

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.5 - phase: 0 /pseudo
(470 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
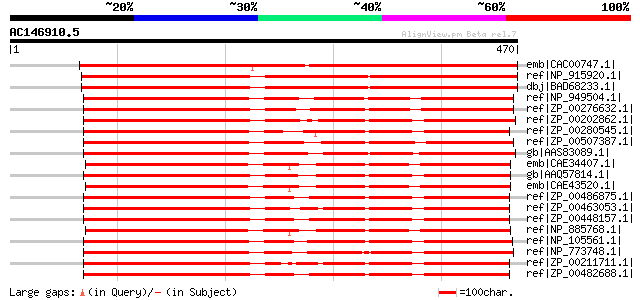
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC00747.1| putative protein [Arabidopsis thaliana] gi|15230... 545 e-153
ref|NP_915920.1| P0468B07.16 [Oryza sativa (japonica cultivar-gr... 537 e-151
dbj|BAD68233.1| dehydrogenase-like protein [Oryza sativa (japoni... 537 e-151
ref|NP_949504.1| NAD binding site:FAD dependent oxidoreductase [... 395 e-108
ref|ZP_00276632.1| COG0579: Predicted dehydrogenase [Ralstonia m... 386 e-106
ref|ZP_00202862.1| COG0579: Predicted dehydrogenase [Ralstonia e... 385 e-105
ref|ZP_00280545.1| COG0579: Predicted dehydrogenase [Burkholderi... 380 e-104
ref|ZP_00507387.1| FAD dependent oxidoreductase [Polaromonas sp.... 379 e-103
gb|AAS83089.1| glycerol-3-P dehydrogenase-like protein [Azospiri... 372 e-101
emb|CAE34407.1| putative conserved exported protein [Bordetella ... 367 e-100
gb|AAQ57814.1| conserved hypothetical protein [Chromobacterium v... 366 e-99
emb|CAE43520.1| putative conserved exported protein [Bordetella ... 364 3e-99
ref|ZP_00486875.1| COG0579: Predicted dehydrogenase [Burkholderi... 364 4e-99
ref|ZP_00463053.1| FAD dependent oxidoreductase [Burkholderia ce... 364 4e-99
ref|ZP_00448157.1| COG0579: Predicted dehydrogenase [Burkholderi... 364 4e-99
ref|NP_885768.1| putative conserved exported protein [Bordetella... 364 4e-99
ref|NP_105561.1| hypothetical protein mll4765 [Mesorhizobium lot... 362 1e-98
ref|NP_773748.1| dehydrogenase [Bradyrhizobium japonicum USDA 11... 362 2e-98
ref|ZP_00211711.1| COG0579: Predicted dehydrogenase [Burkholderi... 361 2e-98
ref|ZP_00482688.1| COG0579: Predicted dehydrogenase [Burkholderi... 360 7e-98
>emb|CAC00747.1| putative protein [Arabidopsis thaliana]
gi|15230145|ref|NP_191243.1| FAD-dependent
oxidoreductase family protein [Arabidopsis thaliana]
gi|11358430|pir||T51272 hypothetical protein T8M16_170 -
Arabidopsis thaliana
Length = 483
Score = 545 bits (1403), Expect = e-153
Identities = 268/411 (65%), Positives = 339/411 (82%), Gaps = 8/411 (1%)
Query: 65 SVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYP 124
++ +ERVD VVIGAGVVG+AVAR L+L+GREV+++++A SFGT TSSRNSEVVHAGIYYP
Sbjct: 74 TIAKERVDTVVIGAGVVGLAVARELSLRGREVLILDAASSFGTVTSSRNSEVVHAGIYYP 133
Query: 125 HHSLKAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGL 184
+SLKA FCV+GRE+LY+YC++++IPH++ GKLIVAT SSEIPKL ++++ G QN V GL
Sbjct: 134 PNSLKAKFCVRGRELLYKYCSEYEIPHKKIGKLIVATGSSEIPKLDLLMHLGTQNRVSGL 193
Query: 185 KMMDGVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLV-----LHIDRNINPMLVQGEA 239
+M++G +AM+MEP+L+CVKA+LSP SGI+D+HS MLSLV + R+ N + +QGEA
Sbjct: 194 RMLEGFEAMRMEPQLRCVKALLSPESGILDTHSFMLSLVEKSFDFMVYRDNNNLRLQGEA 253
Query: 240 ENHGATFTYNSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLS 299
+N+ ATF+YN+ V+ G +E ++ L+V++T+ + + + +L LIP LVVNSAGL
Sbjct: 254 QNNHATFSYNTVVLNGRVEEKKMHLYVADTRFSES---RCEAEAQLELIPNLVVNSAGLG 310
Query: 300 ALALAKRFTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNG 359
A ALAKR GL+++ +P ++YARGCYFTLS KA PF L+YPIPE+GGLGVHVT+DLNG
Sbjct: 311 AQALAKRLHGLDHRFVPSSHYARGCYFTLSGIKAPPFNKLVYPIPEEGGLGVHVTVDLNG 370
Query: 360 QVKFGPDVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIR 419
VKFGPDVEWI+ DD SSF NKFDY V R+EKFYPEIRKYYP+LKDGSLEPGYSGIR
Sbjct: 371 LVKFGPDVEWIECTDDTSSFLNKFDYRVNPQRSEKFYPEIRKYYPDLKDGSLEPGYSGIR 430
Query: 420 PKLSGPCQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFISTKF 470
PKLSGP QSP DFVIQGE+ HGVPGL+NLFGIESPGLTSSLAIA+ I+ KF
Sbjct: 431 PKLSGPKQSPADFVIQGEETHGVPGLVNLFGIESPGLTSSLAIAEHIANKF 481
>ref|NP_915920.1| P0468B07.16 [Oryza sativa (japonica cultivar-group)]
Length = 452
Score = 537 bits (1384), Expect = e-151
Identities = 267/405 (65%), Positives = 324/405 (79%), Gaps = 15/405 (3%)
Query: 67 PRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHH 126
PRE D VV+GAGVVG+AVARALA+ GREV+V+E+APSFGTGTSSRNSEV+HAGIYYP
Sbjct: 24 PREAADAVVVGAGVVGLAVARALAMAGREVVVVEAAPSFGTGTSSRNSEVIHAGIYYPPG 83
Query: 127 SLKAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKM 186
SLKA CV+GREMLY+YCA+ +IPH+Q GKLIVAT +E KL ++L + +NGVD L+M
Sbjct: 84 SLKASLCVRGREMLYKYCAEREIPHKQLGKLIVATGVAETAKLDMLLKNAKENGVDDLQM 143
