

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.9 + phase: 0
(100 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
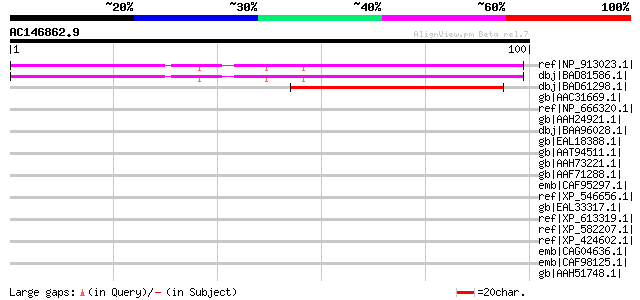
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_913023.1| unnamed protein product [Oryza sativa (japonica... 96 3e-19
dbj|BAD81586.1| putative 3' exoribonuclease [Oryza sativa (japon... 96 3e-19
dbj|BAD61298.1| unknown protein [Oryza sativa (japonica cultivar... 60 1e-08
gb|AAC31669.1| Unknown gene product [Homo sapiens] 39 0.032
ref|NP_666320.1| putative DNA glycosylase [Mus musculus] gi|2447... 39 0.032
gb|AAH24921.1| Putative DNA glycosylase [Mus musculus] 39 0.032
dbj|BAA96028.1| KIAA1504 protein [Homo sapiens] 39 0.032
gb|EAL18388.1| hypothetical protein CNBJ3110 [Cryptococcus neofo... 39 0.041
gb|AAT94511.1| LD04601p [Drosophila melanogaster] gi|22946848|gb... 39 0.041
gb|AAH73221.1| MGC80537 protein [Xenopus laevis] 39 0.041
gb|AAF71288.1| DNA topoisomerase III alpha [Drosophila melanogas... 39 0.041
emb|CAF95297.1| unnamed protein product [Tetraodon nigroviridis] 38 0.054
ref|XP_546656.1| PREDICTED: similar to topoisomerase (DNA) III a... 38 0.070
gb|EAL33317.1| GA10090-PA [Drosophila pseudoobscura] 38 0.070
ref|XP_613319.1| PREDICTED: similar to 4933424N09Rik protein [Bo... 37 0.092
ref|XP_582207.1| PREDICTED: similar to 4933424N09Rik protein [Bo... 37 0.092
ref|XP_424602.1| PREDICTED: similar to RIKEN cDNA 4933424N09 [Ga... 37 0.092
emb|CAG04636.1| unnamed protein product [Tetraodon nigroviridis] 37 0.12
emb|CAF98125.1| unnamed protein product [Tetraodon nigroviridis] 37 0.12
gb|AAH51748.1| Topoisomerase (DNA) III alpha [Homo sapiens] gi|2... 36 0.20
>ref|NP_913023.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|11138061|dbj|BAB17734.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 161
Score = 95.5 bits (236), Expect = 3e-19
Identities = 51/111 (45%), Positives = 66/111 (58%), Gaps = 15/111 (13%)
Query: 1 MHRGFKFSITNSIMWQTADRSLIWKQSPEQPSAYP----HFPFKARDMNTPI-------I 49
MHRGFKFSITNS++WQ+A +S+ + SP S YP H P + M +P+ I
Sbjct: 54 MHRGFKFSITNSLVWQSAPQSITCQSSPAH-SPYPNQSHHKPMEV--MGSPVQVNPYAGI 110
Query: 50 QYHHPC-CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWAS 99
P C+CGV S + +PGP G F+GCGNWT+TRGA C Y+ W S
Sbjct: 111 TVKKPMYCHCGVLSQIKVTYRPGPMHGRYFYGCGNWTSTRGANCDYWVWLS 161
>dbj|BAD81586.1| putative 3' exoribonuclease [Oryza sativa (japonica
cultivar-group)] gi|56783678|dbj|BAD81090.1| putative 3'
exoribonuclease [Oryza sativa (japonica cultivar-group)]
Length = 411
Score = 95.5 bits (236), Expect = 3e-19
Identities = 51/111 (45%), Positives = 66/111 (58%), Gaps = 15/111 (13%)
Query: 1 MHRGFKFSITNSIMWQTADRSLIWKQSPEQPSAYP----HFPFKARDMNTPI-------I 49
MHRGFKFSITNS++WQ+A +S+ + SP S YP H P + M +P+ I
Sbjct: 304 MHRGFKFSITNSLVWQSAPQSITCQSSPAH-SPYPNQSHHKPMEV--MGSPVQVNPYAGI 360
Query: 50 QYHHPC-CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWAS 99
P C+CGV S + +PGP G F+GCGNWT+TRGA C Y+ W S
Sbjct: 361 TVKKPMYCHCGVLSQIKVTYRPGPMHGRYFYGCGNWTSTRGANCDYWVWLS 411
>dbj|BAD61298.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 304
Score = 60.1 bits (144), Expect = 1e-08
Identities = 23/41 (56%), Positives = 28/41 (68%)
Query: 55 CCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYF 95
