

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.7 + phase: 0
(241 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
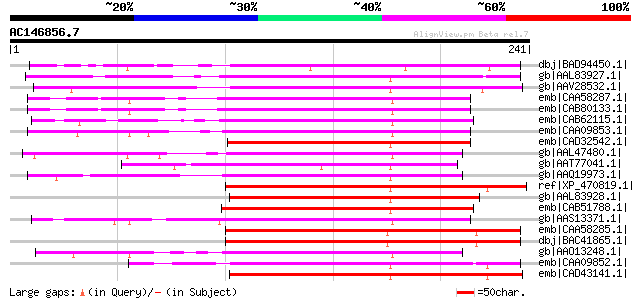
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD94450.1| putative protein [Arabidopsis thaliana] gi|22329... 138 1e-31
gb|AAL83927.1| D-type cyclin [Zea mays] 130 4e-29
gb|AAV28532.1| D-type cyclin [Saccharum officinarum] 125 8e-28
emb|CAA58287.1| cyclin delta-3 [Arabidopsis thaliana] 120 3e-26
emb|CAB80133.1| cyclin delta-3 [Arabidopsis thaliana] gi|2911046... 120 3e-26
emb|CAB62115.1| cyclin D3-like protein [Arabidopsis thaliana] gi... 119 6e-26
emb|CAA09853.1| cyclin D3.1 protein [Nicotiana tabacum] 119 1e-25
emb|CAD32542.1| cyclin D protein [Physcomitrella patens] gi|2619... 119 1e-25
gb|AAL47480.1| cyclin D3 [Helianthus tuberosus] 117 2e-25
gb|AAT77041.1| putative Cyclin [Oryza sativa (japonica cultivar-... 117 3e-25
gb|AAQ19973.1| cyclin D3-1 [Euphorbia esula] 117 4e-25
ref|XP_470819.1| putative cyclin [Oryza sativa (japonica cultiva... 116 5e-25
gb|AAL83928.1| D-type cyclin [Zea mays] 116 5e-25
emb|CAB51788.1| cyclin D3.1 [Lycopersicon esculentum] gi|6434197... 116 5e-25
gb|AAS13371.1| cyclin d3 [Glycine max] 116 6e-25
emb|CAA58285.1| cyclin delta-1 [Arabidopsis thaliana] 115 8e-25
dbj|BAC41865.1| unknown protein [Arabidopsis thaliana] gi|289509... 115 8e-25
gb|AAO13248.1| cyclin D [Populus tremula x Populus tremuloides] 115 8e-25
emb|CAA09852.1| cyclin D2.1 protein [Nicotiana tabacum] 115 1e-24
emb|CAD43141.1| cyclin D2 [Daucus carota] 115 1e-24
>dbj|BAD94450.1| putative protein [Arabidopsis thaliana]
gi|22329219|ref|NP_195478.2| cyclin family protein
[Arabidopsis thaliana] gi|44917541|gb|AAS49095.1|
At4g37630 [Arabidopsis thaliana]
Length = 323
Score = 138 bits (348), Expect = 1e-31
Identities = 93/236 (39%), Positives = 134/236 (56%), Gaps = 37/236 (15%)
Query: 10 SLSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLD-DEEEYIQYLFKQETG 68
SL+ +CHE + S ED+DE+ I S + P F D+E+Y+ L +E
Sbjct: 7 SLALFLCHESESS---LNEDDDET---IERS---DKQEPHFTTTIDDEDYVADLVLKEN- 56
Query: 69 LGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLSI 128
L F ++ + SS RL AIDWI T+ +FGF QTAY++I
Sbjct: 57 LRFETLPS-----------KTTSSSD--------RLIAIDWILTTRTRFGFQHQTAYIAI 97
Query: 129 NYFDRFLSKR--SIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIEYRFENK--VI 184
+YFD FL KR + + + WA++LLSVACLS+AAKMEE+ VP LS+YP ++ F K VI
Sbjct: 98 SYFDLFLHKRFIGLQKDETWAMRLLSVACLSLAAKMEERIVPGLSQYPQDHDFVFKPDVI 157
Query: 185 KNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNGSRS---ETIITKATQHIVTMVK 237
+ ELLILSTL+WKM L TPF Y +YF K + S + ++ +++ ++ + K
Sbjct: 158 RKTELLILSTLDWKMNLITPFHYFNYFLAKISQDNHSVSKDLVLLRSSDSLLALTK 213
>gb|AAL83927.1| D-type cyclin [Zea mays]
Length = 349
Score = 130 bits (326), Expect = 4e-29
Identities = 74/233 (31%), Positives = 122/233 (51%), Gaps = 26/233 (11%)