Query: 187 MDGVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATF 246
M+G +AM+MEPEL+C+KA+LSP +GIVDSHS MLSL+ +AEN G
Sbjct: 144 MEGSEAMEMEPELRCLKALLSPRTGIVDSHSFMLSLL-------------ADAENLGTAI 190
Query: 247 TYNSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKR 306
+YN+TV G++ + LH+SE+K+L+ + S + P+L+L PKL++NSAGLSA LAKR
Sbjct: 191 SYNTTVTNGYIGDEGLELHISESKALENHSVGSPVSPQLILFPKLLINSAGLSAAPLAKR 250
Query: 307 FTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPD 366
F GL +PPAYYARGCYFTLS TK SPF HLIYP+PEDGG+GVHVTLDLNG V+FGPD
Sbjct: 251 FHGLNQVFVPPAYYARGCYFTLSQTK-SPFSHLIYPLPEDGGIGVHVTLDLNGVVRFGPD 309
Query: 367 VEWIDGVDDISS-FQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGP 425
VEWIDG D++S F ++FDYSV R KFYP IRKY+PNLKD SLEP YSGIRPKLSGP
Sbjct: 310 VEWIDGGKDVTSCFLSRFDYSVNPTRCSKFYPVIRKYFPNLKDDSLEPSYSGIRPKLSGP 369
Query: 426 CQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFISTKF 470
Q P DFVIQGEDIHGVPGL+NLFGIESPGLTSSLAIA++I +++
Sbjct: 370 GQPPSDFVIQGEDIHGVPGLVNLFGIESPGLTSSLAIAEYIVSRY 414
>dbj|BAD68233.1| dehydrogenase-like protein [Oryza sativa (japonica cultivar-group)]
Length = 416
Score = 537 bits (1384), Expect = e-151
Identities = 267/405 (65%), Positives = 324/405 (79%), Gaps = 15/405 (3%)
Query: 67 PRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHH 126
PRE D VV+GAGVVG+AVARALA+ GREV+V+E+APSFGTGTSSRNSEV+HAGIYYP
Sbjct: 24 PREAADAVVVGAGVVGLAVARALAMAGREVVVVEAAPSFGTGTSSRNSEVIHAGIYYPPG 83
Query: 127 SLKAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKM 186
SLKA CV+GREMLY+YCA+ +IPH+Q GKLIVAT +E KL ++L + +NGVD L+M
Sbjct: 84 SLKASLCVRGREMLYKYCAEREIPHKQLGKLIVATGVAETAKLDMLLKNAKENGVDDLQM 143
Query: 187 MDGVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATF 246
M+G +AM+MEPEL+C+KA+LSP +GIVDSHS MLSL+ +AEN G
Sbjct: 144 MEGSEAMEMEPELRCLKALLSPRTGIVDSHSFMLSLL-------------ADAENLGTAI 190
Query: 247 TYNSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKR 306
+YN+TV G++ + LH+SE+K+L+ + S + P+L+L PKL++NSAGLSA LAKR
Sbjct: 191 SYNTTVTNGYIGDEGLELHISESKALENHSVGSPVSPQLILFPKLLINSAGLSAAPLAKR 250
Query: 307 FTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPD 366
F GL +PPAYYARGCYFTLS TK SPF HLIYP+PEDGG+GVHVTLDLNG V+FGPD
Sbjct: 251 FHGLNQVFVPPAYYARGCYFTLSQTK-SPFSHLIYPLPEDGGIGVHVTLDLNGVVRFGPD 309
Query: 367 VEWIDGVDDISS-FQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGP 425
VEWIDG D++S F ++FDYSV R KFYP IRKY+PNLKD SLEP YSGIRPKLSGP
Sbjct: 310 VEWIDGGKDVTSCFLSRFDYSVNPTRCSKFYPVIRKYFPNLKDDSLEPSYSGIRPKLSGP 369
Query: 426 CQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFISTKF 470
Q P DFVIQGEDIHGVPGL+NLFGIESPGLTSSLAIA++I +++
Sbjct: 370 GQPPSDFVIQGEDIHGVPGLVNLFGIESPGLTSSLAIAEYIVSRY 414
>ref|NP_949504.1| NAD binding site:FAD dependent oxidoreductase [Rhodopseudomonas
palustris CGA009] gi|39651086|emb|CAE29609.1| NAD
binding site:FAD dependent oxidoreductase
[Rhodopseudomonas palustris CGA009]
Length = 366
Score = 395 bits (1016), Expect = e-108
Identities = 206/399 (51%), Positives = 265/399 (65%), Gaps = 38/399 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
+ V+CVV+GAGVVG+A+AR LA +GREVIV+E A + GTGTSSRNSEV+HAGIYY SL
Sbjct: 2 DEVECVVVGAGVVGLAIARKLAQQGREVIVVEQAEAIGTGTSSRNSEVIHAGIYYRAGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
A FCV GR+ LY YCA H IPH + GKLIVAT ++E KL I H NGVD L+ +D
Sbjct: 62 MAQFCVAGRQALYAYCADHGIPHRKCGKLIVATTAAEADKLESIRAHAAANGVDDLRAID 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
G +A +MEP L CV A++SP +GIVDSH+ ML+L +G+AE GA F +
Sbjct: 122 GAEAQRMEPALSCVAALVSPSTGIVDSHAYMLAL-------------RGDAEAAGAAFAF 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
++ ++ EG + L + + + L +L++N+AGL A A+A+
Sbjct: 169 HAPLLSARSEGEVLALEIGG-------------EMPMTLGCRLLINAAGLGATAVARSIA 215
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
+ ++++P Y A+G YFT S +PF HLIYP+PE GGLGVH+TLDL GQ KFGPDVE
Sbjct: 216 AMPSELVPTPYLAKGNYFTCS--ARAPFSHLIYPVPEPGGLGVHLTLDLGGQAKFGPDVE 273
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID + DY V RA +FYP IRKY+P L DG+L PGYSGIRPK+ P +
Sbjct: 274 WID----------ELDYVVDPARAARFYPAIRKYWPGLPDGALSPGYSGIRPKIVPPSVA 323
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFIS 467
DFV+Q HGVPGLINLFGIESPGLT++LAIAD ++
Sbjct: 324 VQDFVVQSPREHGVPGLINLFGIESPGLTAALAIADHVA 362