CCYCGV S+ G++ PG Q F+GCGNWTA GA C +F
Sbjct: 253 CCYCGVASTGGVMAMPGSTQRRCFYGCGNWTAVSGATCPFF 293
>gb|AAC31669.1| Unknown gene product [Homo sapiens]
Length = 426
Score = 38.9 bits (89), Expect = 0.032
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 12/82 (14%)
Query: 20 RSLIWKQSPEQPSAYPHF--PFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSL 77
RS W++ P ++ + P+K+ M P+ C CG +S R +V GP G +
Sbjct: 301 RSSSWRRLPSILTSTVNLQEPWKSGKMTPPL-------CKCGRRSKRLVVSNNGPNHGKV 353
Query: 78 FFGC--GNWTATRGARCHYFEW 97
F+ C G + R C YF+W
Sbjct: 354 FYCCPIGKYQENRKC-CGYFKW 374
>ref|NP_666320.1| putative DNA glycosylase [Mus musculus] gi|24475412|dbj|BAC22661.1|
putative DNA glycosylase [Mus musculus]
gi|56404613|sp|Q8K203|NEIL3_MOUSE Endonuclease VIII-like
3 (Nei-like 3) (DNA glycosylase FPG2)
gi|21961370|gb|AAH34753.1| Putative DNA glycosylase [Mus
musculus]
Length = 606
Score = 38.9 bits (89), Expect = 0.032
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 9/53 (16%)
Query: 46 TPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
+P+ + HH C V VRK G +G F+ C + RGA+C +FEWA
Sbjct: 505 SPLCKMHHRRCVLRV------VRKDGENKGRQFYAC---SLPRGAQCGFFEWA 548
Score = 31.6 bits (70), Expect = 5.0
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query: 54 PCCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
P C G +S V K GP G FF C + +C++F+WA
Sbjct: 553 PFCRHGKRSIMKTVLKIGPNNGKNFFVC---PLEKKKQCNFFQWA 594
>gb|AAH24921.1| Putative DNA glycosylase [Mus musculus]
Length = 606
Score = 38.9 bits (89), Expect = 0.032
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 9/53 (16%)
Query: 46 TPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
+P+ + HH C V VRK G +G F+ C + RGA+C +FEWA
Sbjct: 505 SPLCKMHHRRCVLRV------VRKDGENKGRQFYAC---SLPRGAQCGFFEWA 548
Score = 31.6 bits (70), Expect = 5.0
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query: 54 PCCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
P C G +S V K GP G FF C + +C++F+WA
Sbjct: 553 PFCKHGKRSIMKTVLKIGPNNGKNFFVC---PLEKEKQCNFFQWA 594
>dbj|BAA96028.1| KIAA1504 protein [Homo sapiens]
Length = 447
Score = 38.9 bits (89), Expect = 0.032
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 12/82 (14%)
Query: 20 RSLIWKQSPEQPSAYPHF--PFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSL 77
RS W++ P ++ + P+K+ M P+ C CG +S R +V GP G +
Sbjct: 322 RSSSWRRLPSILTSTVNLQEPWKSGKMTPPL-------CKCGRRSKRLVVSNNGPNHGKV 374
Query: 78 FFGC--GNWTATRGARCHYFEW 97
F+ C G + R C YF+W
Sbjct: 375 FYCCPIGKYQENRKC-CGYFKW 395
>gb|EAL18388.1| hypothetical protein CNBJ3110 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 1059
Score = 38.5 bits (88), Expect = 0.041
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 3/44 (6%)
Query: 54 PCCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEW 97
P C CG+ ++ V K GP +G F+ C N ARC +F+W
Sbjct: 824 PSCKCGLDAAFATVIKEGPNKGRQFWACPN---NPKARCGFFQW 864
>gb|AAT94511.1| LD04601p [Drosophila melanogaster] gi|22946848|gb|AAF53813.2|
CG10123-PA [Drosophila melanogaster]
gi|24585251|ref|NP_523602.2| CG10123-PA [Drosophila
melanogaster] gi|33860232|sp|Q9NG98|TOP3A_DROME DNA
topoisomerase III alpha
Length = 1250
Score = 38.5 bits (88), Expect = 0.041
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 4/42 (9%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEW 97
