDDPAVDDVISWNDDGSTFVVWRPAEFARDLLPKYFKHNNFSS 242
Query: 77 FVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK-TPQHYQ------------- 122
FVRQLNTYGF+K+V DRWEFAND F++G + LLCEIHRRK TP
Sbjct: 243 FVRQLNTYGFRKIVPDRWEFANDCFRRGERRLLCEIHRRKVTPPAPAATTAAVAAAIPMA 302
Query: 123 ---QQYYEQSPQIFQPDESICWIDSP--------LPSPKSNTDILTA-LSEDNQRLRRKN 170
+ SP + ++ I SP PS + + + + ++N+RLRR+N
Sbjct: 303 LPVTTTRDGSPVLSGEEQVISSSSSPEPPLVLPQAPSGSGSGGVASGDVGDENERLRREN 362
Query: 171 FMLLSELSHMKNLYNDIIYFIQNHVS 196
L ELS M+ L N+I+ + + S
Sbjct: 363 AQLARELSQMRKLCNNILLLMSKYAS 388
>ref|XP_483635.1| putative heat shock factor RHSF2 [Oryza sativa (japonica
cultivar-group)] gi|42408708|dbj|BAD09926.1| putative
heat shock factor RHSF2 [Oryza sativa (japonica
cultivar-group)] gi|42408097|dbj|BAD09238.1| putative
heat shock factor RHSF2 [Oryza sativa (japonica
cultivar-group)]
Length = 390
Score = 187 bits (474), Expect = 2e-46
Identities = 99/206 (48%), Positives = 129/206 (62%), Gaps = 26/206 (12%)
Query: 17 ESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSS 76
+ Q++VP PFLTKTYQLVDDP D ++SW+DD +TFVVWRP EFARDLLP +FKHNNFSS
Sbjct: 40 QQQRTVPTPFLTKTYQLVDDPAVDDVISWNDDGSTFVVWRPAEFARDLLPKYFKHNNFSS 99
Query: 77 FVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK-TPQHYQ------------- 122
FVRQLNTYGF+K+V DRWEFAND F++G + LLCEIHRRK TP
Sbjct: 100 FVRQLNTYGFRKIVPDRWEFANDCFRRGERRLLCEIHRRKVTPPAPAATTAAVAAAIPMA 159
Query: 123 ---QQYYEQSPQIFQPDESICWIDSP--------LPSPKSNTDILTA-LSEDNQRLRRKN 170
+ SP + ++ I SP PS + + + + ++N+RLRR+N
Sbjct: 160 LPVTTTRDGSPVLSGEEQVISSSSSPEPPLVLPQAPSGSGSGGVASGDVGDENERLRREN 219
Query: 171 FMLLSELSHMKNLYNDIIYFIQNHVS 196
L ELS M+ L N+I+ + + S
Sbjct: 220 AQLARELSQMRKLCNNILLLMSKYAS 245
>emb|CAA87079.1| heat shock transcription factor 31 [Glycine max]
gi|2129830|pir||S59540 heat shock transcription factor
HSF31 - soybean (fragment)
Length = 306
Score = 181 bits (459), Expect = 1e-44
Identities = 99/186 (53%), Positives = 118/186 (63%), Gaps = 40/186 (21%)
Query: 59 EFARDLLPNFFKHNNFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTP 118
EFAR+LLPN+FKHNNFSSFVRQLNTYGF+K+V DRWEFAN++FKKG KHLLCEIHRRKT
Sbjct: 1 EFARNLLPNYFKHNNFSSFVRQLNTYGFRKIVPDRWEFANEFFKKGEKHLLCEIHRRKTA 60
Query: 119 QHYQ-----QQYYEQSP-------QIFQP-------------DESICWIDSPLPSPKSNT 153
Q Q ++ SP F P D+ W DSP P+ T
Sbjct: 61 QPQQGIMNHHHHHAHSPLGVNVNVPTFFPFSSRVSISTSNDSDDQSNWCDSP---PRGAT 117
Query: 154 DI------------LTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPASPF 201
+ +TALSEDN+RLRR N ML+SEL+HMK LYNDIIYF+QNHV P +P
Sbjct: 118 SLVNGAAAANYNTSVTALSEDNERLRRSNNMLMSELAHMKKLYNDIIYFVQNHVKPVAPS 177
Query: 202 EQRSNN 207
S++
Sbjct: 178 NSYSSS 183