Query: 8 PFSLSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLDDEEEYIQYLFKQET 67
P + +S++ +D S L+ + D+ T + + + ++D +EEY+ L +E+
Sbjct: 11 PTTPTSILICLEDGSDLLADADDGAGTDLVVA-----RDERLLVVDQDEEYVALLLSKES 65
Query: 68 GLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLS 127
G G E+ED W++ AR + WI T A F F +TAY++
Sbjct: 66 ASGGGGPVE---------EMED--------WMKAARSGCVRWIIKTTAMFRFGGKTAYVA 108
Query: 128 INYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI---EYRFENKVI 184
+NY DRFL++R ++ W +QLL VAC+S+A K+EE P LSE P+ E+ F+ +
Sbjct: 109 VNYLDRFLAQRRVNREHAWGLQLLMVACMSLATKLEEHHAPRLSELPLDACEFAFDRASV 168
Query: 185 KNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNGSRSETIITKATQHIVTMVK 237
MELL+L TLEW+M TPF Y+ F +F R ++ +A + + ++
Sbjct: 169 LRMELLVLGTLEWRMVAVTPFPYISCFAARFRQDER-RAVLVRAVECVFAAIR 220
>gb|AAV28532.1| D-type cyclin [Saccharum officinarum]
Length = 343
Score = 125 bits (315), Expect = 8e-28
Identities = 83/232 (35%), Positives = 116/232 (49%), Gaps = 20/232 (8%)
Query: 12 SSLMCHEQDESTLIFE-EDEDESTFFINSSLDNNNNNPWFLLDDEEEYIQYLFKQETGLG 70
S L+C E S L E E E+E SS + + F + EE ++ + +
Sbjct: 11 SILLCAEDSSSILDLEAEAEEEEVMLARSSRTRGDPSVVFPVPSEECVAGFVEAEAAHMP 70
Query: 71 FGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLSINY 130
L D+ V R+ A+DWI+ A +GF TA L++NY
Sbjct: 71 REDYAERLRGGGMDLRV---------------RMDAVDWIWKVHAYYGFGPLTACLAVNY 115
Query: 131 FDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI---EYRFENKVIKNM 187
DRFLS + E K W QLLSVACLS+AAKMEE VPP + I Y FE K I+ M
Sbjct: 116 LDRFLSLYQLPEGKAWTTQLLSVACLSLAAKMEETYVPPSLDLQIGDARYVFEAKTIQRM 175
Query: 188 ELLILSTLEWKMGLPTPFAYLHYFFTKFCNG-SRSETIITKATQHIVTMVKG 238
ELL+LSTL+W+M TPF+Y+ YF + G + S + ++ + I+ +G
Sbjct: 176 ELLVLSTLKWRMQAVTPFSYIDYFLHRLNGGDAPSRRAVLRSAELILCTARG 227
>emb|CAA58287.1| cyclin delta-3 [Arabidopsis thaliana]
Length = 376
Score = 120 bits (301), Expect = 3e-26
Identities = 77/213 (36%), Positives = 113/213 (52%), Gaps = 35/213 (16%)
Query: 9 FSLSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLDD----EEEYIQYLFK 64
F L +L C E+ + D++ NSSL +++++P+ +L E+E + LF
Sbjct: 17 FLLDALYCEEE-------KWDDEGEEVEENSSL-SSSSSPFVVLQQDLFWEDEDLVTLFS 68
Query: 65 QETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTA 124
+E G C DD +L R A+ WI A +GF+ A
Sbjct: 69 KEEEQGLS------CLDD--------------VYLSTDRKEAVGWILRVNAHYGFSTLAA 108
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIE---YRFEN 181
L+I Y D+F+ S+ KPW +QL+SVACLS+AAK+EE VP L ++ +E Y FE
Sbjct: 109 VLAITYLDKFICSYSLQRDKPWMLQLVSVACLSLAAKVEETQVPLLLDFQVEETKYVFEA 168
Query: 182 KVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
K I+ MELLILSTLEWKM L TP +++ + +
Sbjct: 169 KTIQRMELLILSTLEWKMHLITPISFVDHIIRR 201
>emb|CAB80133.1| cyclin delta-3 [Arabidopsis thaliana] gi|2911046|emb|CAA17556.1|
cyclin delta-3 [Arabidopsis thaliana]
gi|62320771|dbj|BAD95437.1| cyclin delta-3 [Arabidopsis
