

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.6 - phase: 0
(535 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
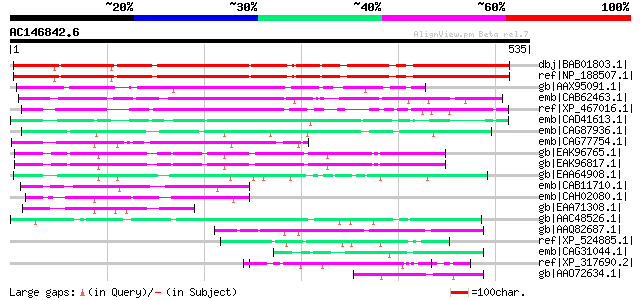
dbj|BAB01803.1| unnamed protein product [Arabidopsis thaliana] 451 e-125
ref|NP_188507.1| expressed protein [Arabidopsis thaliana] 451 e-125
gb|AAX95091.1| expressed protein [Oryza sativa (japonica cultiva... 303 7e-81
emb|CAB62463.1| putative protein [Arabidopsis thaliana] gi|30693... 232 3e-59
ref|XP_467016.1| hypothetical protein [Oryza sativa (japonica cu... 161 6e-38
emb|CAD41613.1| OSJNBa0091D06.22 [Oryza sativa (japonica cultiva... 128 5e-28
emb|CAG87936.1| unnamed protein product [Debaryomyces hansenii C... 97 2e-18
emb|CAG77754.1| unnamed protein product [Yarrowia lipolytica CLI... 94 8e-18
gb|EAK96765.1| hypothetical protein CaO19.2848 [Candida albicans... 92 4e-17
gb|EAK96817.1| hypothetical protein CaO19.10367 [Candida albican... 91 7e-17
gb|EAA64908.1| hypothetical protein AN2076.2 [Aspergillus nidula... 62 5e-08
emb|CAB11710.1| SPAC4F10.07c [Schizosaccharomyces pombe] gi|1911... 61 1e-07
emb|CAH02080.1| unnamed protein product [Kluyveromyces lactis NR... 58 7e-07
gb|EAA71308.1| hypothetical protein FG08491.1 [Gibberella zeae P... 57 1e-06
gb|AAC48526.1| gastric mucin gi|2136504|pir||I47141 gastric muci... 54 2e-05
gb|AAQ82687.1| Epa5p [Candida glabrata] 54 2e-05
ref|XP_524885.1| PREDICTED: hypothetical protein XP_524885 [Pan ... 53 3e-05
emb|CAG31044.1| hypothetical protein [Gallus gallus] 52 5e-05
ref|XP_317690.2| ENSANGP00000010305 [Anopheles gambiae str. PEST... 51 1e-04
gb|AAO72634.1| p67-like superoxide-generating NADPH oxidase [Dic... 51 1e-04
>dbj|BAB01803.1| unnamed protein product [Arabidopsis thaliana]
Length = 653
Score = 451 bits (1160), Expect = e-125
Identities = 272/529 (51%), Positives = 355/529 (66%), Gaps = 50/529 (9%)
Query: 5 SSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFS---TLSSPSSTSSSSSSSVR 61
S++ +E AK EQII EFFAKSLHII+ESR SSRNFS + SPSS SSSSSSSVR
Sbjct: 36 SNNNNSEGAKAEQIIFEFFAKSLHIILESRTPFMSSRNFSGEQMICSPSS-SSSSSSSVR 94
Query: 62 PRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNL------------SFSPKV 109
PRDKWFNLALRECP ALE+ D+ R S+L+P+VVDVVLV R L +FS K
Sbjct: 95 PRDKWFNLALRECPAALESFDIGRRSSLEPLVVDVVLVVRPLVGDQSGKRELIRNFSGK- 153
Query: 110 RSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLY 169
+ N ++ G +NE+I+ERWV+QY+++KI++ + ++RRSS+ LQ +Y
Sbjct: 154 -DYQSGWNSDQDELGCETKNEQIIERWVVQYDNRKIRE----SVTTSSRRSSSNKLQVMY 208
Query: 170 KKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFV 229
KK+TLLLRSL+ VRLLPA+KIF+EL+SS + F L RV + EPFT K+EAEM KF
Sbjct: 209 KKATLLLRSLFVMVRLLPAYKIFRELNSSGQIFKFKLVPRVPSIVEPFTRKEEAEMQKFS 268
Query: 230 FTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRM 289
FTPV+T G+LCLSV+YR +SDVS E +TP+SP ITDYVGSPL D L RFPSLP+
Sbjct: 269 FTPVETICGRLCLSVLYR-SLSDVSCEHSTPMSPTFITDYVGSPLADPLKRFPSLPL--- 324
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL 349
S+G SP LPF RRHSWS++ +ASPP+V+ SPSPT S+SH LVS+ S+ L
Sbjct: 325 -SYG-SPPLLPFQRRHSWSFDRYKASPPSVS-CSPSPTRSDSHALVSHPCSR------HL 375
Query: 350 PPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP 409
PPHPS++ +K +EY P +FSP PSPS+ G + R+ESAPV IP
Sbjct: 376 PPHPSDIPTGRRKESYPEEYSPCQDFSPPPSPSAPKHAVPRG------ITRTESAPVRIP 429
Query: 410 NAEVTYSPGHTSRQN-LPPSTPIRISRCTS-ETERSRNLMQSCTTPEKMFSIGKES-QKY 466
+P S++N + PS +++SR S + R+ +S +K+F G++ ++
Sbjct: 430 ------APTFQSKENVVAPSAHLKLSRHASLKPVRNLGPGESGAAIDKLFLYGRDDFRRP 483
Query: 467 SGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTTDPGSR 515
SG + + +SSP+ISFSRSSSRS+QD+FDD DF CPFDV+ DD TD SR
Sbjct: 484 SGVRPSSSSSPRISFSRSSSRSFQDDFDDPDFPCPFDVEYDDITDRNSR 532
>ref|NP_188507.1| expressed protein [Arabidopsis thaliana]
Length = 625
Score = 451 bits (1160), Expect = e-125