>ref|ZP_00276632.1| COG0579: Predicted dehydrogenase [Ralstonia metallidurans CH34]
Length = 368
Score = 386 bits (991), Expect = e-106
Identities = 203/399 (50%), Positives = 268/399 (66%), Gaps = 37/399 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E+VDCVVIGAGVVG+AVARALA +GREVI++E+ +FGT TS+RNSEV+HAGIYYP SL
Sbjct: 2 EQVDCVVIGAGVVGLAVARALAPQGREVIILEAENAFGTITSARNSEVIHAGIYYPAGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV+G+ MLY+YCA H + H++ GKLIVAT ++++ L I NGV ++++
Sbjct: 62 KAELCVRGKTMLYDYCASHHVAHQRCGKLIVATSTAQVATLEGIRAKAAANGVHDMQLLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A MEP+L+C A+LSP +GI+DSH LM +L+ G+AE GA
Sbjct: 122 RDEARAMEPQLECEAALLSPSTGIIDSHGLMTALL-------------GDAERAGAMLAV 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V+ G + + I L V S VL+ + VVNSAGL+A LA+R
Sbjct: 169 QSPVLSGAVTPDGIRLEV------------GSDDGSTVLLARTVVNSAGLTAPDLARRID 216
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
G+ + IPP YYA+GCYFTL+ +PF LIYP+PE GLGVH+TLD+ GQ +FGP+V
Sbjct: 217 GIPPEHIPPQYYAKGCYFTLAG--RAPFSRLIYPVPEAAGLGVHLTLDMGGQARFGPNVR 274
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID + +YSV + A+ FY E+R+Y+P L DG+L+PGY+GIRPK+SGP +
Sbjct: 275 WID----------EIEYSVPPHDADSFYDEVRRYWPGLADGALQPGYAGIRPKISGPTEV 324
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFIS 467
DF I G HGVPGL+NLFGIESPGLTSSLAIA+ ++
Sbjct: 325 AADFRIDGPRTHGVPGLVNLFGIESPGLTSSLAIAERVA 363
>ref|ZP_00202862.1| COG0579: Predicted dehydrogenase [Ralstonia eutropha JMP134]
Length = 368
Score = 385 bits (988), Expect = e-105
Identities = 205/401 (51%), Positives = 267/401 (66%), Gaps = 36/401 (8%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E VDCVVIGAGVVG+AVARALA GREVI++E+ +FGT TS+RNSEV+HAGIYYP SL
Sbjct: 2 ENVDCVVIGAGVVGLAVARALAQNGREVIILEAENAFGTITSARNSEVIHAGIYYPAGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV+G+EMLYEYCA H + H++ GKLIVAT +++ L I NGV ++++
Sbjct: 62 KAELCVRGKEMLYEYCASHHVAHQRCGKLIVATSEAQVATLEGIRAKAAANGVHDMQLLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A +EP+LQC A+LSP +GIVDSH LM +L+ G+AEN GA
Sbjct: 122 RAEAQALEPQLQCHAALLSPSTGIVDSHGLMTALL-------------GDAENAGAMLAV 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V G + I L V +G S+ L+ + VVNSAGL+A LA+R
Sbjct: 169 QSPVRRGAITPAGIELEVGSE------DGGST-----TLLARSVVNSAGLTAPELARRID 217
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
G+ IPP YYA+GCYFTL+ +PF LIYP+PE GLGVH+T+D+ GQ +FGP+V
Sbjct: 218 GMPEDHIPPQYYAKGCYFTLAG--RAPFSRLIYPVPEAAGLGVHLTIDMGGQARFGPNVR 275
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID + +YSV A+ FY E+R+Y+P L DG+L+PGY+GIRPK+SGP +
Sbjct: 276 WIDDI----------EYSVDPADADSFYDEVRRYWPALADGALQPGYAGIRPKISGPHEV 325
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFISTK 469
DF I G +HGVPGL++LFGIESPGLTSSLAIA+ ++ +
Sbjct: 326 AADFRIDGPAVHGVPGLVHLFGIESPGLTSSLAIAERVAAE 366
>ref|ZP_00280545.1| COG0579: Predicted dehydrogenase [Burkholderia fungorum LB400]
Length = 368
Score = 380 bits (975), Expect = e-104
Identities = 200/399 (50%), Positives = 262/399 (65%), Gaps = 46/399 (11%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
++++CVVIGAGVVG+AVARALA +GREVIV+E+A + G GTSSRNSEV+HAGIYYP SL
Sbjct: 2 DQIECVVIGAGVVGLAVARALAARGREVIVLEAAEAIGVGTSSRNSEVIHAGIYYPRGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV+GREMLY+YC + +PH + GKL+VAT ++IP+L I+ G NGV L +
Sbjct: 62 KAALCVRGREMLYDYCVERKVPHSRCGKLLVATSRNQIPQLENIMAKGRDNGVLDLMRIT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
G A +EP L+CV+A+ SP +GIVDSH ML+L QG+AE GA +
Sbjct: 122 GDQAQALEPALECVEAVFSPQTGIVDSHQFMLAL-------------QGDAERDGAVCAF 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQ----PELVLIPKLVVNSAGLSALALA 304
++ V G ++ NG+ ++Q + V+NSAGL A ALA
Sbjct: 169 HAPVQG-----------------IEASNGRFNIQVGGDAPTTISAACVINSAGLQANALA 211
Query: 305 KRFTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFG 364
++ GL+ + +PP Y ARG YF++S +PF LIYP+P + GLGVH+T+DL GQ +FG
Sbjct: 212 RKIRGLDARHVPPLYLARGNYFSISG--RAPFSRLIYPMPNEAGLGVHLTIDLGGQARFG 269
Query: 365 PDVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSG 424
PDVEW+D + +Y V RAE FY IR Y+P L D +L+P Y+GIRPKLSG
Sbjct: 270 PDVEWVDAI----------NYDVDPRRAESFYAAIRAYWPALPDNALQPAYAGIRPKLSG 319
Query: 425 PCQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