thaliana] gi|15235254|ref|NP_195142.1| cyclin delta-3
(CYCD3) [Arabidopsis thaliana] gi|7484884|pir||T05420
cyclin delta-3 - Arabidopsis thaliana
gi|59802919|sp|P42753|CCND3_ARATH Cyclin delta-3
Length = 376
Score = 120 bits (301), Expect = 3e-26
Identities = 77/213 (36%), Positives = 113/213 (52%), Gaps = 35/213 (16%)
Query: 9 FSLSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLDD----EEEYIQYLFK 64
F L +L C E+ + D++ NSSL +++++P+ +L E+E + LF
Sbjct: 17 FLLDALYCEEE-------KWDDEGEEVEENSSL-SSSSSPFVVLQQDLFWEDEDLVTLFS 68
Query: 65 QETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTA 124
+E G C DD +L R A+ WI A +GF+ A
Sbjct: 69 KEEEQGLS------CLDD--------------VYLSTDRKEAVGWILRVNAHYGFSTLAA 108
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIE---YRFEN 181
L+I Y D+F+ S+ KPW +QL+SVACLS+AAK+EE VP L ++ +E Y FE
Sbjct: 109 VLAITYLDKFICSYSLQRDKPWMLQLVSVACLSLAAKVEETQVPLLLDFQVEETKYVFEA 168
Query: 182 KVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
K I+ MELLILSTLEWKM L TP +++ + +
Sbjct: 169 KTIQRMELLILSTLEWKMHLITPISFVDHIIRR 201
>emb|CAB62115.1| cyclin D3-like protein [Arabidopsis thaliana]
gi|21593092|gb|AAM65041.1| cyclin D3-like protein
[Arabidopsis thaliana] gi|17380632|gb|AAL36079.1|
AT3g50070/F3A4_150 [Arabidopsis thaliana]
gi|15450595|gb|AAK96569.1| AT3g50070/F3A4_150
[Arabidopsis thaliana] gi|15229665|ref|NP_190576.1|
cyclin family protein [Arabidopsis thaliana]
gi|11357237|pir||T45860 cyclin D3-like protein -
Arabidopsis thaliana
Length = 361
Score = 119 bits (299), Expect = 6e-26
Identities = 78/214 (36%), Positives = 114/214 (52%), Gaps = 38/214 (17%)
Query: 11 LSSLMCHEQDESTLIFEEDED------ESTFFINSSLDNNNNNPWFLLDDEEEYIQYLFK 64
L L C E+ E F E D E F+N L +++ +L D++E + K
Sbjct: 17 LDGLFCEEESE----FHEQVDLCDESVEKFPFLNLGLSDHD-----MLWDDDELSTLISK 67
Query: 65 QETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTA 124
QE L YD E+ DD+ +L R A+DWIF ++ +GF TA
Sbjct: 68 QEPCL----------YD----EILDDE------FLVLCREKALDWIFKVKSHYGFNSLTA 107
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIE---YRFEN 181
L++NYFDRF++ R KPW QL ++ACLS+AAK+EE VP L ++ +E Y FE
Sbjct: 108 LLAVNYFDRFITSRKFQTDKPWMSQLTALACLSLAAKVEEIRVPFLLDFQVEEARYVFEA 167
Query: 182 KVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKF 215
K I+ MELL+LSTL+W+M TP ++ + ++
Sbjct: 168 KTIQRMELLVLSTLDWRMHPVTPISFFDHIIRRY 201
>emb|CAA09853.1| cyclin D3.1 protein [Nicotiana tabacum]
Length = 373
Score = 119 bits (297), Expect = 1e-25
Identities = 79/222 (35%), Positives = 116/222 (51%), Gaps = 35/222 (15%)
Query: 9 FSLSSLMCHEQDESTLIFEEDE-------DESTFFINSSLDNNNNNPWFLLDD----EEE 57
F L +L C E++E +DE E T ++ N+ P LL+ E+E
Sbjct: 15 FLLDALYCEEEEEKWGDLVDDETIITPLSSEVTTTTTTTTKPNSLLPLLLLEQDLFWEDE 74
Query: 58 YIQYLF--KQETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQA 115
+ LF ++ET F S DDS L +AR+ +++WI
Sbjct: 75 ELLSLFSKEKETHCWFNSF--------------QDDSL-----LCSARVDSVEWILKVNG 115