C CG +S+ VRK GP QG F+ C TR C +F+W
Sbjct: 1152 CNCGQLASQLTVRKDGPNQGRPFYAC----PTREKSCGFFKW 1189
Score = 35.4 bits (80), Expect = 0.35
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 12/93 (12%)
Query: 13 IMWQTADRSLIWKQSPEQP----SAYPHFPFKARDMNTPIIQYHHP---CCYCGVKSSRG 65
++W T +R+ + +P P + P + RD P C C + +
Sbjct: 986 MLWGTRERASLGTAAPTPPPKPAAKRPRWDSVERDSTPPSSVPESETVLCTGCQQPARQN 1045
Query: 66 MVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
VRK GP G L++ C + C++F+WA
Sbjct: 1046 TVRKNGPNLGRLYYKC-----PKPDECNFFQWA 1073
>gb|AAH73221.1| MGC80537 protein [Xenopus laevis]
Length = 1022
Score = 38.5 bits (88), Expect = 0.041
Identities = 18/43 (41%), Positives = 23/43 (52%), Gaps = 6/43 (13%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
C CGV + + VRK GP QG F+ C G C++F WA
Sbjct: 827 CNCGVAAVQLTVRKEGPNQGRPFYKC------NGGACNFFLWA 863
>gb|AAF71288.1| DNA topoisomerase III alpha [Drosophila melanogaster]
Length = 1250
Score = 38.5 bits (88), Expect = 0.041
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 4/42 (9%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEW 97
C CG +S+ VRK GP QG F+ C TR C +F+W
Sbjct: 1152 CNCGQLASQLTVRKDGPNQGRPFYAC----PTREKSCGFFKW 1189
Score = 35.4 bits (80), Expect = 0.35
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 12/93 (12%)
Query: 13 IMWQTADRSLIWKQSPEQP----SAYPHFPFKARDMNTPIIQYHHP---CCYCGVKSSRG 65
++W T +R+ + +P P + P + RD P C C + +
Sbjct: 986 MLWGTRERASLGTAAPTPPPKPAAKRPRWDSVERDSTPPSSVPESETVLCTGCQQPARQN 1045
Query: 66 MVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
VRK GP G L++ C + C++F+WA
Sbjct: 1046 TVRKNGPNLGRLYYKC-----PKPDECNFFQWA 1073
>emb|CAF95297.1| unnamed protein product [Tetraodon nigroviridis]
Length = 984
Score = 38.1 bits (87), Expect = 0.054
Identities = 23/74 (31%), Positives = 32/74 (43%), Gaps = 11/74 (14%)
Query: 25 KQSPEQPSAYPHFPFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSLFFGCGNW 84
+ P QP P +R+ ++ I C CG ++ VRK GP QG F+ C
Sbjct: 787 RPGPPQPGPPPQALSASRNSDSDAIM-----CNCGQQAVLLTVRKDGPNQGRQFYKC--- 838
Query: 85 TATRGARCHYFEWA 98
G C +F WA
Sbjct: 839 ---NGGTCAFFLWA 849
Score = 35.8 bits (81), Expect = 0.27
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
C C + V+K GP +G +F CG TR +C +F+WA
Sbjct: 886 CNCQETAVTRTVQKDGPNKGRMFHTCGK---TREQQCGFFQWA 925
>ref|XP_546656.1| PREDICTED: similar to topoisomerase (DNA) III alpha [Canis
familiaris]
Length = 1033
Score = 37.7 bits (86), Expect = 0.070
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 6/45 (13%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWASA 100
C CG ++ VRK GP QG F+ CG G C +F WA +
Sbjct: 845 CNCGQEAVLLTVRKEGPNQGRQFYKCG------GGGCDFFLWADS 883
>gb|EAL33317.1| GA10090-PA [Drosophila pseudoobscura]
Length = 1204
Score = 37.7 bits (86), Expect = 0.070
Identities = 18/42 (42%), Positives = 22/42 (51%), Gaps = 4/42 (9%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEW 97
C CG +SR VR GP QG F+ C TR C +F+W
Sbjct: 1107 CNCGQLASRLTVRTEGPNQGRPFYAC----PTREKSCGFFKW 1144
Score = 34.3 bits (77), Expect = 0.78
Identities = 21/86 (24%), Positives = 37/86 (42%), Gaps = 6/86 (6%)
Query: 13 IMWQTADRSLIWKQSPEQPSAYPHFPFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGP 72
++W T +++ P +P+A + + TP C C + + VR+ GP