P + DF+IQG HGV GL+NLFGIESPGLT+SLAIA
Sbjct: 320 PGEPAADFLIQGPAAHGVRGLVNLFGIESPGLTASLAIA 358
>ref|ZP_00507387.1| FAD dependent oxidoreductase [Polaromonas sp. JS666]
gi|67779075|gb|EAM38695.1| FAD dependent oxidoreductase
[Polaromonas sp. JS666]
Length = 364
Score = 379 bits (973), Expect = e-103
Identities = 200/399 (50%), Positives = 263/399 (65%), Gaps = 39/399 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
+++DCVVIGAGVVG+AVARALAL+GREV+V+E+A + G+GTSSRNSEV+HAGIYY SL
Sbjct: 2 DQIDCVVIGAGVVGLAVARALALQGREVMVLEAADAIGSGTSSRNSEVIHAGIYYAQGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV+G+++LY YCA+ I H + GKLIVA+ ++ +L I++ NGV L ++
Sbjct: 62 KARLCVEGKDLLYAYCAERGIGHSRCGKLIVASAEPQVAQLRGIIDKAAANGVRDLVLLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A +EP+L CV A+ SP +GIVDSH LML+L QG+ EN G
Sbjct: 122 RDEAQSLEPQLACVAAVHSPSTGIVDSHGLMLAL-------------QGDLENAGGLVVL 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
NS + + I L + L+ K VVN+AGL A +LA+RF
Sbjct: 169 NSPLDRAVCSQDAITLIAEDGTELQ---------------AKTVVNAAGLHAQSLARRFA 213
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL +PP++YA+G YFTLS SPF LIYP+PE GLGVH+T+DL GQ KFGPDV+
Sbjct: 214 GLAGHHVPPSHYAKGNYFTLSG--RSPFSRLIYPVPEAAGLGVHLTIDLGGQAKFGPDVQ 271
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
W++ DD+ V R E FY E+RKY+P L+DG+L PGY+GIRPK+ P +
Sbjct: 272 WVNSPDDL---------VVDPARGEAFYAEVRKYWPALQDGALIPGYAGIRPKIQAPDEP 322
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFIS 467
DF+IQG D+HG+PGL+NLFGIESPGLTSSLAI ++S
Sbjct: 323 ARDFLIQGPDVHGIPGLVNLFGIESPGLTSSLAIGSYVS 361
>gb|AAS83089.1| glycerol-3-P dehydrogenase-like protein [Azospirillum brasilense]
Length = 373
Score = 372 bits (956), Expect = e-101
Identities = 196/401 (48%), Positives = 258/401 (63%), Gaps = 32/401 (7%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
ERVDCVV+GAGV+G+AVAR LA GREV+V+E+A + GTGTSSRNSEV+HAGIYYP SL
Sbjct: 2 ERVDCVVVGAGVIGLAVARRLARAGREVVVLEAADAIGTGTSSRNSEVIHAGIYYPTGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
+A CV GR+ LY YCA H + H++ GKLIVAT +++P+L I NGV L
Sbjct: 62 RARLCVAGRDALYAYCAAHGVEHKRIGKLIVATEEAQLPRLEAIRAQAAANGVGDLVAFS 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
AM +EP L+CV A+LSP +GI+DSH LML+L +G+AE GA +
Sbjct: 122 ASKAMALEPSLRCVGALLSPSTGIIDSHGLMLAL-------------RGDAEAAGAMLAF 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S + + + L V + ++ G VVN+AGL A +A+
Sbjct: 169 LSPLERAYRAADGFELEVGGAEPMRVACG-------------TVVNAAGLGAWGVARNLG 215
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
G + +PP A+G Y+ L + SPF L+YP+P +GGLGVH+TLDL GQ +FGPDVE
Sbjct: 216 GFPAEHVPPRVLAKGNYYALGQGR-SPFARLVYPVPVEGGLGVHLTLDLAGQARFGPDVE 274
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
W+D D ++ DY V RA+ FY E+R+Y+P+L DG+L P YSG+RPKLSGP Q
Sbjct: 275 WLDPAD-----YDRPDYRVDPKRADGFYAEVRRYWPDLPDGALVPAYSGVRPKLSGPGQP 329
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFISTK 469
DF+IQG + HGV GL+NLFG+ESPGLTS LAIAD ++ +
Sbjct: 330 QADFLIQGPETHGVAGLVNLFGMESPGLTSCLAIADAVAER 370
>emb|CAE34407.1| putative conserved exported protein [Bordetella bronchiseptica
RB50] gi|33603018|ref|NP_890578.1| putative conserved
exported protein [Bordetella bronchiseptica RB50]
Length = 369
Score = 367 bits (941), Expect = e-100
Identities = 194/397 (48%), Positives = 253/397 (62%), Gaps = 43/397 (10%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKA 130
+DC+VIGAGVVG+A+ARALA G EV+V E+A GTGTSSRNSEV+HAGIYYP SLKA
Sbjct: 5 IDCIVIGAGVVGLAIARALAAGGHEVLVAEAAEGIGTGTSSRNSEVIHAGIYYPADSLKA 64
Query: 131 IFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDGV 190
CV+G+ +LYEYCA +PH++ GKLIVAT +E +L I NGVD L+ +DG
Sbjct: 65 RLCVRGKHLLYEYCAARGVPHQRLGKLIVATSDAEASQLDSIARRAGANGVDDLQHIDGA 124
Query: 191 DAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTYNS 250
A ++EP L C A++SP +GIVDSH+LML+ QG+AEN GA +++
Sbjct: 125 AARRLEPALHCTAALVSPSTGIVDSHALMLA-------------YQGDAENDGAQLVFHT 171
Query: 251 TVIGGHME---GNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRF 307
+I G + G E+ +E + L ++++N+AGL A LA+R
Sbjct: 172 PLIAGRVRPEGGFELDFGGAE---------------PMTLSCRVLINAAGLHAPGLARRI 216
Query: 308 TGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDV 367
G+ IPP Y +G YFTL+ +PF LIYP+P+ GLGVH+TLDL GQ KFGPD