Identities = 272/529 (51%), Positives = 355/529 (66%), Gaps = 50/529 (9%)
Query: 5 SSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFS---TLSSPSSTSSSSSSSVR 61
S++ +E AK EQII EFFAKSLHII+ESR SSRNFS + SPSS SSSSSSSVR
Sbjct: 8 SNNNNSEGAKAEQIIFEFFAKSLHIILESRTPFMSSRNFSGEQMICSPSS-SSSSSSSVR 66
Query: 62 PRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNL------------SFSPKV 109
PRDKWFNLALRECP ALE+ D+ R S+L+P+VVDVVLV R L +FS K
Sbjct: 67 PRDKWFNLALRECPAALESFDIGRRSSLEPLVVDVVLVVRPLVGDQSGKRELIRNFSGK- 125
Query: 110 RSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLY 169
+ N ++ G +NE+I+ERWV+QY+++KI++ + ++RRSS+ LQ +Y
Sbjct: 126 -DYQSGWNSDQDELGCETKNEQIIERWVVQYDNRKIRE----SVTTSSRRSSSNKLQVMY 180
Query: 170 KKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFV 229
KK+TLLLRSL+ VRLLPA+KIF+EL+SS + F L RV + EPFT K+EAEM KF
Sbjct: 181 KKATLLLRSLFVMVRLLPAYKIFRELNSSGQIFKFKLVPRVPSIVEPFTRKEEAEMQKFS 240
Query: 230 FTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRM 289
FTPV+T G+LCLSV+YR +SDVS E +TP+SP ITDYVGSPL D L RFPSLP+
Sbjct: 241 FTPVETICGRLCLSVLYR-SLSDVSCEHSTPMSPTFITDYVGSPLADPLKRFPSLPL--- 296
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL 349
S+G SP LPF RRHSWS++ +ASPP+V+ SPSPT S+SH LVS+ S+ L
Sbjct: 297 -SYG-SPPLLPFQRRHSWSFDRYKASPPSVS-CSPSPTRSDSHALVSHPCSR------HL 347
Query: 350 PPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP 409
PPHPS++ +K +EY P +FSP PSPS+ G + R+ESAPV IP
Sbjct: 348 PPHPSDIPTGRRKESYPEEYSPCQDFSPPPSPSAPKHAVPRG------ITRTESAPVRIP 401
Query: 410 NAEVTYSPGHTSRQN-LPPSTPIRISRCTS-ETERSRNLMQSCTTPEKMFSIGKES-QKY 466
+P S++N + PS +++SR S + R+ +S +K+F G++ ++
Sbjct: 402 ------APTFQSKENVVAPSAHLKLSRHASLKPVRNLGPGESGAAIDKLFLYGRDDFRRP 455
Query: 467 SGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTTDPGSR 515
SG + + +SSP+ISFSRSSSRS+QD+FDD DF CPFDV+ DD TD SR
Sbjct: 456 SGVRPSSSSSPRISFSRSSSRSFQDDFDDPDFPCPFDVEYDDITDRNSR 504
>gb|AAX95091.1| expressed protein [Oryza sativa (japonica cultivar-group)]
Length = 601
Score = 303 bits (777), Expect = 7e-81
Identities = 187/437 (42%), Positives = 253/437 (57%), Gaps = 66/437 (15%)
Query: 8 AQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWF 67
A E EQ+ITEFFAKSLHII+ESR+ SSRNF+ S PSS S S +PRD+WF
Sbjct: 3 AAAEPPMVEQVITEFFAKSLHIILESRSPYESSRNFTRPSPPSSPLSGS----QPRDRWF 58
Query: 68 NLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSE 127
NLALR+CP LEN DLWR SNL+P+V+D+VL+ R+ + + G G
Sbjct: 59 NLALRDCPAVLENFDLWRQSNLEPLVIDIVLLCRDSTSNTAA-------------GSG-- 103
Query: 128 QNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQN------LYKKSTLLLRSLYA 181
KI+ERWVIQYE++K + + + N R+S N+ ++ Y+ ST+LLRSL+
Sbjct: 104 ---KIIERWVIQYEARKSGGGNGNGSKNNGRKSRNSSAEDHSLYRATYQGSTVLLRSLHL 160
Query: 182 TVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLC 241
VRLLPA+ +F+EL+SS + +L+H++S+F EPFT ++AEM + F P++T G+L
Sbjct: 161 LVRLLPAYSLFRELNSSGRIRPLNLSHKISSFVEPFTRAEDAEMKHYAFAPIETLFGRLS 220
Query: 242 LSVMYRPCVSDV-SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLP 300
LSV Y P + V +SEPT+P+ P++ITDYVGSP TD L +F SL PS G +P+
Sbjct: 221 LSVSYVPVLEVVAASEPTSPMPPEIITDYVGSPTTDFLKKFNSL-----PSAGIAPACAA 275
Query: 301 FSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLH 360
+RRHSWS E+ + + SPSPT ++S P +P H S + H
Sbjct: 276 MTRRHSWSIEHGAGTSVSP---SPSPTKAQSRG----------SPQLGVPLHVSLKTCSH 322
Query: 361 KKNVN---------FDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNA 411
+N + F+E YPSP SPSPS S S+ + L R ESAPV IP
Sbjct: 323 PQNASSSGQKKYTPFEECYPSPPLSPSPSQSPSA------NYPKNPLFRYESAPVTIP-- 374
Query: 412 EVTYSPGHTSRQNLPPS 428
T G LPPS
Sbjct: 375 --TLKSGGGGGSGLPPS 389
>emb|CAB62463.1| putative protein [Arabidopsis thaliana]
gi|30693211|ref|NP_850673.1| expressed protein
[Arabidopsis thaliana] gi|15229172|ref|NP_190528.1|
expressed protein [Arabidopsis thaliana]