>dbj|BAA83710.1| heat shock factor [Nicotiana tabacum]
Length = 292
Score = 173 bits (439), Expect = 3e-42
Identities = 93/204 (45%), Positives = 126/204 (61%), Gaps = 11/204 (5%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
SQ++VPAPFLTKTYQLVDD TD +VSW++ TTFVVW+ EFA+DL+P +FKHNNFSSF
Sbjct: 2 SQRTVPAPFLTKTYQLVDDATTDDVVSWNESGTTFVVWKTAEFAKDLVPTYFKHNNFSSF 61
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK----TPQHYQQQYYEQSPQIF 133
VRQLNTYGF+K+V D+WEFAN+ FK+G K LL I RRK TP + S
Sbjct: 62 VRQLNTYGFRKIVPDKWEFANENFKRGQKELLTAIRRRKTVTPTPAGGKSVVPGTSASPD 121
Query: 134 QPDESICWIDSPLPSPKSNTDILTA-------LSEDNQRLRRKNFMLLSELSHMKNLYND 186
E + + P K+ + T LS++N++L++ N ML SEL+ K ++
Sbjct: 122 NSGEDLGSSSTSSPDSKNPGSVDTPGKSQFADLSDENEKLKKDNQMLSSELAQAKKQCDE 181
Query: 187 IIYFIQNHVSPASPFEQRSNNSAT 210
++ F+ +V A R + T
Sbjct: 182 LVAFLNQYVKVAPDMINRIISQGT 205
>gb|AAP40470.1| putative heat shock factor 6 [Arabidopsis thaliana]
gi|30793833|gb|AAP40369.1| putative heat shock factor 6
[Arabidopsis thaliana] gi|6624616|emb|CAB63802.1| heat
shock factor 6 [Arabidopsis thaliana]
gi|10176919|dbj|BAB10163.1| heat shock factor 6
[Arabidopsis thaliana] gi|30697614|ref|NP_201008.2| heat
shock factor protein, putative (HSF6) / heat shock
transcription factor, putative (HTSF6) [Arabidopsis
thaliana] gi|11386851|sp|Q9SCW4|HSF6_ARATH Heat shock
factor protein 6 (HSF 6) (Heat shock transcription
factor 6) (HSTF 6)
Length = 299
Score = 171 bits (433), Expect = 1e-41
Identities = 90/196 (45%), Positives = 121/196 (60%), Gaps = 18/196 (9%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
SQ+S+P PFLTKT+ LV+D D ++SW++D ++F+VW P +FA+DLLP FKHNNFSSF
Sbjct: 16 SQRSIPTPFLTKTFNLVEDSSIDDVISWNEDGSSFIVWNPTDFAKDLLPKHFKHNNFSSF 75
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQ----------QQYYE 127
VRQLNTYGFKKVV DRWEF+ND+FK+G K LL EI RRK +Q Q
Sbjct: 76 VRQLNTYGFKKVVPDRWEFSNDFFKRGEKRLLREIQRRKITTTHQTVVAPSSEQRNQTMV 135
Query: 128 QSPQIFQPDESICWIDSPLPS--------PKSNTDILTALSEDNQRLRRKNFMLLSELSH 179
SP D + + S PS N + L E+N++LR +N L EL+
Sbjct: 136 VSPSNSGEDNNNNQVMSSSPSSWYCHQTKTTGNGGLSVELLEENEKLRSQNIQLNRELTQ 195
Query: 180 MKNLYNDIIYFIQNHV 195
MK++ ++I + N+V
Sbjct: 196 MKSICDNIYSLMSNYV 211
>emb|CAA39034.1| heat stress transcription factor [Lycopersicon peruvianum]
gi|100267|pir||S12361 heat shock transcription factor
HSF24 - Peruvian tomato gi|123684|sp|P22335|HSF24_LYCPE
Heat shock factor protein HSF24 (Heat shock
transcription factor 24) (HSTF 24) (Heat stress
transcription factor)
Length = 301
Score = 171 bits (432), Expect = 2e-41
Identities = 92/205 (44%), Positives = 123/205 (59%), Gaps = 12/205 (5%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