Query: 116 KFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI 175
+GF+ TA L+INYFDRFL+ + KPW IQL +V CLS+AAK+EE VP L ++ +
Sbjct: 116 YYGFSALTAVLAINYFDRFLTSLHYQKDKPWMIQLAAVTCLSLAAKVEETQVPLLLDFQV 175
Query: 176 E---YRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
E Y FE K I+ MELL+LS+L+W+M TP ++L + +
Sbjct: 176 EDAKYVFEAKTIQRMELLVLSSLKWRMNPVTPLSFLDHIIRR 217
>emb|CAD32542.1| cyclin D protein [Physcomitrella patens]
gi|26190151|emb|CAD21955.1| cyclin D [Physcomitrella
patens]
Length = 360
Score = 119 bits (297), Expect = 1e-25
Identities = 59/116 (50%), Positives = 79/116 (67%), Gaps = 3/116 (2%)
Query: 102 ARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAK 161
ARL AI+WI + + ++ T L++NY DRFLS+ E K W +QLLSVAC+S+AAK
Sbjct: 82 ARLAAIEWILKVHSFYNYSPLTVALAVNYMDRFLSRYYFPEGKEWMLQLLSVACISLAAK 141
Query: 162 MEEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
MEE VP L ++ + E+ FE I+ MELL+LSTLEW+M TPF+Y+ YFF K
Sbjct: 142 MEESDVPILLDFQVEQEEHIFEAHTIQRMELLVLSTLEWRMSGVTPFSYVDYFFHK 197
>gb|AAL47480.1| cyclin D3 [Helianthus tuberosus]
Length = 357
Score = 117 bits (294), Expect = 2e-25
Identities = 81/220 (36%), Positives = 115/220 (51%), Gaps = 37/220 (16%)
Query: 7 TPFS---LSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLD---DEEEYIQ 60
+P+S L +L C+EQ + E EDE T + + + P LD + EE +
Sbjct: 5 SPYSSSFLDTLFCNEQQDHEYHEYEYEDEFTQTTLTDSSDLHLPPLDQLDLSWEHEELVS 64
Query: 61 YLFKQETG-------LGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNT 113
K++ L FG +S S+F AR A+DWI
Sbjct: 65 LFTKEQEQQKQTPCTLSFGK------------------TSPSVF---AARKEAVDWILKV 103
Query: 114 QAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEY 173
++ +GFT TA L+INY DRFLS E KPW IQL++V+CLS+AAK+EE VP L +
Sbjct: 104 KSCYGFTPLTAILAINYLDRFLSSLHFQEDKPWMIQLVAVSCLSLAAKVEETQVPLLLDL 163
Query: 174 PIE---YRFENKVIKNMELLILSTLEWKMGLPTPFAYLHY 210
+E Y FE K I+ MELL++STL+W+M TP ++L +
Sbjct: 164 QVEDTKYLFEAKNIQKMELLVMSTLKWRMNPVTPISFLDH 203
>gb|AAT77041.1| putative Cyclin [Oryza sativa (japonica cultivar-group)]
Length = 367
Score = 117 bits (293), Expect = 3e-25
Identities = 69/165 (41%), Positives = 94/165 (56%), Gaps = 11/165 (6%)
Query: 53 DDEEEYIQYLFKQETGLGFGSIT---HFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDW 109
++ EEY+ +L +E+ S + C+ D E ++ + W AR + W
Sbjct: 58 EEVEEYMDHLVSKESSFCSSSSSTSSSSCCFSD--AGGESAAAAAPMDWFALARRATVKW 115
Query: 110 IFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDES-KPWAIQLLSVACLSIAAKMEEQSVP 168
I T+ FGF +TAYL+I YFDRF +R ID S PWA +LL+VAC+S+AAKMEE P
Sbjct: 116 ILETRGCFGFCHRTAYLAIAYFDRFCLRRCIDRSVMPWAARLLAVACVSLAAKMEEYRAP 175
Query: 169 PLSEYPI-----EYRFENKVIKNMELLILSTLEWKMGLPTPFAYL 208
LSE+ Y F I+ MELL+LSTL+W+M TPF YL
Sbjct: 176 ALSEFRAGVGDDGYEFSCVCIRRMELLVLSTLDWRMAAVTPFDYL 220
>gb|AAQ19973.1| cyclin D3-1 [Euphorbia esula]
Length = 350
Score = 117 bits (292), Expect = 4e-25