gi|11285730|pir||T46236 hypothetical protein T9C5.180 -
Arabidopsis thaliana
Length = 603
Score = 232 bits (591), Expect = 3e-59
Identities = 180/531 (33%), Positives = 259/531 (47%), Gaps = 80/531 (15%)
Query: 10 TEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNL 69
++ + EQI++ FF K+LHI++ SR S SR + S +VR DKWFNL
Sbjct: 9 SDIGRLEQIVSHFFPKALHIVLNSRIPSLQSRG-------RTRERLSGLNVRKSDKWFNL 61
Query: 70 ALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQN 129
+ + P ALE W + L +++D++LVH P + + + + S
Sbjct: 62 VMGDRPAALEKLHSWHRNILDSMIIDIILVH------PISNDNLDDDDDHSDSVVRSA-- 113
Query: 130 EKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAF 189
E ++ERWV+QYE+ I SS++A T Q +YKKS +LLRSLYA RLLPA+
Sbjct: 114 ETVIERWVVQYENPLIMSPQSSDSA--------TRYQKVYKKSIILLRSLYAQTRLLPAY 165
Query: 190 KIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPC 249
++ ++LSSS S + L ++VS+F + F+ M +F F PV+ G+LC SV YR
Sbjct: 166 RVSRQLSSSLASSGYDLIYKVSSFSDIFSGPVTETMKEFRFAPVEVPPGRLCASVTYRSD 225
Query: 250 VSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSH------GSSP-SSLPFS 302
+SD + L P++ITDYVGSP TD + FPS P + H G P +
Sbjct: 226 LSDFNLGAHITLPPRIITDYVGSPATDPMRFFPS-PGRSVEGHSFTGRAGRPPLTGSSAE 284
Query: 303 RRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKK 362
R HSW+ R PPA + +P +Q + P S P +
Sbjct: 285 RPHSWTSGFHR--PPA-QFATP---------------NQSFSPAQSHQLSPGLHDFHWSR 326
Query: 363 NVNF-DEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIP-----NAEVTYS 416
F D + SP FSPS SP S+P Y G + + +R +APV IP N V+ +
Sbjct: 327 TDAFGDNHQLSPPFSPSGSP--STPRYISGGNSPRINVRPGTAPVTIPSSATLNRYVSSN 384
Query: 417 PGHTSRQNLPPSTP---------------IRISRCTSETERSRNLMQSCTTPEKMFSIGK 461
R LPP +P I + R + E LM T + + K
Sbjct: 385 FSEPGRNPLPPFSPKSTRRSPSSQDSLPGIALYRSSRSGESPSGLMNQYPTQK----LSK 440
Query: 462 ESQKYSG---GKIAPNSSPQISFSRSSSR-SYQDEFDDTDFTCPFDVDDDD 508
+S+ SG G ++ + SP+ +FSRS SR S QD+ DD D +CPFD DD D
Sbjct: 441 DSKYDSGRFSGVLSSSDSPRFAFSRSPSRLSSQDDLDDPDCSCPFDFDDVD 491
>ref|XP_467016.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|49388657|dbj|BAD25792.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 544
Score = 161 bits (407), Expect = 6e-38
Identities = 151/523 (28%), Positives = 214/523 (40%), Gaps = 114/523 (21%)
Query: 13 AKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALR 72
A E ++ +F K+LH I+ RA P + + + ++S R RD+WF+L L
Sbjct: 16 AGAELMVPQFHLKALHAILAVRA-----------PRPLAAAPAPAASFRRRDRWFHLPLH 64
Query: 73 ECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQNEKI 132
P L S +P+VVDV L GG E +
Sbjct: 65 APPPPASAEHLPEPSPGEPLVVDVYLTP----------------------SGGGGGAEAV 102
Query: 133 VERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIF 192
VERW + E + + YK+ LLRS+Y +RLLPA+++F
Sbjct: 103 VERWTVSCEPWS---AGARGGGGAAASGEGLAVNRAYKRCITLLRSVYTALRLLPAYRVF 159
Query: 193 KELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSD 252
+ L +S + + RV +F PFT +EA M F PV+T G+L +SV Y P ++
Sbjct: 160 RLLCASGQAYNYEMGFRVGSFAAPFTRAEEAAMSTRRFAPVETQLGRLVVSVQYLPSLAA 219
Query: 253 VSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENC 312
+ E + +ITDYVGSP D + FP+ + SS + P R +SW
Sbjct: 220 FNLEICSLAPAMLITDYVGSPAADPMRAFPA----SLTEAASSAPAFPPRRPNSW----- 270
Query: 313 RASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPS 372
A PA Y+P + PP P+
Sbjct: 271 -APSPAPWPYTP-------------GQQAKFSPP------------------------PA 292
Query: 373 PNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQN-------L 425
SP+PSP P ++ G L S+ L E+AP+ IP P H + L
Sbjct: 293 LYASPTPSP----PTFAGGYLQSR--LSGETAPMIIPGG--GRGPVHNRNMSDPVRGFML 344
Query: 426 PPSTPIRI------SRCTSETERSRNLMQSCTT-----PEKMFSIGKESQKYSGGKIAPN 474
PP +P I ET R+ M T P+ ++ G + +
Sbjct: 345 PPPSPKNIRGDSGGHETPMETGRTGIRMADLYTNLPSVPKIKIKDSRDESGRFSGVFSSS 404
Query: 475 SSPQISFSRSSSR-SYQDEFDDTDFTCPFDVDDDDTTD--PGS 514
SP++ FSRSSSR S QD+ DD DF PF VDD DT D PGS