SQ++ PAPFL KTYQLVDD TD ++SW++ TTFVVW+ EFA+DLLP +FKHNNFSSF
Sbjct: 2 SQRTAPAPFLLKTYQLVDDAATDDVISWNEIGTTFVVWKTAEFAKDLLPKYFKHNNFSSF 61
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK----TPQHYQQQYYEQSPQIF 133
VRQLNTYGF+K+V D+WEFAN+ FK+G K LL I RRK TP + S
Sbjct: 62 VRQLNTYGFRKIVPDKWEFANENFKRGQKELLTAIRRRKTVTSTPAGGKSVAAGASASPD 121
Query: 134 QPDESICWIDSPLPSPKSNTDI--------LTALSEDNQRLRRKNFMLLSELSHMKNLYN 185
+ I + P K+ + T LS++N++L++ N ML SEL K N
Sbjct: 122 NSGDDIGSSSTSSPDSKNPGSVDTPGKLSQFTDLSDENEKLKKDNQMLSSELVQAKKQCN 181
Query: 186 DIIYFIQNHVSPASPFEQRSNNSAT 210
+++ F+ +V A R + T
Sbjct: 182 ELVAFLSQYVKVAPDMINRIMSQGT 206
>gb|AAP37830.1| At4g11660 [Arabidopsis thaliana] gi|7267866|emb|CAB78209.1| heat
shock transcription factor-like protein [Arabidopsis
thaliana] gi|4539457|emb|CAB39937.1| heat shock
transcription factor-like protein [Arabidopsis thaliana]
gi|20260614|gb|AAM13205.1| heat shock transcription
factor-like protein [Arabidopsis thaliana]
gi|7485003|pir||T04213 heat shock transcription factor
homolog T5C23.90 - Arabidopsis thaliana
gi|15234264|ref|NP_192903.1| heat shock factor protein 7
(HSF7) / heat shock transcription factor 7 (HSTF7)
[Arabidopsis thaliana] gi|12643858|sp|Q9T0D3|HSF7_ARATH
Heat shock factor protein 7 (HSF 7) (Heat shock
transcription factor 7) (HSTF 7)
Length = 377
Score = 171 bits (432), Expect = 2e-41
Identities = 95/213 (44%), Positives = 123/213 (57%), Gaps = 36/213 (16%)
Query: 17 ESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSS 76
+SQ+S+P PFLTKTYQLV+DP+ D ++SW++D TTF+VWRP EFARDLLP +FKHNNFSS
Sbjct: 51 DSQRSIPTPFLTKTYQLVEDPVYDELISWNEDGTTFIVWRPAEFARDLLPKYFKHNNFSS 110
Query: 77 FVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQ----------------- 119
FVRQLNTYGF+KVV DRWEF+ND FK+G K LL +I RRK Q
Sbjct: 111 FVRQLNTYGFRKVVPDRWEFSNDCFKRGEKILLRDIQRRKISQPAMAAAAAAAAAAVAAS 170
Query: 120 -----------HYQQQYYEQSPQIFQPDESICWIDSPL------PSPKSNTDILTA--LS 160
H Q+ + S + + S + T TA L
Sbjct: 171 AVTVAAVPVVAHIVSPSNSGEEQVISSNSSPAAAAAAIGGVVGGGSLQRTTSCTTAPELV 230
Query: 161 EDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
E+N+RLR+ N L E++ +K LY +I + N
Sbjct: 231 EENERLRKDNERLRKEMTKLKGLYANIYTLMAN 263
>emb|CAB63803.1| heat shock factor 7 [Arabidopsis thaliana]
Length = 328
Score = 171 bits (432), Expect = 2e-41
Identities = 95/213 (44%), Positives = 123/213 (57%), Gaps = 36/213 (16%)
Query: 17 ESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSS 76
+SQ+S+P PFLTKTYQLV+DP+ D ++SW++D TTF+VWRP EFARDLLP +FKHNNFSS
Sbjct: 2 DSQRSIPTPFLTKTYQLVEDPVYDELISWNEDGTTFIVWRPAEFARDLLPKYFKHNNFSS 61
Query: 77 FVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQ----------------- 119