Identities = 73/209 (34%), Positives = 106/209 (49%), Gaps = 29/209 (13%)
Query: 9 FSLSSLMCHEQD----ESTLIFEEDEDESTFFINSSLDNNNNNPWFLLDDEEEYIQYLFK 64
F +L C E+D E IF E ED+ N+S+ N++P L +E+E K
Sbjct: 8 FLYDALYCSEEDNWEGEVVDIFHEQEDQGE---NTSVFPQNSSPVDLNWEEDELTSVFSK 64
Query: 65 QETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTA 124
QE + + C L +R A+DW+ A + FT T+
Sbjct: 65 QEQNQLYKKLEINPC-------------------LAKSRRDAVDWMMKVNAHYSFTALTS 105
Query: 125 YLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI---EYRFEN 181
L++N+ DRFL + KPW QL +VACLS+AAK+EE VP L + + +Y FE
Sbjct: 106 VLAVNFLDRFLFSFDLQTEKPWMTQLTAVACLSLAAKVEETQVPLLLDLQVVDSKYVFEA 165
Query: 182 KVIKNMELLILSTLEWKMGLPTPFAYLHY 210
K I+ MELL+LSTL+W+M TP +++ Y
Sbjct: 166 KTIQRMELLVLSTLQWRMNPVTPLSFIDY 194
>ref|XP_470819.1| putative cyclin [Oryza sativa (japonica cultivar-group)]
gi|40539012|gb|AAR87269.1| putative cyclin [Oryza sativa
(japonica cultivar-group)]
Length = 358
Score = 116 bits (291), Expect = 5e-25
Identities = 59/144 (40%), Positives = 88/144 (60%), Gaps = 4/144 (2%)
Query: 101 NARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAA 160
+ R+ AIDWI QA + F AYL++NY DRFLS PW QLL VACLS+AA
Sbjct: 102 SCRIAAIDWICKVQAYYSFGPLCAYLAVNYLDRFLSSVEFSNDMPWMQQLLIVACLSLAA 161
Query: 161 KMEEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCN 217
KMEE + P + + EY F+ + I ME+++L+TL+W+M TPF Y+ +F K
Sbjct: 162 KMEETAAPGTLDLQVCNPEYVFDAETIHRMEIIVLTTLKWRMQAVTPFTYIGHFLDKINE 221
Query: 218 GSR-SETIITKATQHIVTMVKGNI 240
G+R + +I++ T+ I++ +K +
Sbjct: 222 GNRITSELISRCTEIILSTMKATV 245
>gb|AAL83928.1| D-type cyclin [Zea mays]
Length = 390
Score = 116 bits (291), Expect = 5e-25
Identities = 56/119 (47%), Positives = 75/119 (62%), Gaps = 3/119 (2%)
Query: 103 RLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKM 162
R A+DWI+ F TAYL++NY DRFLS + + K W QLL+VAC+S+AAKM
Sbjct: 92 RREAVDWIWKAYTHHRFRPLTAYLAVNYLDRFLSLSEVPDGKDWMTQLLAVACVSLAAKM 151
Query: 163 EEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNG 218
EE +VP + + Y FE K ++ MELL+L+TL W+M TPF+Y+ YF K NG
Sbjct: 152 EETAVPQCLDLQVGDARYVFEAKTVQRMELLVLTTLNWRMHAVTPFSYVDYFLNKLSNG 210
>emb|CAB51788.1| cyclin D3.1 [Lycopersicon esculentum] gi|6434197|emb|CAB60836.1|
CycD3;1 [Lycopersicon esculentum]
Length = 359
Score = 116 bits (291), Expect = 5e-25
Identities = 58/120 (48%), Positives = 78/120 (64%), Gaps = 3/120 (2%)
Query: 99 LRNARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSI 158
L AR A+DW+ A +GFT TA L++NYFDRF+S + KPW QL +VACLSI
Sbjct: 86 LMGARKEALDWMLRVIAYYGFTATTAVLAVNYFDRFVSGWCFQKDKPWMSQLAAVACLSI 145
Query: 159 AAKMEEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKF 215
AAK+EE VP L + + + FE K I+ MELL+LSTL+WKM L TP +++ + +F
Sbjct: 146 AAKVEETQVPLLLDLQVADSRFVFEAKTIQRMELLVLSTLKWKMNLVTPLSFIDHIMRRF 205
>gb|AAS13371.1| cyclin d3 [Glycine max]
Length = 396
Score = 116 bits (290), Expect = 6e-25
