

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.10 - phase: 0
(124 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
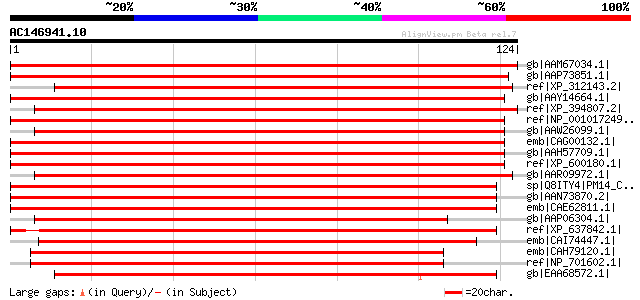
gb|AAM67034.1| unknown [Arabidopsis thaliana] gi|9759386|dbj|BAB... 229 1e-59
gb|AAP73851.1| putative RNA binding protein [Oryza sativa (japon... 211 3e-54
ref|XP_312143.2| ENSANGP00000022192 [Anopheles gambiae str. PEST... 173 7e-43
gb|AAY14664.1| unknown [Homo sapiens] gi|57098153|ref|XP_532889.... 171 3e-42
ref|XP_394807.2| PREDICTED: similar to CG13298-PA [Apis mellifera] 171 3e-42
ref|NP_001017249.1| hypothetical protein LOC550003 [Xenopus trop... 171 4e-42
gb|AAW26099.1| unknown [Schistosoma japonicum] 170 6e-42
emb|CAG00132.1| unnamed protein product [Tetraodon nigroviridis] 170 6e-42
gb|AAH57709.1| MGC68842 protein [Xenopus laevis] 169 1e-41
ref|XP_600180.1| PREDICTED: similar to RIKEN cDNA 0610009D07 [Bo... 169 1e-41
gb|AAR09972.1| similar to Drosophila melanogaster CG13298 [Droso... 166 9e-41
sp|Q8ITY4|PM14_CAEEL Pre-mRNA branch site p14-like protein gi|32... 157 4e-38
gb|AAN73870.2| Hypothetical protein C50D2.5 [Caenorhabditis eleg... 157 4e-38
emb|CAE62811.1| Hypothetical protein CBG06987 [Caenorhabditis br... 157 6e-38
gb|AAP06304.1| similar to GenBank Accession Number AE003563 CG13... 153 1e-36
ref|XP_637842.1| hypothetical protein DDB0186914 [Dictyostelium ... 152 1e-36
emb|CAI74447.1| RNA binding protein, putative [Theileria annulata] 152 2e-36
emb|CAH79120.1| conserved hypothetical protein [Plasmodium chaba... 142 2e-33
ref|NP_701602.1| hypothetical protein PFL1200c [Plasmodium falci... 138 3e-32
gb|EAA68572.1| hypothetical protein FG00536.1 [Gibberella zeae P... 128 4e-29
>gb|AAM67034.1| unknown [Arabidopsis thaliana] gi|9759386|dbj|BAB10037.1| unnamed
protein product [Arabidopsis thaliana]
gi|28394033|gb|AAO42424.1| putative RRM-containing
protein [Arabidopsis thaliana]
gi|27754399|gb|AAO22648.1| putative RRM-containing
protein [Arabidopsis thaliana]
gi|15239901|ref|NP_196780.1| RNA recognition motif
(RRM)-containing protein [Arabidopsis thaliana]
gi|32129916|sp|Q9FMP4|PM14_ARATH Pre-mRNA branch site
p14-like protein
Length = 124
Score = 229 bits (584), Expect = 1e-59
Identities = 113/124 (91%), Positives = 117/124 (94%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
MTTISLRK NTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIG +K T+GTA
Sbjct: 1 MTTISLRKSNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGCDKATKGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAK AVDHLSGFNVANRYLIVLYYQ AKMSKKFDQKK EDEI ++QEKYGVS
Sbjct: 61 FVVYEDIYDAKNAVDHLSGFNVANRYLIVLYYQHAKMSKKFDQKKSEDEITKLQEKYGVS 120
Query: 121 TKDK 124
TKDK
Sbjct: 121 TKDK 124
>gb|AAP73851.1| putative RNA binding protein [Oryza sativa (japonica
cultivar-group)] gi|50919311|ref|XP_470052.1| putative
RNA binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 130
Score = 211 bits (537), Expect = 3e-54
Identities = 103/122 (84%), Positives = 110/122 (89%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M +SLRK N RLPPEVNRVLYVRNLPFNI+SEEMYDIFGKYGAIRQIR+G KDTRGTA
Sbjct: 1 MAAVSLRKSNARLPPEVNRVLYVRNLPFNISSEEMYDIFGKYGAIRQIRLGNAKDTRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAK AVDHLSGFNVANRYLIVLYYQ AKMSKK D KKKE+EI R+QEKYG+