FVRQLNTYGF+KVV DRWEF+ND FK+G K LL +I RRK Q
Sbjct: 62 FVRQLNTYGFRKVVPDRWEFSNDCFKRGEKILLRDIQRRKISQPAMAAAAAAAAAAVAAS 121
Query: 120 -----------HYQQQYYEQSPQIFQPDESICWIDSPL------PSPKSNTDILTA--LS 160
H Q+ + S + + S + T TA L
Sbjct: 122 AVTVAAVPVVAHIVSPSNSGEEQVISSNSSPAAAAAAIGGVVGGGSLQRTTSCTTAPELV 181
Query: 161 EDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
E+N+RLR+ N L E++ +K LY +I + N
Sbjct: 182 EENERLRKDNERLRKEMTKLKGLYANIYTLMAN 214
>dbj|BAD46358.1| putative heat shock factor [Oryza sativa (japonica cultivar-group)]
Length = 414
Score = 170 bits (430), Expect = 3e-41
Identities = 73/101 (72%), Positives = 88/101 (86%)
Query: 16 MESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFS 75
+ Q+S+P PFLTKTYQLV+DP D ++SW++D +TFVVWRP EFARDLLP +FKHNNFS
Sbjct: 29 LAGQRSLPTPFLTKTYQLVEDPAVDDVISWNEDGSTFVVWRPAEFARDLLPKYFKHNNFS 88
Query: 76 SFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK 116
SFVRQLNTYGF+K+V DRWEFAND F++G K LLC+IHRRK
Sbjct: 89 SFVRQLNTYGFRKIVPDRWEFANDCFRRGEKRLLCDIHRRK 129
Score = 38.1 bits (87), Expect = 0.16
Identities = 16/38 (42%), Positives = 25/38 (65%)
Query: 159 LSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVS 196
+ E+N+RLRR+N L EL HMK L N+I+ + + +
Sbjct: 218 MGEENERLRRENARLTRELGHMKKLCNNILLLMSKYAA 255
>dbj|BAD46357.1| putative heat shock factor [Oryza sativa (japonica cultivar-group)]
Length = 454
Score = 170 bits (430), Expect = 3e-41
Identities = 73/101 (72%), Positives = 88/101 (86%)
Query: 16 MESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFS 75
+ Q+S+P PFLTKTYQLV+DP D ++SW++D +TFVVWRP EFARDLLP +FKHNNFS
Sbjct: 29 LAGQRSLPTPFLTKTYQLVEDPAVDDVISWNEDGSTFVVWRPAEFARDLLPKYFKHNNFS 88
Query: 76 SFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK 116
SFVRQLNTYGF+K+V DRWEFAND F++G K LLC+IHRRK
Sbjct: 89 SFVRQLNTYGFRKIVPDRWEFANDCFRRGEKRLLCDIHRRK 129
Score = 38.1 bits (87), Expect = 0.16
Identities = 16/38 (42%), Positives = 25/38 (65%)
Query: 159 LSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVS 196
+ E+N+RLRR+N L EL HMK L N+I+ + + +
Sbjct: 218 MGEENERLRRENARLTRELGHMKKLCNNILLLMSKYAA 255
>emb|CAA87077.1| heat shock transcription factor 34 [Glycine max]
gi|2129831|pir||S59538 heat shock transcription factor
HSF34 - soybean
Length = 282
Score = 164 bits (416), Expect = 1e-39
Identities = 87/193 (45%), Positives = 123/193 (63%), Gaps = 15/193 (7%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
SQ+SVPAPFLTKTYQLV+D TD ++SW + TFVVW+ +FA+DLLP +FKHNNFSSF
Sbjct: 2 SQRSVPAPFLTKTYQLVEDQGTDQVISWGESGNTFVVWKHADFAKDLLPKYFKHNNFSSF 61
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKT--------PQHYQQQYYEQS 129