Identities = 73/222 (32%), Positives = 113/222 (50%), Gaps = 29/222 (13%)
Query: 11 LSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNN---PWFLLDD----EEEYIQYLF 63
L +L C E +E++E+E + S + N P +L+ E+E + LF
Sbjct: 20 LDALYCDEGK-----WEDEEEEEEXYEESEVTTNTGTSLFPLLMLEQDLFWEDEELNSLF 74
Query: 64 KQETGLGFGSITHFLCYDDHDVEVEDDDSSKSL--------FWLRNARLHAIDWIFNTQA 115
+E + H YD +++ +D+ + S L R A++WI A
Sbjct: 75 SKE------KVQHEEAYDYNNLNSDDNSNDHSNNNNNVLSDSCLSQPRREAVEWILKVNA 128
Query: 116 KFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI 175
+GF+ TA L++ Y DRFL KPW IQL++V C+S+AAK+EE VP L + +
Sbjct: 129 HYGFSALTATLAVTYLDRFLLSFHFQREKPWMIQLVAVTCISLAAKVEETQVPLLLDLQV 188
Query: 176 E---YRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
+ Y FE K I+ MELL+LSTL+WKM TP ++L + +
Sbjct: 189 QDTKYVFEAKTIQRMELLVLSTLKWKMHPVTPLSFLDHIIRR 230
>emb|CAA58285.1| cyclin delta-1 [Arabidopsis thaliana]
Length = 335
Score = 115 bits (289), Expect = 8e-25
Identities = 62/141 (43%), Positives = 91/141 (63%), Gaps = 4/141 (2%)
Query: 101 NARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAA 160
+AR ++ WI QA + F TAYL++NY DRFL R + E+ W +QLL+VACLS+AA
Sbjct: 80 SAREDSVAWILKVQAYYNFQPLTAYLAVNYMDRFLYARRLPETSGWPMQLLAVACLSLAA 139
Query: 161 KMEEQSVPPLSEYP---IEYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKF-C 216
KMEE VP L ++ ++Y FE K IK MELL+LS L+W++ TPF ++ +F K
Sbjct: 140 KMEEILVPSLFDFQVAGVKYLFEAKTIKRMELLVLSVLDWRLRSVTPFDFISFFAYKIDP 199
Query: 217 NGSRSETIITKATQHIVTMVK 237
+G+ I+ AT+ I++ +K
Sbjct: 200 SGTFLGFFISHATEIILSNIK 220
>dbj|BAC41865.1| unknown protein [Arabidopsis thaliana] gi|28950911|gb|AAO63379.1|
At1g70210 [Arabidopsis thaliana]
gi|15223075|ref|NP_177178.1| cyclin delta-1 (CYCD1)
[Arabidopsis thaliana] gi|25406041|pir||A96725
hypothetical protein F20P5.7 [imported] - Arabidopsis
thaliana gi|2194121|gb|AAB61096.1| Strong similarity to
Arabidopsis cyclin delta-1 (gb|ATCD1). EST gb|ATTS4338
comes from this gene. [Arabidopsis thaliana]
gi|59802916|sp|P42751|CCND1_ARATH Cyclin delta-1
Length = 339
Score = 115 bits (289), Expect = 8e-25
Identities = 62/141 (43%), Positives = 91/141 (63%), Gaps = 4/141 (2%)
Query: 101 NARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAA 160
+AR ++ WI QA + F TAYL++NY DRFL R + E+ W +QLL+VACLS+AA
Sbjct: 80 SAREDSVAWILKVQAYYNFQPLTAYLAVNYMDRFLYARRLPETSGWPMQLLAVACLSLAA 139
Query: 161 KMEEQSVPPLSEYP---IEYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKF-C 216
KMEE VP L ++ ++Y FE K IK MELL+LS L+W++ TPF ++ +F K
Sbjct: 140 KMEEILVPSLFDFQVAGVKYLFEAKTIKRMELLVLSVLDWRLRSVTPFDFISFFAYKIDP 199
Query: 217 NGSRSETIITKATQHIVTMVK 237
+G+ I+ AT+ I++ +K
Sbjct: 200 SGTFLGFFISHATEIILSNIK 220
>gb|AAO13248.1| cyclin D [Populus tremula x Populus tremuloides]
Length = 376
Score = 115 bits (289), Expect = 8e-25
Identities = 75/207 (36%), Positives = 111/207 (53%), Gaps = 27/207 (13%)
Query: 13 SLMCHEQDESTLIFEE---DEDESTFFINSSLDNNNNNPWFLLDD---EEEYIQYLFKQE 66