Sbjct: 61 FVVYEDIYDAKNAVDHLSGFNVANRYLIVLYYQPAKMSKKSDVKKKEEEITRLQEKYGLG 120
Query: 121 TK 122
+K
Sbjct: 121 SK 122
>ref|XP_312143.2| ENSANGP00000022192 [Anopheles gambiae str. PEST]
gi|55241995|gb|EAA07781.2| ENSANGP00000022192 [Anopheles
gambiae str. PEST]
Length = 115
Score = 173 bits (439), Expect = 7e-43
Identities = 81/112 (72%), Positives = 95/112 (84%)
Query: 12 RLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAK 71
RLPPEVNRVLYVRNLP+ ITS+EMYDIFGKYGAIRQIR+G +TRGTAFVVYEDI+DAK
Sbjct: 2 RLPPEVNRVLYVRNLPYKITSDEMYDIFGKYGAIRQIRVGNTPETRGTAFVVYEDIFDAK 61
Query: 72 TAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKD 123
A DHLSGFNV NRYL+VLYYQ K K+ D KK+DE+ +M+ KY +ST++
Sbjct: 62 NARDHLSGFNVCNRYLVVLYYQSTKAFKQLDVDKKQDELDQMKAKYNISTEE 113
>gb|AAY14664.1| unknown [Homo sapiens] gi|57098153|ref|XP_532889.1| PREDICTED:
hypothetical protein XP_532889 [Canis familiaris]
gi|56207570|emb|CAI21137.1| novel protein similar to
vertebrate pre-mRNA branch site protein p14 (P14) [Danio
rerio] gi|55595200|ref|XP_515324.1| PREDICTED:
hypothetical protein XP_515324 [Pan troglodytes]
gi|15930037|gb|AAH15463.1| Splicing factor 3B, 14 kDa
subunit [Homo sapiens] gi|61815688|ref|XP_585719.1|
PREDICTED: similar to RIKEN cDNA 0610009D07 [Bos taurus]
gi|47939375|gb|AAH71385.1| Hypothetical protein
LOC415157 [Danio rerio] gi|13384692|ref|NP_079599.1|
splicing factor 3B, 14 kDa subunit [Mus musculus]
gi|50745107|ref|XP_419986.1| PREDICTED: similar to RIKEN
cDNA 0610009D07 [Gallus gallus]
gi|4929689|gb|AAD34105.1| CGI-110 protein [Homo sapiens]
gi|18044537|gb|AAH19535.1| RIKEN cDNA 0610009D07 [Mus
musculus] gi|12832311|dbj|BAB22052.1| unnamed protein
product [Mus musculus] gi|15278118|gb|AAK94041.1|
pre-mRNA branch site protein p14 [Homo sapiens]
gi|12585536|sp|Q9Y3B4|PM14_HUMAN Pre-mRNA branch site
protein p14 (SF3B 14 kDa subunit)
gi|6841570|gb|AAF29138.1| HSPC175 [Homo sapiens]
gi|31563064|sp|P59708|PM14_MOUSE Pre-mRNA branch site
protein p14 (SF3B 14 kDa subunit)
gi|50344788|ref|NP_001002067.1| hypothetical protein
LOC415157 [Danio rerio] gi|9963797|gb|AAG09698.1| ht006
protein [Homo sapiens] gi|7706326|ref|NP_057131.1|
splicing factor 3B, 14 kDa subunit [Homo sapiens]
gi|12834501|dbj|BAB22936.1| unnamed protein product [Mus
musculus]
Length = 125
Score = 171 bits (434), Expect = 3e-42
Identities = 77/121 (63%), Positives = 98/121 (80%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVNR+LY+RNLP+ IT+EEMYDIFGKYG IRQIR+G +TRGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+VVYEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVVYEDIFDAKNACDHLSGFNVCNRYLVVLYYNANRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>ref|XP_394807.2| PREDICTED: similar to CG13298-PA [Apis mellifera]
Length = 125
Score = 171 bits (434), Expect = 3e-42
Identities = 80/118 (67%), Positives = 96/118 (80%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYED 66
R+ N RLPPEVNRVLY+RNLP+ IT+EEMYDIFGKYGAIRQIR+G +TRGTAFVVYED
Sbjct: 8 RRANVRLPPEVNRVLYIRNLPYKITAEEMYDIFGKYGAIRQIRVGNTAETRGTAFVVYED 67
Query: 67 IYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKDK 124
I+DAK A DHLSGFNV NRYL+VLYYQ K K+ D KK +EI +++ KY ++ + K
Sbjct: 68 IFDAKNACDHLSGFNVCNRYLVVLYYQSNKAFKRVDVDKKMEEIDKLKTKYNLNEEKK 125