Sbjct: 954 MLWGTREQAAAAPTPPPKPAA-KRARRDSAEWGTPAAVPAVICSGCHQPARQHTVRREGP 1012
Query: 73 KQGSLFFGCGNWTATRGARCHYFEWA 98
QG F+ C + C +F+WA
Sbjct: 1013 NQGRHFYKC-----PKPNECQFFQWA 1033
>ref|XP_613319.1| PREDICTED: similar to 4933424N09Rik protein [Bos taurus]
Length = 825
Score = 37.4 bits (85), Expect = 0.092
Identities = 21/62 (33%), Positives = 30/62 (47%), Gaps = 10/62 (16%)
Query: 38 PFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSLFFGC--GNWTATRGARCHYF 95
P+K+R + P+ C CG +S R V GP G +F+ C G + R C YF
Sbjct: 720 PWKSRKITPPL-------CKCGRRSKRLTVSNNGPNHGKVFYCCPVGKYQEKRKC-CGYF 771
Query: 96 EW 97
+W
Sbjct: 772 KW 773
>ref|XP_582207.1| PREDICTED: similar to 4933424N09Rik protein [Bos taurus]
Length = 689
Score = 37.4 bits (85), Expect = 0.092
Identities = 21/62 (33%), Positives = 30/62 (47%), Gaps = 10/62 (16%)
Query: 38 PFKARDMNTPIIQYHHPCCYCGVKSSRGMVRKPGPKQGSLFFGC--GNWTATRGARCHYF 95
P+K+R + P+ C CG +S R V GP G +F+ C G + R C YF
Sbjct: 584 PWKSRKITPPL-------CKCGRRSKRLTVSNNGPNHGKVFYCCPVGKYQEKRKC-CGYF 635
Query: 96 EW 97
+W
Sbjct: 636 KW 637
>ref|XP_424602.1| PREDICTED: similar to RIKEN cDNA 4933424N09 [Gallus gallus]
Length = 645
Score = 37.4 bits (85), Expect = 0.092
Identities = 18/49 (36%), Positives = 25/49 (50%), Gaps = 3/49 (6%)
Query: 54 PCCYCGVKSSRGMVRKPGPKQGSLFFGC--GNWTATRGARCHYFEWASA 100
P C CG ++ R V GP G FF C G ++ + C YF+W +A
Sbjct: 556 PLCNCGQRAKRRYVSNAGPNHGKAFFCCPVGRHEGSKRS-CGYFKWENA 603
>emb|CAG04636.1| unnamed protein product [Tetraodon nigroviridis]
Length = 533
Score = 37.0 bits (84), Expect = 0.12
Identities = 17/47 (36%), Positives = 24/47 (50%), Gaps = 3/47 (6%)
Query: 52 HHPCCYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWA 98
H P C+ G + V K GP G FF C + +G +C +F+WA
Sbjct: 478 HFPTCHHGKRCLMRTVLKLGPNNGRNFFTC---SFQKGKQCDFFQWA 521
>emb|CAF98125.1| unnamed protein product [Tetraodon nigroviridis]
Length = 58
Score = 37.0 bits (84), Expect = 0.12
Identities = 19/53 (35%), Positives = 26/53 (48%), Gaps = 8/53 (15%)
Query: 54 PCCYCGVKSSRGMVRKPGPKQGSLFFGC------GNWTATRGARCHYFEWASA 100
P C CG ++ R +V GP G F+ C G T +G C +F+W SA
Sbjct: 5 PLCSCGRRAKRQVVSNGGPNHGRGFYCCAVRRSGGAGTVQKG--CQFFQWESA 55
>gb|AAH51748.1| Topoisomerase (DNA) III alpha [Homo sapiens]
gi|2501242|sp|Q13472|TOP3A_HUMAN DNA topoisomerase III
alpha gi|1292912|gb|AAB03694.1| DNA topoisomerase III
gi|10835218|ref|NP_004609.1| topoisomerase (DNA) III
alpha [Homo sapiens]
Length = 1001
Score = 36.2 bits (82), Expect = 0.20
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 6/45 (13%)
Query: 56 CYCGVKSSRGMVRKPGPKQGSLFFGCGNWTATRGARCHYFEWASA 100
C CG ++ VRK GP +G FF C G C++F WA +
Sbjct: 813 CNCGQEAVLLTVRKEGPNRGRQFFKC------NGGSCNFFLWADS 851
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.137 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 199,648,827
Number of Sequences: 2540612
Number of extensions: 7350452
Number of successful extensions: 17368
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 17304
Number of HSP's gapped (non-prelim): 92
length of query: 100
length of database: 863,360,394
effective HSP length: 76
effective length of query: 24
effective length of database: 670,273,882
effective search space: 16086573168
effective search space used: 16086573168
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146862.9