Sbjct: 405 GSPRLGFSRSSSRLSMQDDTDDLDF--PFAVDDVDTPDSRPGS 445
>emb|CAD41613.1| OSJNBa0091D06.22 [Oryza sativa (japonica cultivar-group)]
gi|50926951|ref|XP_473344.1| OSJNBa0091D06.22 [Oryza
sativa (japonica cultivar-group)]
Length = 520
Score = 128 bits (321), Expect = 5e-28
Identities = 131/528 (24%), Positives = 211/528 (39%), Gaps = 117/528 (22%)
Query: 1 MTTTSSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSV 60
M++ S E ++ +F K LH ++ R P ++S+S
Sbjct: 1 MSSMSDSGTGGRGGAELMVEQFHLKVLHAVLAVRG-------------PRPLQPAASASF 47
Query: 61 RPRDKWFNLALREC--PTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNP 118
R RD+WF+L L + P A E + +P+VVD++L H
Sbjct: 48 RRRDRWFHLPLHDPQPPPAAEGVEAPEAG--EPLVVDILLAHAAAGG------------- 92
Query: 119 FEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRS 178
GGG ++VERW + E D ++ R YK+ +LRS
Sbjct: 93 ----GGGGGAGGEVVERWTVVCEPWP--DAAAGEGIPVNRA---------YKRCMTMLRS 137
Query: 179 LYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSG 238
+YAT+R LPA+++F+ L ++ + + + + HRV +F P + +EA M + F PV+T G
Sbjct: 138 VYATLRFLPAYRVFRLLCANQSYN-YEMVHRVGSFAVPLSRDEEAAMRSYQFVPVETQHG 196
Query: 239 KLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFP-SLPVVRMPSHGSSPS 297
+L +SV Y P ++ + E ++ +I DYVGSP + + FP SL + + S
Sbjct: 197 RLVVSVQYLPSLAAFNLEISSLSPSMLIADYVGSPAAEPMRAFPASLTGATGSAFPQALS 256
Query: 298 SLPFSRRHSWS---------YENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSS 348
+ P R HSW+ + R SPP + SP+P+ + + +G P S
Sbjct: 257 NQP-QRPHSWATPALWPQAPRQQARFSPPHLLNASPTPSPPNFPSGYLQSRPKGGSAPMS 315
Query: 349 LPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNI 408
+P S +H+ + P S + SP S R++ +
Sbjct: 316 IPQVGDRRSPIHRP-ITLPPTSPRRVGETGTSSAQQSPSERCPSFG-----RADGFRIMD 369
Query: 409 PNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSG 468
P A + SPG R + T + + + SC +P
Sbjct: 370 PYASL--SPG-------------RKGKDTKDESGRFSALSSCDSPR-------------- 400
Query: 469 GKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDT--TDPGS 514
QD+ DD D+ PF VDD DT + PGS
Sbjct: 401 ---------------------QDDIDDADY--PFAVDDVDTPSSQPGS 425
>emb|CAG87936.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50422275|ref|XP_459700.1| unnamed protein product
[Debaryomyces hansenii]
Length = 837
Score = 96.7 bits (239), Expect = 2e-18
Identities = 119/520 (22%), Positives = 208/520 (39%), Gaps = 67/520 (12%)
Query: 13 AKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALR 72
AK Q+I FF+KS+ I+++SR S S L + SS +KWFNL +
Sbjct: 25 AKLTQVIQNFFSKSVQIVLQSRIQSESHNKEEQLKVGGDVGGHNVSS--KINKWFNLHMY 82
Query: 73 ECPTALENTDLWRHSN----LQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQ 128
E LW++ N + P++++V L R L+ V NP+ GGS++
Sbjct: 83 NDNLPKEELKLWKNINDVSQMPPMIIEVYLDLRQLTAIQTVILRDDNGNPWTVAKGGSKK 142
Query: 129 NEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPA 188
+E ++ERW+I+++ ANT + L +YK++ +L RSLY RL+P
Sbjct: 143 HEVVLERWLIEFD------------ANTVSGTIVDELPLIYKQAIILFRSLYGYTRLMPT 190
Query: 189 FKIFKELSSSANVSAFSLAHRVSTFFEPFTTK----------------QEAEMLKFVFTP 232
FK+ K L N S ++ ++ +P ++K E+ M F P
Sbjct: 191 FKLKKNL----NKSNLNIGCKILDGKQPISSKGRIGLSKSIIPHQMLTTESHMSHKHFLP 246
Query: 233 VDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVIT---DYVGSPLTDRLMRF-PSLPVVR 288
+ T G L +S+ YR + LS + + S +T + F L + R
Sbjct: 247 IQTTLGTLKISIAYRNHHEFSIHDNEELLSTHFVNIDDNDKDSEITPVEIEFKEELKIDR 306
Query: 289 MPSHGSSPSSLPFSRRH----SWSYENCRASPPAVNYYSPSP--THSESHTLVSNASSQG 342
+ + H S E+ ++ P ++ + + S +SN +S
Sbjct: 307 DSTTNEKVNEPEDDELHHADVESSQEHLQSEEPDESFEQDAKHISGSRKKFSISNNASMS 366
Query: 343 YCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSE 402
P SS P +E S H K PS N +P S + + +GS+++ S
Sbjct: 367 LSPCSSGPQTVTEDSPSHNK--------PSANTTPIVSQRPTINPFKVGSIST-----SP 413
Query: 403 SAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISR----CTSETERSRNLMQSCTTPEKMF- 457