>ref|NP_001017249.1| hypothetical protein LOC550003 [Xenopus tropicalis]
Length = 125
Score = 171 bits (433), Expect = 4e-42
Identities = 77/121 (63%), Positives = 97/121 (79%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVNR+LY+RNLP+ IT EEMYDIFGKYG IRQIR+G +TRGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNRILYIRNLPYKITGEEMYDIFGKYGPIRQIRVGNTPETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+VVYEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVVYEDIFDAKNACDHLSGFNVCNRYLVVLYYNANRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>gb|AAW26099.1| unknown [Schistosoma japonicum]
Length = 123
Score = 170 bits (431), Expect = 6e-42
Identities = 79/115 (68%), Positives = 97/115 (83%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYED 66
RK TRLPPEVNR+L V+NLP+NI++EEMYDIFGKYGAIRQIRIG +T+GTA V+YED
Sbjct: 5 RKNATRLPPEVNRILLVKNLPYNISAEEMYDIFGKYGAIRQIRIGNTPETKGTALVIYED 64
Query: 67 IYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVST 121
I+DAK A DHLSGFNV NRYL+VLYYQ+ K KK D KK++E+ R++ KYG++T
Sbjct: 65 IFDAKNACDHLSGFNVCNRYLVVLYYQRNKQFKKTDVDKKKEELDRLKAKYGLNT 119
>emb|CAG00132.1| unnamed protein product [Tetraodon nigroviridis]
Length = 125
Score = 170 bits (431), Expect = 6e-42
Identities = 78/121 (64%), Positives = 97/121 (79%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVNR+LYVRNLP+ IT+EEMYDIFGKYG IRQIR G +TRGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNRILYVRNLPYKITAEEMYDIFGKYGPIRQIRTGNTPETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+VVYEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVVYEDIFDAKNACDHLSGFNVCNRYLVVLYYNANRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>gb|AAH57709.1| MGC68842 protein [Xenopus laevis]
Length = 125
Score = 169 bits (429), Expect = 1e-41
Identities = 76/121 (62%), Positives = 97/121 (79%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVNR+LY+RNLP+ IT EEMYDIFGKYG IRQIR+G ++RGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNRILYIRNLPYKITGEEMYDIFGKYGPIRQIRVGNTPESRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+VVYEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVVYEDIFDAKNACDHLSGFNVCNRYLVVLYYNPNRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>ref|XP_600180.1| PREDICTED: similar to RIKEN cDNA 0610009D07 [Bos taurus]
Length = 125
Score = 169 bits (429), Expect = 1e-41
Identities = 75/121 (61%), Positives = 98/121 (80%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVN++LY+RNLP+ IT+EEMYDIFGKYG IRQIR+G +TRGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNQILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+V+YEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVIYEDIFDAKNACDHLSGFNVCNRYLVVLYYNANRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>gb|AAR09972.1| similar to Drosophila melanogaster CG13298 [Drosophila yakuba]
gi|7295357|gb|AAF50675.1| CG13298-PA [Drosophila
melanogaster] gi|19528453|gb|AAL90341.1| RE19804p
[Drosophila melanogaster] gi|21357707|ref|NP_648037.1|
CG13298-PA [Drosophila melanogaster]
gi|54642523|gb|EAL31270.1| GA12181-PA [Drosophila
pseudoobscura] gi|32699626|sp|Q9VRV7|PM14_DROME Pre-mRNA
branch site p14-like protein
Length = 121
Score = 166 bits (421), Expect = 9e-41
Identities = 76/117 (64%), Positives = 95/117 (80%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYED 66