VRQLNTYGF+K+V D+WEFAN++FK+G K LL EI RRKT P+ + S
Sbjct: 62 VRQLNTYGFRKIVPDKWEFANEHFKRGQKELLSEIKRRKTVPQSSAHPPEAGKSGGDGNS 121
Query: 130 PQIFQPDESICWIDSPLP-------SPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKN 182
P D++ S S ++NT LS +N++L++ N L EL+ +
Sbjct: 122 PLNSGSDDAGSTSTSSSSSGSKNQGSVETNTTPSHQLSSENEKLKKDNETLSCELARARK 181
Query: 183 LYNDIIYFIQNHV 195
++++ F+++ +
Sbjct: 182 QCDELVAFLRDRL 194
>emb|CAA53761.1| heat shock factor [Arabidopsis thaliana]
Length = 483
Score = 162 bits (409), Expect = 8e-39
Identities = 89/191 (46%), Positives = 117/191 (60%), Gaps = 7/191 (3%)
Query: 15 TMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNF 74
T+ + S+P PFL+KTY +V+DP TD IVSWS +F+VW PPEF+RDLLP +FKHNNF
Sbjct: 42 TLLNANSLPPPFLSKTYDMVEDPATDAIVSWSPTNNSFIVWDPPEFSRDLLPKYFKHNNF 101
Query: 75 SSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQ-HYQQQYYEQSPQIF 133
SSFVRQLNTYGF+KV DRWEFAN+ F +G KHLL +I RRK+ Q H QS Q+
Sbjct: 102 SSFVRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLS 161
Query: 134 QPDESICWIDSPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
Q S+ + S + K L E+ ++L+R +L+ EL ++ +Q
Sbjct: 162 QGQGSMAALSSCVEVGK------FGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQV 215
Query: 194 HVSPASPFEQR 204
V EQR
Sbjct: 216 MVKHLQVMEQR 226
>emb|CAB10555.1| heat shock transcription factor HSF1 [Arabidopsis thaliana]
gi|7268528|emb|CAB78778.1| heat shock transcription
factor HSF1 [Arabidopsis thaliana]
gi|7428781|pir||S52641 heat shock transcription factor
HSF1 - Arabidopsis thaliana gi|15236631|ref|NP_193510.1|
heat shock factor protein 1 (HSF1) / heat shock
transcription factor 1 (HSTF1) [Arabidopsis thaliana]
gi|12644262|sp|P41151|HSF1_ARATH Heat shock factor
protein 1 (HSF 1) (Heat shock transcription factor 1)
(HSTF 1)
Length = 495
Score = 161 bits (408), Expect = 1e-38
Identities = 89/191 (46%), Positives = 117/191 (60%), Gaps = 7/191 (3%)
Query: 15 TMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNF 74
T+ + S+P PFL+KTY +V+DP TD IVSWS +F+VW PPEF+RDLLP +FKHNNF
Sbjct: 42 TLLNANSLPPPFLSKTYDMVEDPATDAIVSWSPTNNSFIVWDPPEFSRDLLPKYFKHNNF 101
Query: 75 SSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQ-HYQQQYYEQSPQIF 133
SSFVRQLNTYGF+KV DRWEFAN+ F +G KHLL +I RRK+ Q H QS Q+
Sbjct: 102 SSFVRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLS 161
Query: 134 QPDESICWIDSPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
Q S+ + S + K L E+ ++L+R +L+ EL ++ +Q
Sbjct: 162 QGQGSMAALSSCVEVGK------FGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQV 215
Query: 194 HVSPASPFEQR 204
V EQR
Sbjct: 216 LVKHLQVMEQR 226
>emb|CAA47869.1| heat shock transcription factor 8 [Lycopersicon peruvianum]