A N + + TS ++ ++ + R TS T + N+ + +
Sbjct: 414 PATTNFGGSSLERKVSITSNKSASNASLAAMLRNPRSSTSSTNTTANIPIANNNSNNQYN 473
Query: 458 -SIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDT 496
+ + G +A ++ + FS + S F +
Sbjct: 474 STFPRSVSSSHGSNLAHDNDNLLGFSNPDNTSNTPRFSSS 513
>emb|CAG77754.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50555077|ref|XP_504947.1| hypothetical protein
[Yarrowia lipolytica]
Length = 715
Score = 94.4 bits (233), Expect = 8e-18
Identities = 94/333 (28%), Positives = 142/333 (42%), Gaps = 56/333 (16%)
Query: 3 TTSSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRP 62
T+S + + +K Q++ FF+K+ +++++R +P S
Sbjct: 6 TSSGSSHSSKSKLNQVVQNFFSKATQVVVQARL------------NPRLREQRQSGDKHK 53
Query: 63 RDKWFNLALRECPTALENTDLWRH------SNLQPIVVDVVLVHRNLSFSPKV---RSFV 113
+KWFNL E E+ LWR + P+VV+ L R+LS + + +F
Sbjct: 54 LNKWFNLETDELEAYREDLKLWRTIDIYNMDKMPPVVVETYLDMRHLSPNQTLVLEDAFG 113
Query: 114 KERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKST 173
K N FGG + E ++ERWVI+ I+ TT +S+T L YKK
Sbjct: 114 KRWNA--SFGG--RKTEVVLERWVIE-----IQRPDPQAGGATTSPASSTELPMAYKKCI 164
Query: 174 LLLRSLYATVRLLPAFKIFKELS-SSANVSAFSLAHRVSTFFEPFTTKQEAEMLK----- 227
LL RSLY RLLPA+ + K LS S + S + RV +P +++ + K
Sbjct: 165 LLFRSLYTYCRLLPAWTLQKRLSKSKLSTSPLRIGCRVLNGSQPISSRGRVGLSKLIAGS 224
Query: 228 --------FVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLM 279
F F PV+ SG +SV YR + + LS Q + P+
Sbjct: 225 SESQHLQTFSFDPVEIPSGVFKISVSYRVNCNFAVDDSEAMLSSQFLRIDEEKPVV---- 280
Query: 280 RFPSLPVVRMPSHGSSP----SSLPFSRRHSWS 308
P P V+ PS + + LP RRHS S
Sbjct: 281 --PPSPPVKHPSMAAPNLHYITGLP--RRHSHS 309
>gb|EAK96765.1| hypothetical protein CaO19.2848 [Candida albicans SC5314]
Length = 761
Score = 92.0 bits (227), Expect = 4e-17
Identities = 111/465 (23%), Positives = 196/465 (41%), Gaps = 56/465 (12%)
Query: 6 SHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDK 65
+ + + AK Q+I +FF K+ II+ESRA + ++PS + SS +K
Sbjct: 25 TQTKLQVAKLTQVIQKFFTKAAQIILESRA-------YPETTTPSLYPTKEESS--KINK 75
Query: 66 WFNLALRECPTAL-ENTDLWRHSNLQ---PIVVDVVLVHRNLSFSPKVRSFVKERNPFEE 121
WFNL + P + ++ LW+ +L P++++ + RNL + E++P+
Sbjct: 76 WFNLYMTNIPDSCKDDLKLWKGVDLTTIPPMIIETYIDLRNLPADQTLVLMDDEKHPWTV 135
Query: 122 FGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYA 181
++ E ++ERW+I++E NT + T L YK++ +L RS+Y
Sbjct: 136 AKSRGKKQEVVLERWLIEFEP---------NTTDATVMVEELPLS--YKQAIVLFRSIYG 184
Query: 182 TVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTK--------------QEAEMLK 227
RL+PAFK+ K L + L +++ +P ++K E+ M +
Sbjct: 185 FTRLMPAFKVKKNLQNK-----LPLGNKILDGNQPISSKGRIGLSKPIINTRTNESHMTQ 239
Query: 228 FVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVV 287
F PV T+ G L +SV YR + SE + ++++ + + + S V
Sbjct: 240 KYFQPVHTSLGTLKISVAYR-----MDSEFCLHENEELLSSHFHKRDEEETKKKVSSSVS 294
Query: 288 RMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSP---THSESHTLVSNASSQGYC 344
+ S S + R+ S R P V S SP + S S + +Q
Sbjct: 295 PLSSGTSLKETSTSPRK---SQPPIRIQPFKVGSMSTSPPVQSPSISQPGTAPIQNQPSV 351
Query: 345 PPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESA 404
P SSL S S N + + + S +PS +++ PI + ++ ++ RS S+
Sbjct: 352 PSSSLERRVSITSNKSTSNASLAAFLRNAR-SSTPS-ANNIPIINANPISGTSVPRSFSS 409
Query: 405 PVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQS 449
++ ++ S R SR S T + QS
Sbjct: 410 STGHEDSIFVNPDSASNTPRFASSFGSRASRRYSSTSIRQQTPQS 454
>gb|EAK96817.1| hypothetical protein CaO19.10367 [Candida albicans SC5314]
Length = 761
Score = 91.3 bits (225), Expect = 7e-17
Identities = 111/465 (23%), Positives = 196/465 (41%), Gaps = 56/465 (12%)
Query: 6 SHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDK 65
+ + + AK Q+I +FF K+ II+ESRA + S+PS + SS +K
Sbjct: 25 TQTKLQVAKLTQVIQKFFTKAAQIILESRA-------YPETSTPSLYPTKEESS--KINK 75