Sbjct: 217 EGIPRDSIPPEYLCKGSYFTLAG--RAPFSRLIYPVPQHAGLGVHLTLDLGGQAKFGPDT 274
Query: 368 EWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQ 427
EWI DY++ RA+ FY +R Y+P L DG+L PGY+GIRPK+SGP +
Sbjct: 275 EWI----------ATEDYTLDPRRADVFYAAVRSYWPALPDGALAPGYTGIRPKISGPHE 324
Query: 428 SPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIAD 464
DF I G HGV GL+NL+GIESPGLT+SLAIA+
Sbjct: 325 PAADFAISGPASHGVAGLVNLYGIESPGLTASLAIAE 361
>gb|AAQ57814.1| conserved hypothetical protein [Chromobacterium violaceum ATCC
12472] gi|34495590|ref|NP_899805.1| hypothetical protein
CV0135 [Chromobacterium violaceum ATCC 12472]
Length = 364
Score = 366 bits (939), Expect = e-99
Identities = 197/396 (49%), Positives = 251/396 (62%), Gaps = 41/396 (10%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
ERV+CVVIGAGVVG+AVARA A GREV+++E+ P+ G SSRNSEV+HAG+YYP S+
Sbjct: 2 ERVECVVIGAGVVGLAVARAQAQAGREVVIVEAEPAMGLHASSRNSEVIHAGLYYPAGSM 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV GR+ LY YCA+ IPH + GKLIVA+R S++ +L + NGV ++ +
Sbjct: 62 KARLCVAGRDALYRYCAERAIPHRRLGKLIVASRESQLARLDALEAQARANGVADIRRLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A +EP L C A+LSP +GIVDSH+LMLSL+ +AE GA
Sbjct: 122 ASEARALEPALDCAAALLSPSTGIVDSHALMLSLL-------------ADAEAAGAQLAL 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V GG + + + L V+ + L+ K VVN+AGL A +A+
Sbjct: 169 ASPVEGGEVTPDGVALRVA----------------GMALLAKRVVNAAGLFAPDVARAIA 212
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL IP A YARG YF+L +PF L+YP+PE GGLG H+TLDL GQ +FGPDVE
Sbjct: 213 GLRADSIPQARYARGVYFSLQG--RAPFSRLVYPLPEAGGLGSHLTLDLAGQARFGPDVE 270
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
W+DGV DY V RA+ FY +R ++P L DG+L PGY+GIR K++GP Q
Sbjct: 271 WVDGV----------DYRVDPARADAFYRAVRAWWPQLPDGALAPGYAGIRAKIAGPGQP 320
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIAD 464
DFVIQG +HG PGL+NLFGIESPGLTS LAIAD
Sbjct: 321 DADFVIQGPAVHGAPGLVNLFGIESPGLTSCLAIAD 356
>emb|CAE43520.1| putative conserved exported protein [Bordetella pertussis Tohama I]
gi|33594157|ref|NP_881801.1| putative conserved exported
protein [Bordetella pertussis Tohama I]
Length = 369
Score = 364 bits (935), Expect = 3e-99
Identities = 193/397 (48%), Positives = 253/397 (63%), Gaps = 43/397 (10%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKA 130
+DC+VIGAGVVG+A+ARALA G EV+V E+A GTGTSSRNSEV+HAGIYYP SLKA
Sbjct: 5 IDCIVIGAGVVGLAIARALAAGGHEVLVAEAAEGIGTGTSSRNSEVIHAGIYYPADSLKA 64
Query: 131 IFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDGV 190
CV+G+ +LYEYCA +PH++ GKLIVAT +E +L I NGVD L+ +DG
Sbjct: 65 RLCVRGKHLLYEYCAARGVPHQRLGKLIVATSDAEASQLDSIARRAGANGVDDLQHIDGA 124
Query: 191 DAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTYNS 250
A ++EP L C A++SP +GIVDSH+LML+ QG+AE+ GA +++
Sbjct: 125 AARRLEPALHCTAALVSPSTGIVDSHALMLA-------------YQGDAESDGAQLVFHT 171
Query: 251 TVIGGHME---GNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRF 307
+I G + G E+ +E + L ++++N+AGL A LA+R
Sbjct: 172 PLIAGRVRPEGGFELDFGGAE---------------PMTLSCRVLINAAGLHAPGLARRI 216
Query: 308 TGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDV 367
G+ IPP Y +G YFTL+ +PF LIYP+P+ GLGVH+TLDL GQ KFGPD
Sbjct: 217 EGIPRDSIPPEYLCKGSYFTLAG--RAPFSRLIYPVPQHAGLGVHLTLDLGGQAKFGPDT 274
Query: 368 EWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQ 427
EWI DY++ RA+ FY +R Y+P L DG+L PGY+GIRPK+SGP +
Sbjct: 275 EWI----------ATEDYTLDPRRADVFYAAVRSYWPALPDGALAPGYTGIRPKISGPHE 324
Query: 428 SPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIAD 464
DF I G HGV GL+NL+GIESPGLT+SLAIA+
Sbjct: 325 PAADFAIAGPASHGVAGLVNLYGIESPGLTASLAIAE 361
>ref|ZP_00486875.1| COG0579: Predicted dehydrogenase [Burkholderia pseudomallei 668]
Length = 373
Score = 364 bits (934), Expect = 4e-99
Identities = 189/395 (47%), Positives = 252/395 (62%), Gaps = 38/395 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
ER+DCVVIGAGVVG+A+AR LA +GRE +++E+A + GTG SSRNSEV+HAG+YYP SL
Sbjct: 4 ERIDCVVIGAGVVGLAIARELAARGRETLILEAADAIGTGMSSRNSEVIHAGLYYPRGSL 63
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA C+ GR++LY++C H +PH + GKL+VAT ++ +L I +NGV L +
Sbjct: 64 KASLCLHGRDLLYDFCETHGVPHRRCGKLVVATSPAQAKQLKAIAARAEENGVLDLLTLS 123
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+ +EPEL+C++A+ SP +GI+DSH LML+L+ G+AE GA+