++ + RLPPEVNR+LYVRNLP+ ITS+EMYDIFGK+GAIRQIR+G +TRGTAFVVYED
Sbjct: 3 KRNHIRLPPEVNRLLYVRNLPYKITSDEMYDIFGKFGAIRQIRVGNTPETRGTAFVVYED 62
Query: 67 IYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKD 123
I+DAK A DHLSGFNV NRYL+VLYYQ K K+ D KK++E+ ++ KY + T +
Sbjct: 63 IFDAKNACDHLSGFNVCNRYLVVLYYQSNKAFKRVDMDKKQEELNNIKAKYNLKTPE 119
>sp|Q8ITY4|PM14_CAEEL Pre-mRNA branch site p14-like protein gi|32564829|ref|NP_493659.2|
RNA-binding region RNP-1 (2A389) [Caenorhabditis
elegans]
Length = 188
Score = 157 bits (398), Expect = 4e-38
Identities = 69/119 (57%), Positives = 95/119 (78%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + + +LPPEVNR+LY++NLP+ IT+EEMY+IFGK+GA+RQIR+G +TRGTA
Sbjct: 51 MAMANRQNRGAKLPPEVNRILYIKNLPYKITTEEMYEIFGKFGAVRQIRVGNTAETRGTA 110
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGV 119
FVVYEDI+DAKTA +HLSG+NV+NRYL+VLYYQ K K+ D +K ++ ++E+YG+
Sbjct: 111 FVVYEDIFDAKTACEHLSGYNVSNRYLVVLYYQATKAWKRMDTEKARTKLEDIKERYGI 169
>gb|AAN73870.2| Hypothetical protein C50D2.5 [Caenorhabditis elegans]
Length = 138
Score = 157 bits (398), Expect = 4e-38
Identities = 69/119 (57%), Positives = 95/119 (78%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + + +LPPEVNR+LY++NLP+ IT+EEMY+IFGK+GA+RQIR+G +TRGTA
Sbjct: 1 MAMANRQNRGAKLPPEVNRILYIKNLPYKITTEEMYEIFGKFGAVRQIRVGNTAETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGV 119
FVVYEDI+DAKTA +HLSG+NV+NRYL+VLYYQ K K+ D +K ++ ++E+YG+
Sbjct: 61 FVVYEDIFDAKTACEHLSGYNVSNRYLVVLYYQATKAWKRMDTEKARTKLEDIKERYGI 119
>emb|CAE62811.1| Hypothetical protein CBG06987 [Caenorhabditis briggsae]
Length = 138
Score = 157 bits (397), Expect = 6e-38
Identities = 69/119 (57%), Positives = 95/119 (78%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + + +LPPEVNR+LY++NLP+ IT+EEMY+IFGK+GA+RQIR+G +TRGTA
Sbjct: 1 MAMANRQNRGAKLPPEVNRILYIKNLPYKITTEEMYEIFGKFGAVRQIRVGNTAETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGV 119
FVVYEDI+DAKTA +HLSG+NV+NRYL+VLYYQ K K+ D +K ++ ++E+YG+
Sbjct: 61 FVVYEDIFDAKTACEHLSGYNVSNRYLVVLYYQATKAWKRMDTEKARAKLEDIKERYGI 119
>gb|AAP06304.1| similar to GenBank Accession Number AE003563 CG13298 gene product
in Drosophila melanogaster [Schistosoma japonicum]
Length = 110
Score = 153 bits (386), Expect = 1e-36
Identities = 71/101 (70%), Positives = 85/101 (83%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYED 66
RK TRLPPEVNR+L V+NLP+NI++EEMYDIFGKYGAIRQIRIG +T+GTA V+YED
Sbjct: 5 RKNATRLPPEVNRILLVKNLPYNISAEEMYDIFGKYGAIRQIRIGNTPETKGTALVIYED 64
Query: 67 IYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKE 107
I+DAK A DHLSGFNV NRYL+VLYYQ+ K KK D +++
Sbjct: 65 IFDAKNACDHLSGFNVCNRYLVVLYYQRNKQFKKTDVDRRK 105
>ref|XP_637842.1| hypothetical protein DDB0186914 [Dictyostelium discoideum]
gi|60466262|gb|EAL64324.1| hypothetical protein
DDB0186914 [Dictyostelium discoideum]
Length = 117
Score = 152 bits (385), Expect = 1e-36
Identities = 71/119 (59%), Positives = 94/119 (78%), Gaps = 3/119 (2%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
MTT+ K ++LPPE+NR+LYV+NLPF I+SEE+Y IF YGAIRQIR+G +T GTA
Sbjct: 1 MTTL---KKVSKLPPEINRILYVKNLPFKISSEELYGIFSNYGAIRQIRLGNTTETMGTA 57
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGV 119
FVVYEDI+DA+ A +HLSG+NV NRYL+VLYYQQ K KK D + K+ E+ +++K+G+