Query: 66 WFNLALRECPTAL-ENTDLWRHSNLQ---PIVVDVVLVHRNLSFSPKVRSFVKERNPFEE 121
WFNL + P + ++ LW+ +L P++++ + R+L + E++P+
Sbjct: 76 WFNLYMTNIPDSCKDDLKLWKGVDLTTIPPMIIETYIDLRSLPADQTLVLMDDEKHPWTV 135
Query: 122 FGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYA 181
++ E ++ERW+I++E NT + T L YK++ +L RS+Y
Sbjct: 136 AKSRGKKQEVVLERWLIEFEP---------NTTDATVMVEELPLS--YKQAIVLFRSIYG 184
Query: 182 TVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTK--------------QEAEMLK 227
RL+PAFK+ K L + L +++ +P ++K E+ M +
Sbjct: 185 FTRLMPAFKVKKNLQNK-----LPLGNKILDGNQPISSKGRIGLSKPIINTRTNESHMTQ 239
Query: 228 FVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVV 287
F PV T+ G L +SV YR + SE + ++++ + + + S V
Sbjct: 240 KYFQPVHTSLGTLKISVAYR-----MDSEFCLHENEELLSSHFHKRDEEETKKKVSSSVS 294
Query: 288 RMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSP---THSESHTLVSNASSQGYC 344
+ S S + R+ S R P V S SP + S S + +Q
Sbjct: 295 PLSSGTSLKETSTSPRK---SQPPIRIQPFKVGSMSTSPPVQSPSISQPGTAPIQNQPSV 351
Query: 345 PPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESA 404
P SSL S S N + + + S +PS +++ PI + ++ ++ RS S+
Sbjct: 352 PSSSLERRVSITSNKSTSNASLAAFLRNAR-SSTPS-ANNIPIINANPISGTSVPRSFSS 409
Query: 405 PVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQS 449
++ ++ S R SR S T + QS
Sbjct: 410 STGHEDSIFVNPDSASNTPRFASSFGSRASRRYSSTSIRQQTPQS 454
>gb|EAA64908.1| hypothetical protein AN2076.2 [Aspergillus nidulans FGSC A4]
gi|67523239|ref|XP_659680.1| hypothetical protein
AN2076_2 [Aspergillus nidulans FGSC A4]
gi|49088868|ref|XP_406213.1| hypothetical protein
AN2076.2 [Aspergillus nidulans FGSC A4]
Length = 974
Score = 62.0 bits (149), Expect = 5e-08
Identities = 122/557 (21%), Positives = 203/557 (35%), Gaps = 107/557 (19%)
Query: 5 SSHAQTEA-AKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPR 63
SS EA +K QI+T + K+ II+ SR S + +S+V
Sbjct: 56 SSQPGPEAISKVNQIVTNYHTKAALIILHSRV-------------ELPPSYAKNSNVPRV 102
Query: 64 DKWFNLALRECPTALENTDLWRHSNLQ-----PIVVDVVLVHRNLSFSPKV--------- 109
++WFN+ L E + WR + P++++ L L+ + +
Sbjct: 103 NRWFNVELEETDALKDQLKTWRTCDATDNRPPPMIIETYLDTAGLTNNQTLVALDDNGKR 162
Query: 110 ----RSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFL 165
S ++ S + I+ERW ++ K S L
Sbjct: 163 WDVTESLAASQSSRPAKASSSRAVDVILERWRVELGDMPGKLPSDLGA----------IL 212
Query: 166 QNLYKKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEM 225
+YKKS +L RSL+ + LPA+K K S A + +R+ + +KQ+
Sbjct: 213 PTVYKKSIILFRSLFTYSKFLPAWKFIKRNGRSRAHPALRVKYRIFSGHARDLSKQDHLT 272
Query: 226 L-----------KFVFTPVDTNSGKLCLSVMYRPC------------------VSDVSSE 256
+ + F D+ +G + V YR C D +
Sbjct: 273 MPLFEGDTKVVDTYSFGVTDSPAGPFSVQVTYRTCCDFRVDDSEALLSSQFMGADDEIFQ 332
Query: 257 PTTP---LSPQVITDYVGSPLTDRLMRFPSLPVVR---------MPSHGSSPSSLPFSRR 304
P+ P L +V + +PLT R + P L P+ G+SP S + R
Sbjct: 333 PSLPSGGLDARVTPEVGSAPLTRRTVEDPDLSRAYGSLSTFHHVGPTTGASPISALRAAR 392
Query: 305 HSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNV 364
+ RAS P+ P+ +H S + AS G + +P ++ + ++
Sbjct: 393 EA------RASSPS----PPTSSHRNSFA-AARASPVGRAATLANDTNP---NVARRPSI 438
Query: 365 NFDEYYPSPNFSPSPS----PSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHT 420
+F + + +P S SPS P S+SP + S + R P ++ S G
Sbjct: 439 SF-QPFKAPPLSASPSLVDPPLSASPRTTGAGRTSLSDSRHMPPPSVTTSSRKPPSFGPD 497
Query: 421 SRQNLPPST---PIRISRCTS--ETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNS 475
+ + P S P +SR +S R R E S G+ S SG +
Sbjct: 498 NANSSPNSASPRPTPMSRYSSAFSHRRGRPSSGGINKLEDDNSSGRASATSSGAQPGSGL 557
Query: 476 SPQISFSRSSSRSYQDE 492
+I+ + S S DE
Sbjct: 558 LAEITGTSSDSIHADDE 574
>emb|CAB11710.1| SPAC4F10.07c [Schizosaccharomyces pombe]
gi|19115662|ref|NP_594750.1| hypothetical protein
SPAC4F10.07c [Schizosaccharomyces pombe 972h-]
gi|48474255|sp|O36019|YEK7_SCHPO Protein C4F10.07c in
chromosome I
Length = 758
Score = 60.8 bits (146), Expect = 1e-07