Sbjct: 124 RDEVQALEPELECLEALFSPSTGIIDSHQLMLALL-------------GDAERDGASCAL 170
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V G + ++ + + V+NSAGL A ALAKR
Sbjct: 171 RSPVESIDAAGGRFVV-------------RTGGEAPAAIAAACVINSAGLDAQALAKRIR 217
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL+++ +PP Y ARG YF+LS +PF HL+YP+P+ GLGVH+TLDL G +FGPDVE
Sbjct: 218 GLDSRWVPPLYLARGNYFSLSG--RAPFAHLVYPVPDRAGLGVHLTLDLAGDARFGPDVE 275
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID + Y V RAE FY IR Y+P L +GSL+P Y+GIR K+SGP +
Sbjct: 276 WIDAL----------RYDVDPRRAESFYTSIRAYWPGLPEGSLQPAYAGIRAKVSGPGEP 325
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
DFVIQG HGV GL+NLFGIESPGLT++LAIA
Sbjct: 326 AADFVIQGTAQHGVRGLVNLFGIESPGLTAALAIA 360
>ref|ZP_00463053.1| FAD dependent oxidoreductase [Burkholderia cenocepacia HI2424]
gi|67658338|ref|ZP_00455711.1| FAD dependent
oxidoreductase [Burkholderia cenocepacia AU 1054]
gi|67100602|gb|EAM17754.1| FAD dependent oxidoreductase
[Burkholderia cenocepacia HI2424]
gi|67094179|gb|EAM11715.1| FAD dependent oxidoreductase
[Burkholderia cenocepacia AU 1054]
Length = 369
Score = 364 bits (934), Expect = 4e-99
Identities = 194/395 (49%), Positives = 254/395 (64%), Gaps = 37/395 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E+ DCVVIGAGVVG+A+AR LA +GRE IV+E+A + GTGTSSRNSEV+HAG+YYP SL
Sbjct: 2 EQTDCVVIGAGVVGLAIARELAARGRETIVLEAADAIGTGTSSRNSEVIHAGLYYPRGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV GR++LYE+C H +PH +TGKL+VAT ++++ +L I +NGV L +
Sbjct: 62 KATSCVHGRDLLYEFCDTHHVPHRRTGKLLVATSAAQLKQLKAIAARAAENGVLDLLPLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A +EP L+CV+A+ SP +GIVDSH LML+L+ G+AE GA
Sbjct: 122 RAEAQTLEPALECVEALFSPSTGIVDSHQLMLALL-------------GDAERDGAVCAL 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V + G + + G + + + V+NSAGL A ALA+R
Sbjct: 169 QSPVESIDVLGG--------GRFIVRTGGAAPTEIDAA----CVINSAGLGAQALARRTR 216
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL+ + +PP Y ARG YF+LS +PF HLIYP+PE GLGVH+T DL GQ +FGPDVE
Sbjct: 217 GLDPRWVPPLYLARGNYFSLSG--RAPFSHLIYPMPERAGLGVHLTFDLGGQARFGPDVE 274
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
W D + Y V RA FY IR ++P L D +L+P Y+GIRPKL+GP +
Sbjct: 275 WCDSL----------RYEVDPARAGAFYTSIRAFWPGLPDDALQPAYAGIRPKLAGPGEP 324
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
P DF++QG HGV GL+NLFGIESPGLT+SLA+A
Sbjct: 325 PADFIVQGAAQHGVRGLVNLFGIESPGLTASLALA 359
>ref|ZP_00448157.1| COG0579: Predicted dehydrogenase [Burkholderia mallei SAVP1]
gi|67649006|ref|ZP_00447232.1| COG0579: Predicted
dehydrogenase [Burkholderia mallei NCTC 10247]
gi|67641512|ref|ZP_00440289.1| COG0579: Predicted
dehydrogenase [Burkholderia mallei GB8 horse 4]
gi|67635581|ref|ZP_00434535.1| COG0579: Predicted
dehydrogenase [Burkholderia mallei 10399]
gi|53724881|ref|YP_104769.1| oxidoreductase, FAD-binding
family protein [Burkholderia mallei ATCC 23344]
gi|52428304|gb|AAU48897.1| oxidoreductase, FAD-binding
family protein [Burkholderia mallei ATCC 23344]
Length = 373
Score = 364 bits (934), Expect = 4e-99
Identities = 189/395 (47%), Positives = 252/395 (62%), Gaps = 38/395 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
ER+DCVVIGAGVVG+A+AR LA +GRE +++E+A + GTG SSRNSEV+HAG+YYP SL
Sbjct: 4 ERIDCVVIGAGVVGLAIARELATRGRETLILEAADAIGTGMSSRNSEVIHAGLYYPRGSL 63
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA C+ GR++LY++C H +PH + GKL+VAT ++ +L I +NGV L +
Sbjct: 64 KASLCLHGRDLLYDFCETHGVPHRRCGKLVVATSPAQAKQLKAIAARAEENGVLDLLTLS 123
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+ +EPEL+C++A+ SP +GI+DSH LML+L+ G+AE GA+
Sbjct: 124 RDEVQALEPELECLEALFSPSTGIIDSHQLMLALL-------------GDAERDGASCAL 170
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V G + ++ + + V+NSAGL A ALAKR
Sbjct: 171 RSPVESIDAAGGRFVV-------------RTGGEAPAAIAAACVINSAGLDAQALAKRIR 217
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL+++ +PP Y ARG YF+LS +PF HL+YP+P+ GLGVH+TLDL G +FGPDVE
Sbjct: 218 GLDSRWVPPLYLARGNYFSLSG--RAPFAHLVYPVPDRAGLGVHLTLDLAGDARFGPDVE 275
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID + Y V RAE FY IR Y+P L +GSL+P Y+GIR K+SGP +
Sbjct: 276 WIDAL----------RYDVDPRRAESFYTSIRAYWPGLPEGSLQPAYAGIRAKVSGPGEP 325
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
DFVIQG HGV GL+NLFGIESPGLT++LAIA