gi|100264|pir||S25481 heat shock transcription factor
HSF8 - Peruvian tomato gi|729775|sp|P41153|HSF8_LYCPE
Heat shock factor protein HSF8 (Heat shock transcription
factor 8) (HSTF 8) (Heat stress transcription factor)
Length = 527
Score = 157 bits (398), Expect = 1e-37
Identities = 85/165 (51%), Positives = 105/165 (63%), Gaps = 11/165 (6%)
Query: 21 SVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQ 80
+ P PFL KTY +VDDP TD IVSWS +FVVW PPEFA+DLLP +FKHNNFSSFVRQ
Sbjct: 37 NAPPPFLVKTYDMVDDPSTDKIVSWSPTNNSFVVWDPPEFAKDLLPKYFKHNNFSSFVRQ 96
Query: 81 LNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQH----YQQQYYEQSPQIFQPD 136
LNTYGF+KV DRWEFAN+ F +G KHLL I RRK P H QQQ + + Q QP
Sbjct: 97 LNTYGFRKVDPDRWEFANEGFLRGQKHLLKSISRRK-PAHGHAQQQQQPHGHAQQQMQPP 155
Query: 137 ESICWIDSPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMK 181
+ + + K L E+ +RL+R +L+ EL ++
Sbjct: 156 GHSASVGACVEVGK------FGLEEEVERLKRDKNVLMQELVRLR 194
>emb|CAA47868.1| heat stress transcription factor 8 [Lycopersicon esculentum]
gi|100225|pir||S25478 heat shock transcription factor
HSF8 - tomato gi|11386827|sp|Q40152|HSF8_LYCES Heat
shock factor protein HSF8 (Heat shock transcription
factor 8) (HSTF 8) (Heat stress transcription factor)
Length = 527
Score = 157 bits (398), Expect = 1e-37
Identities = 85/165 (51%), Positives = 105/165 (63%), Gaps = 11/165 (6%)
Query: 21 SVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQ 80
+ P PFL KTY +VDDP TD IVSWS +FVVW PPEFA+DLLP +FKHNNFSSFVRQ
Sbjct: 35 NAPPPFLVKTYDMVDDPSTDKIVSWSPTNNSFVVWDPPEFAKDLLPKYFKHNNFSSFVRQ 94
Query: 81 LNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQH----YQQQYYEQSPQIFQPD 136
LNTYGF+KV DRWEFAN+ F +G KHLL I RRK P H QQQ + + Q QP
Sbjct: 95 LNTYGFRKVDPDRWEFANEGFLRGQKHLLKSISRRK-PAHGHAQQQQQPHGNAQQQMQPP 153
Query: 137 ESICWIDSPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMK 181
+ + + K L E+ +RL+R +L+ EL ++
Sbjct: 154 GHSASVGACVEVGK------FGLEEEVERLKRDKNVLMQELVRLR 192
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 392,917,286
Number of Sequences: 2540612
Number of extensions: 17162571
Number of successful extensions: 42889
Number of sequences better than 10.0: 310
Number of HSP's better than 10.0 without gapping: 275
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 42455
Number of HSP's gapped (non-prelim): 361
length of query: 211
length of database: 863,360,394
effective HSP length: 122
effective length of query: 89
effective length of database: 553,405,730
effective search space: 49253109970
effective search space used: 49253109970
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146941.18