+L C E++ + E+ DE E + +++ + N P L D E+E + LF +E
Sbjct: 25 ALYCSEENWVEEVREDWFQDELEGESYCSNNSNKLNTFPILLEQDLSWEDEELSSLFAKE 84
Query: 67 TGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYL 126
+ LC D +E + S L AR A++WI + FT TA L
Sbjct: 85 E-------QNQLCKD-----LETNPS------LARARCEAVEWILKVNEHYSFTALTAVL 126
Query: 127 SINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIE---YRFENKV 183
++NY DRFL + + KPW QL +V+CLS+AAK+EE VP L ++ +E Y FE K
Sbjct: 127 AVNYLDRFLFSVHLQKEKPWMAQLAAVSCLSLAAKVEETQVPLLLDFQVEDSKYVFEAKT 186
Query: 184 IKNMELLILSTLEWKMGLPTPFAYLHY 210
I+ ME+L+LSTL+WKM TP ++L Y
Sbjct: 187 IQRMEILVLSTLKWKMNPVTPISFLDY 213
>emb|CAA09852.1| cyclin D2.1 protein [Nicotiana tabacum]
Length = 354
Score = 115 bits (288), Expect = 1e-24
Identities = 70/187 (37%), Positives = 106/187 (56%), Gaps = 20/187 (10%)
Query: 56 EEYIQYLFKQETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQA 115
EE + ++ ++E FL DD+ + D S+ R A+DWI
Sbjct: 63 EECLSFMVQREM--------EFLPKDDYVERLRSGDLDLSV------RKEALDWILKAHM 108
Query: 116 KFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI 175
+GF + LSINY DRFLS + SK W +QLL+VACLS+AAKMEE +VP + +
Sbjct: 109 HYGFGELSFCLSINYLDRFLSLYELPRSKTWTVQLLAVACLSLAAKMEEINVPLTVDLQV 168
Query: 176 ---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNGSR--SETIITKATQ 230
++ FE K I+ MELL+LSTL+W+M TP+ ++ YF K NG + S +I+ + Q
Sbjct: 169 GDPKFVFEGKTIQRMELLVLSTLKWRMQAYTPYTFIDYFMRKM-NGDQIPSRPLISGSMQ 227
Query: 231 HIVTMVK 237
I+++++
Sbjct: 228 LILSIIR 234
>emb|CAD43141.1| cyclin D2 [Daucus carota]
Length = 382
Score = 115 bits (288), Expect = 1e-24
Identities = 60/141 (42%), Positives = 87/141 (61%), Gaps = 5/141 (3%)
Query: 103 RLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKM 162
R A+DWI+ A + F + L++NY DRFLS + K W +QLL+VACLS+AAKM
Sbjct: 95 RKEALDWIYKAHAHYNFGALSVCLAVNYLDRFLSLYELPSGKKWTVQLLAVACLSLAAKM 154
Query: 163 EEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNGS 219
EE +VP + + ++ FE K IK MELL+LSTL+W+M TP +++ YF K N
Sbjct: 155 EEVNVPLTVDLQVADPKFVFEAKTIKRMELLVLSTLKWRMQACTPCSFIDYFLRKINNAD 214
Query: 220 R--SETIITKATQHIVTMVKG 238
S ++I ++ Q I+ +KG
Sbjct: 215 ALPSGSLIDRSIQFILKTMKG 235
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 421,607,945
Number of Sequences: 2540612
Number of extensions: 18212767
Number of successful extensions: 70095
Number of sequences better than 10.0: 874
Number of HSP's better than 10.0 without gapping: 375
Number of HSP's successfully gapped in prelim test: 499
Number of HSP's that attempted gapping in prelim test: 68841
Number of HSP's gapped (non-prelim): 918
length of query: 241
length of database: 863,360,394
effective HSP length: 124
effective length of query: 117
effective length of database: 548,324,506
effective search space: 64153967202
effective search space used: 64153967202
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146856.7