Sbjct: 326 AADFVIQGTAQHGVRGLVNLFGIESPGLTAALAIA 360
>ref|NP_885768.1| putative conserved exported protein [Bordetella parapertussis
12822] gi|33566683|emb|CAE38893.1| putative conserved
exported protein [Bordetella parapertussis]
Length = 369
Score = 364 bits (934), Expect = 4e-99
Identities = 192/397 (48%), Positives = 253/397 (63%), Gaps = 43/397 (10%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKA 130
+DC+VIGAGVVG+A+ARALA G EV+V E+A GTGTSSRNSEV+HAGIYYP SLKA
Sbjct: 5 IDCIVIGAGVVGLAIARALAAGGHEVLVAEAAEGIGTGTSSRNSEVIHAGIYYPADSLKA 64
Query: 131 IFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDGV 190
CV+G+ +LYEYCA +PH++ GKLIVAT +E +L I NGVD L+ +DG
Sbjct: 65 RLCVRGKHLLYEYCAARGVPHQRLGKLIVATSDAEASQLDSIARRAGANGVDDLQHIDGA 124
Query: 191 DAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTYNS 250
A ++EP L C A++SP +GIVDSH+LML+ QG+AEN GA +++
Sbjct: 125 AARRLEPALHCTAALVSPSTGIVDSHALMLA-------------YQGDAENDGAQLVFHT 171
Query: 251 TVIGGHME---GNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRF 307
+I G + G E+ +E + L ++++N+AGL A LA+R
Sbjct: 172 PLIAGRVRPEGGFELDFGGAE---------------PMTLSCRVLINAAGLHAPGLARRI 216
Query: 308 TGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDV 367
G+ IPP Y +G YFTL+ +PF LIYP+P+ GLGVH+TL+L GQ KFGPD
Sbjct: 217 EGIPQDSIPPEYLCKGSYFTLAG--RAPFSRLIYPVPQHAGLGVHLTLELGGQAKFGPDT 274
Query: 368 EWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQ 427
EWI DY++ RA+ FY +R Y+P L DG+L PGY+GIRPK+SGP +
Sbjct: 275 EWI----------TTEDYTLDPRRADVFYAAVRSYWPALPDGALAPGYTGIRPKISGPHE 324
Query: 428 SPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIAD 464
DF I G HGV GL+NL+GI+SPGLT+SLAIA+
Sbjct: 325 PAADFAIAGPASHGVAGLVNLYGIKSPGLTASLAIAE 361
>ref|NP_105561.1| hypothetical protein mll4765 [Mesorhizobium loti MAFF303099]
gi|14024744|dbj|BAB51347.1| mll4765 [Mesorhizobium loti
MAFF303099]
Length = 370
Score = 362 bits (930), Expect = 1e-98
Identities = 199/398 (50%), Positives = 255/398 (64%), Gaps = 36/398 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
+ +DCVV GAGVVGIAVARALA+ GREV+V+E A + GT TSSRNSEV+HAG+YY SL
Sbjct: 2 QTIDCVVAGAGVVGIAVARALAVSGREVVVVEQADAIGTVTSSRNSEVIHAGLYYAPGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA CV+GR+ LY YCA+HDI H++TGKLIVA+ + L I + ++GVD L+ +
Sbjct: 62 KARLCVEGRQRLYAYCAEHDIAHKRTGKLIVASEPGQTDGLRKIEANARRSGVDDLQFLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A +EP L C A+LSP +GIVDSH+LMLSL +GEAE GA+F +
Sbjct: 122 RDEAENLEPALTCAGALLSPSTGIVDSHALMLSL-------------RGEAEAAGASFAF 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
+ V G +E + I + + + + L +N+AGL A ALA R
Sbjct: 169 LTAVAGATIEADGIRI-----------DTRDATGETFALEAGAFINAAGLEAQALAGRIE 217
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
G IP + ARG YF L SPF LIYP+P +GGLGVH+TLDL G+ +FGPDVE
Sbjct: 218 GFPRNFIPRQWLARGNYFALPG--RSPFSRLIYPVPVEGGLGVHLTLDLGGRARFGPDVE 275
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID V DY+V R+ FY IR+Y+P+L DG+L+P Y+GIRPKLSGP Q
Sbjct: 276 WIDHV----------DYTVDPGRSVVFYEAIRRYWPDLADGALQPAYAGIRPKLSGPGQP 325
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFI 466
DF+IQG HG ++NLFGIESPGLT+SLAIAD +
Sbjct: 326 AADFIIQGPADHGAGRIVNLFGIESPGLTASLAIADHV 363
>ref|NP_773748.1| dehydrogenase [Bradyrhizobium japonicum USDA 110]
gi|27355390|dbj|BAC52373.1| dehydrogenase
[Bradyrhizobium japonicum USDA 110]
Length = 367
Score = 362 bits (928), Expect = 2e-98
Identities = 200/399 (50%), Positives = 254/399 (63%), Gaps = 38/399 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
+RVDC+VIGAGVVG+AVAR LA GREVIV+E A + GT TSSRNSEV+HAGIYY S
Sbjct: 2 DRVDCIVIGAGVVGLAVARKLAQAGREVIVLEEAEAIGTITSSRNSEVIHAGIYYRAGSW 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
A CV G+ LY YC + IPH+ GKLIVAT + E KL I H NGV ++++
Sbjct: 62 MARMCVNGKHALYRYCGERGIPHKNCGKLIVATNAKETEKLQSIKAHAEANGVLDMQLLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
G A +EP L C A+LSP +GI+DSH+ MLSL +GEAE GA F +
Sbjct: 122 GEAARALEPALACDAALLSPSTGIIDSHAYMLSL-------------RGEAEEAGAAFAF 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
++ +I I ++ + + L L+VN+AGL+A +A+
Sbjct: 169 HTPLIRAKAAAGMI-------------EVEAGGEAPMALQCSLLVNAAGLAATTVARNID 215
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
G+ IPPAY A+G YF+ N KA PF HLIYP+PE GGLGVH+TLD+ GQ +FGPDVE