Sbjct: 58 FVVYEDIFDARNACEHLSGYNVNNRYLVVLYYQQQKQQKKMDHQAKKKELDLLKQKFGL 116
>emb|CAI74447.1| RNA binding protein, putative [Theileria annulata]
Length = 122
Score = 152 bits (383), Expect = 2e-36
Identities = 69/107 (64%), Positives = 91/107 (84%)
Query: 8 KGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDI 67
K N RLPPEV+R+LY+RNLP+ ITSEE+YDIFGKYG++RQIR G + T+GTAFVVY+DI
Sbjct: 9 KKNIRLPPEVSRILYLRNLPYKITSEELYDIFGKYGSVRQIRKGNSATTKGTAFVVYDDI 68
Query: 68 YDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQ 114
+DAK A+DHLSGFNVA RYL+VLYY A+++K+ D K+E ++ R++
Sbjct: 69 FDAKNALDHLSGFNVAGRYLVVLYYNPARVNKRKDLDKQEADLKRIR 115
>emb|CAH79120.1| conserved hypothetical protein [Plasmodium chabaudi]
Length = 109
Score = 142 bits (358), Expect = 2e-33
Identities = 65/101 (64%), Positives = 82/101 (80%)
Query: 6 LRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYE 65
+ + N RLP EV+R+LYVRNLP+ IT+EE+YDIFGKYG +RQIR G + TRGT+FVVY+
Sbjct: 1 MSRRNIRLPAEVSRILYVRNLPYKITAEELYDIFGKYGTVRQIRKGNAEGTRGTSFVVYD 60
Query: 66 DIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKK 106
DIYDAK A+DHLSGFNVA RYL++LYY K KK + ++K
Sbjct: 61 DIYDAKNALDHLSGFNVAGRYLVILYYDPVKAQKKKELQEK 101
>ref|NP_701602.1| hypothetical protein PFL1200c [Plasmodium falciparum 3D7]
gi|23496772|gb|AAN36326.1| hypothetical protein,
conserved [Plasmodium falciparum 3D7]
Length = 106
Score = 138 bits (348), Expect = 3e-32
Identities = 63/101 (62%), Positives = 82/101 (80%)
Query: 6 LRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYE 65
+ + N RLP EV+R+LYVRNLP+ I+++E+YDIFGKYG +RQIR G + T+GT+FVVY+
Sbjct: 1 MSRRNIRLPAEVSRILYVRNLPYKISADELYDIFGKYGTVRQIRKGNAEGTKGTSFVVYD 60
Query: 66 DIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKK 106
DIYDAK A+DHLSGFNVA RYL+VLYY K KK + ++K
Sbjct: 61 DIYDAKNALDHLSGFNVAGRYLVVLYYDPVKAQKKKEIQEK 101
>gb|EAA68572.1| hypothetical protein FG00536.1 [Gibberella zeae PH-1]
gi|46107306|ref|XP_380712.1| hypothetical protein
FG00536.1 [Gibberella zeae PH-1]
Length = 173
Score = 128 bits (321), Expect = 4e-29
Identities = 58/109 (53%), Positives = 86/109 (78%), Gaps = 1/109 (0%)
Query: 12 RLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAK 71
+L PEVNR L+V+NL +N+T EE++D+FGK+G IRQ+R G +T+GTAFVVYED+ DAK
Sbjct: 64 KLAPEVNRALFVKNLSYNVTPEELFDLFGKFGPIRQVRQGIANNTKGTAFVVYEDVTDAK 123
Query: 72 TAVDHLSGFNVANRYLIVLYYQQAKMSK-KFDQKKKEDEIARMQEKYGV 119
A D L+GFN NRYL+VLY+Q KM+K K D + + + +A++++++G+
Sbjct: 124 QACDKLNGFNFQNRYLVVLYHQPDKMAKSKEDLEARRESLAQLKQQHGI 172
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.136 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 193,061,359
Number of Sequences: 2540612
Number of extensions: 7003566
Number of successful extensions: 26620
Number of sequences better than 10.0: 3602
Number of HSP's better than 10.0 without gapping: 1117
Number of HSP's successfully gapped in prelim test: 2490
Number of HSP's that attempted gapping in prelim test: 22571
Number of HSP's gapped (non-prelim): 5361
length of query: 124
length of database: 863,360,394
effective HSP length: 100
effective length of query: 24
effective length of database: 609,299,194
effective search space: 14623180656
effective search space used: 14623180656
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146941.10