Identities = 63/252 (25%), Positives = 106/252 (42%), Gaps = 44/252 (17%)
Query: 12 AAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLAL 71
+AK Q+I F K+ II+ESR N S P +S ++ KWFNL +
Sbjct: 37 SAKLGQVIHHCFYKTGLIILESRL------NVFGTSRPRESSKNN--------KWFNLEI 82
Query: 72 RECPTALENTDLWRHSNLQPI-VVDVVLVHRNLSFSPKVRS---FVKERNPFEEFGGGSE 127
E E +W++ L P + +++H L S ++ V + +
Sbjct: 83 VETELYAEQFKIWKNIELSPSRKIPPMVLHTYLDISDLSKNQTLSVSDGTHSHAINFNNM 142
Query: 128 QNEKIV-ERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLL 186
KIV ERW++ + + + S+ L LYKK +L RSLY L+
Sbjct: 143 STMKIVLERWIVNLDGEAL--------------STPLELAVLYKKLVVLFRSLYTYTHLM 188
Query: 187 PAFKIFKELSS-SANVSAFSLAHRVST----------FFEPFTTKQEAEMLKFVFTPVDT 235
P +K+ ++ A+ ++ + +ST P ++ + + F F+PV T
Sbjct: 189 PLWKLKSKIHKLRAHGTSLKVGCALSTDDVLSNDFLPISAPISSSLGSSIATFSFSPVGT 248
Query: 236 NSGKLCLSVMYR 247
+G +SV YR
Sbjct: 249 PAGDFRISVQYR 260
>emb|CAH02080.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50303491|ref|XP_451687.1| unnamed protein product
[Kluyveromyces lactis]
Length = 684
Score = 58.2 bits (139), Expect = 7e-07
Identities = 59/256 (23%), Positives = 103/256 (40%), Gaps = 52/256 (20%)
Query: 17 QIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALRECPT 76
++I FF KS ++ +S+ + NF T ++R + FN+ R P
Sbjct: 10 ELINNFFLKSALLLEQSKV----AHNFDT-----------EEALRDGNHLFNIETRGDPL 54
Query: 77 ALENTDLWRH----SNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQNEKI 132
W + P+V++ L R L + V + NP+ GG ++ E +
Sbjct: 55 LEAQIQPWITFDGVKTMPPLVIETYLDLRALQPNHMVYLHDADGNPWMVCKGG-KKTEIV 113
Query: 133 VERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIF 192
+ERW+++ + + I D SN +NL+K+ LL R LY +LLPA I
Sbjct: 114 LERWLVELDKQTIDDSIDSNDP-----------ENLHKQLVLLFRYLYTLTQLLPANDII 162
Query: 193 KELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFV---------------------FT 231
+ +S + ++ R+ +P +K + K + T
Sbjct: 163 TKPHNSQQPALINVQTRLLDGSKPILSKGRVGLSKPIIASYSNTMNETNIASHLEQRKIT 222
Query: 232 PVDTNSGKLCLSVMYR 247
P+ T G L ++V YR
Sbjct: 223 PIKTTFGSLRITVSYR 238
>gb|EAA71308.1| hypothetical protein FG08491.1 [Gibberella zeae PH-1]
gi|46128227|ref|XP_388667.1| hypothetical protein
FG08491.1 [Gibberella zeae PH-1]
Length = 927
Score = 57.4 bits (137), Expect = 1e-06
Identities = 46/195 (23%), Positives = 82/195 (41%), Gaps = 43/195 (22%)
Query: 14 KTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALRE 73
K +QII F+AK+ ++++SR S +R ++ R +KWF + E
Sbjct: 70 KLDQIIQNFYAKAAVLVLDSRIKSRPARG--------------ANGARKPNKWFQIETDE 115
Query: 74 CPTALENTDLWRH-----SNLQPIVVDVVLVHRNLSFSP---------KVRSFVKERNPF 119
+ +W++ + P+V++V L L S K +++ N +
Sbjct: 116 IDDFRDELKIWKNCGSLDNRPPPMVIEVYLDASRLKDSQSLVIVDENGKRWDVMEQLNSY 175
Query: 120 ----EEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLL 175
+ G NE ++ERW ++ + + T L +YKK+ +
Sbjct: 176 GSSTDSSGASRRNNEVVIERWQVELKH-----------SGMTSVDFGPILPTVYKKAIVF 224
Query: 176 LRSLYATVRLLPAFK 190
RSL+ T RLLPA+K
Sbjct: 225 FRSLFITTRLLPAWK 239
>gb|AAC48526.1| gastric mucin gi|2136504|pir||I47141 gastric mucin (clone PGM-2A) -
pig (fragment)
Length = 528
Score = 53.5 bits (127), Expect = 2e-05
Identities = 110/508 (21%), Positives = 187/508 (36%), Gaps = 53/508 (10%)
Query: 2 TTTSSHAQTEAAKTEQIITEFFAK----------SLHIIIESRALSASSRNFSTLSSPSS 51
TT+++ QT ++ + I + + S + S + SA + +++ S SS
Sbjct: 46 TTSATSVQTSSSSSPPISSTISVQTSSSSSVPTTSTTSVQPSSSSSAPTTRATSVQSSSS 105
Query: 52 TSS--SSSSSVRPRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKV 109
+S+ SS++SV+P PT T + S+ V + S SP +
Sbjct: 106 SSAPISSTTSVQPSSSG------SVPTT-SATSVQSSSSSSAPTTSATSVQPSSSSSPPI 158
Query: 110 RSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLY 169
S V + S + +Q S SS+ + T+ SS
Sbjct: 159 SSTVSVQP-------SSSSSAPTTSATSVQPSSSSSPPISSTVSVQTSSSSSVPTTS--- 208