Sbjct: 216 GMPLDRIPPAYLAKGNYFS-CNAKA-PFSHLIYPVPEPGGLGVHLTLDMAGQARFGPDVE 273
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WI+ + +Y V +RAE+FYP IR+Y+P L DG+L P YSGIRPK+ P +
Sbjct: 274 WIETI----------NYEVDPSRAERFYPAIRRYWPTLPDGALMPSYSGIRPKIVPPAVA 323
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFIS 467
DF++QG HGV GLINLFGIESPGLTSSLAIAD ++
Sbjct: 324 TQDFLMQGPRDHGVAGLINLFGIESPGLTSSLAIADHVA 362
>ref|ZP_00211711.1| COG0579: Predicted dehydrogenase [Burkholderia cepacia R18194]
Length = 369
Score = 361 bits (927), Expect = 2e-98
Identities = 191/395 (48%), Positives = 258/395 (64%), Gaps = 37/395 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E++DCVVIGAGVVG+A+AR LA +GRE +V+E+A + GTGTSSRNSEV+HAG+YYP SL
Sbjct: 2 EQMDCVVIGAGVVGLAIARELAARGRETLVLEAADAIGTGTSSRNSEVIHAGLYYPRGSL 61
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA+ CV GR++LYE+C +PH + GKL+VAT ++++ +L I +NGV L +
Sbjct: 62 KAMSCVHGRDLLYEFCETRHVPHRRAGKLLVATSAAQVKQLKAIAVRATENGVLDLMPLT 121
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+A +EP L+CV+A+ SP +GIVDSH LML+L+ G+AE GA
Sbjct: 122 RAEAQTLEPALECVEALFSPSTGIVDSHQLMLALL-------------GDAERDGAVCAL 168
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V E ++ + + + G + + E V+NSAGL A ALA+R
Sbjct: 169 KSPV-----ESIDV---LRGGRFVVRTGGDAPTEIEAA----CVINSAGLGAQALARRTR 216
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL+ + +PP Y ARG YF+LS +PF HLIYP+P+ GLGVH+TLDL GQ +FGPDVE
Sbjct: 217 GLDPRWVPPLYLARGNYFSLSG--RAPFSHLIYPMPDRAGLGVHLTLDLGGQARFGPDVE 274
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
W+D + Y V RA FY IR ++P L D +L+P Y+GIRPK++GP +
Sbjct: 275 WVDAL----------RYEVDPARATAFYASIRAFWPGLPDDALQPAYAGIRPKVAGPGEP 324
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
P DF++QG HGV GL+NLFGIESPGLT+SLA+A
Sbjct: 325 PADFIVQGAAQHGVRGLVNLFGIESPGLTASLALA 359
>ref|ZP_00482688.1| COG0579: Predicted dehydrogenase [Burkholderia pseudomallei 1710b]
gi|52208301|emb|CAH34234.1| putative FAD dependent
oxidoreductase [Burkholderia pseudomallei K96243]
gi|53717887|ref|YP_106873.1| putative FAD dependent
oxidoreductase [Burkholderia pseudomallei K96243]
Length = 373
Score = 360 bits (923), Expect = 7e-98
Identities = 188/395 (47%), Positives = 250/395 (62%), Gaps = 38/395 (9%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
ER+DCVVIGAGVVG+A+AR LA +GRE +++E+A + GTG SSRNSEV+HAG+YYP SL
Sbjct: 4 ERIDCVVIGAGVVGLAIARELAARGRETLILEAADAIGTGMSSRNSEVIHAGLYYPRGSL 63
Query: 129 KAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMD 188
KA C+ GR++LY++C H +PH + GKL+VAT ++ +L I +NGV L +
Sbjct: 64 KASLCLHGRDLLYDFCETHGVPHRRCGKLVVATSPAQAKQLKAIAARAEENGVLDLLTLS 123
Query: 189 GVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
+ +EPEL+C++A+ SP +GI+DSH LML+L+ G+AE GA+
Sbjct: 124 RDEVQALEPELECLEALFSPSTGIIDSHQLMLALL-------------GDAERDGASCAL 170
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
S V G + ++ + + V+NSAGL A ALAKR
Sbjct: 171 RSPVESIDAAGGRFVV-------------RTGGEAPAAIAAACVINSAGLDAQALAKRIR 217
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQVKFGPDVE 368
GL+++ +PP Y ARG YF+LS +PF HL+YP+P+ GLGVH+TLDL G +FGPDVE
Sbjct: 218 GLDSRWVPPLYLARGNYFSLSG--RAPFAHLVYPVPDRAGLGVHLTLDLAGDARFGPDVE 275
Query: 369 WIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRPKLSGPCQS 428
WID + Y V RAE F IR Y+P L +GSL P Y+GIR K+SGP +
Sbjct: 276 WIDAL----------RYDVDPRRAESFCTSIRAYWPGLPEGSLRPAYAGIRAKVSGPGEP 325
Query: 429 PVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
DFVIQG HGV GL+NLFGIESPGLT++LAIA
Sbjct: 326 AADFVIQGTAQHGVRGLVNLFGIESPGLTAALAIA 360
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 812,783,319
Number of Sequences: 2540612
Number of extensions: 34894197
Number of successful extensions: 109458
Number of sequences better than 10.0: 1195
Number of HSP's better than 10.0 without gapping: 596
Number of HSP's successfully gapped in prelim test: 601
Number of HSP's that attempted gapping in prelim test: 107971
Number of HSP's gapped (non-prelim): 1438
length of query: 470
length of database: 863,360,394
effective HSP length: 132
effective length of query: 338
effective length of database: 527,999,610
effective search space: 178463868180
effective search space used: 178463868180
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146910.5