Query: 170 KKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFV 229
+T + S ++V A + SSS + + + S+ P T+ +
Sbjct: 209 --TTSVQPSSSSSVPTTSATSVRSSSSSSTPIPSTTSVQPSSSSSAPTTSATSVQPSSSS 266
Query: 230 FTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRM 289
TP+ + + S P S S +P++ SP + + P + S V+
Sbjct: 267 STPIPSTTSVQPSSSSSAPTTSATSVQPSSSSSPPISSTISVQPSSSSSSPTTSTTSVQP 326
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNA----SSQGYCP 345
S GS+P++ S + S S +SPP + S P+ S S S SS G P
Sbjct: 327 SSSGSAPTTSATSVQPSSS-----SSPPISSTISVQPSSSSSSPTTSTTSVQPSSSGSAP 381
Query: 346 PSSL----PPHPSEMSLLHKKNVNFDEYYPSP---NFSPSPSPSSSSPIYSLGSLASKTL 398
+S P S + +V +P S PS SSS P S S+ +
Sbjct: 382 TTSATSVQPSSSSSVPTTSATSVRSSSSSSTPIPTTTSVQPSSSSSVPTTSATSVQT--- 438
Query: 399 LRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFS 458
S S+ IP+ + P +S +T ++ S +S S +Q ++ +
Sbjct: 439 --SSSSSTPIPST-TSVQPSSSSSAPTTSATSVQPSSSSSPPISSTISVQPSSSSSSPTT 495
Query: 459 IGKESQKYSGGKIAPNSSPQISFSRSSS 486
Q S G S+ + S SSS
Sbjct: 496 STTSVQPSSSGSAPTTSATSVQPSSSSS 523
Score = 38.1 bits (87), Expect = 0.71
Identities = 58/236 (24%), Positives = 90/236 (37%), Gaps = 23/236 (9%)
Query: 251 SDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYE 310
S SS PTT + + P+ PS V+ S GS+P++ S + S S
Sbjct: 7 SSSSSSPTTSTTSVQSSSSSSVPI-------PSTTSVQPSSSGSAPTTSATSVQTSSS-- 57
Query: 311 NCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYY 370
+SPP + S + S S S S Q PSS P+ + + + +
Sbjct: 58 ---SSPPISSTISVQTSSSSSVPTTSTTSVQ----PSSSSSAPTTRATSVQSSSSSSAPI 110
Query: 371 PSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTP 430
S S PS S S P S S+ S + S SAP + + P +S + +
Sbjct: 111 SSTT-SVQPSSSGSVPTTSATSVQSSS---SSSAPT---TSATSVQPSSSSSPPISSTVS 163
Query: 431 IRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSS 486
++ S +S S +Q ++ S Q S + S+ + S SSS
Sbjct: 164 VQPSSSSSAPTTSATSVQPSSSSSPPISSTVSVQTSSSSSVPTTSTTSVQPSSSSS 219
>gb|AAQ82687.1| Epa5p [Candida glabrata]
Length = 1218
Score = 53.5 bits (127), Expect = 2e-05
Identities = 76/291 (26%), Positives = 117/291 (40%), Gaps = 28/291 (9%)
Query: 212 TFFEPFTTKQEAEMLKFVFTPVDTNSGKLCL-SVMYRPCVSDVSSEPTTPLSPQVITDYV 270
+F F T++ + + F+ D G+ C +V Y +DV S T S I
Sbjct: 226 SFTTEFDTERVHDFTSYFFSASD---GEQCPGNVNYNYHCADVKSS-TILTSSTTIDKQD 281
Query: 271 GS--PLTDRLMRF-----PSLPVVRMPSHGSSP-----SSLPFSRRHSWSYENCRASPPA 318
G+ P+T + P P + P +P LP + S + SP +
Sbjct: 282 GNIVPITKTIYEIGVPCNPDQPTQKCPGEFYNPIRDVCEPLPTPSQDINSSSSSSPSPSS 341
Query: 319 VNYYSPSPTHSESHTLVSNASSQGYCPPSSLP-PHPSEMSLLHKKNVNFDEYYPSPNFSP 377
+ S S + S S + S++SS SS P PS S + + S + SP
Sbjct: 342 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSS-----SSSSSSP 396
Query: 378 SPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCT 437
SPS SSSS S S +S + S S+ + ++ + SP +S + S S +
Sbjct: 397 SPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSSSSSSSSS-----SSSS 451
Query: 438 SETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRS 488
S + S + S ++P S S S + +SSP S S SSS S
Sbjct: 452 SSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSS 502
Score = 45.8 bits (107), Expect = 0.003
Identities = 43/143 (30%), Positives = 65/143 (45%), Gaps = 7/143 (4%)
Query: 291 SHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP 350
S SS SS S S S + +S + + SPSP+ S S + S++SS SS
Sbjct: 401 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSSSSSSSSSSSSSSSSSSSSSS 460
Query: 351 PH----PSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPV 406
PS S + + S + SPSPS SSSS S S +S ++ S PV
Sbjct: 461 SSSSSSPSPSSSSSSSSSSSSSSSSSSSSSPSPSSSSSS---SSSSSSSSPSIQPSSKPV 517
Query: 407 NIPNAEVTYSPGHTSRQNLPPST 429
+ A+ + +P + ++ PS+
Sbjct: 518 DPSPADPSNNPSSVNPSSVNPSS 540
Score = 42.4 bits (98), Expect = 0.037
Identities = 36/131 (27%), Positives = 54/131 (40%)
Query: 371 PSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTP 430
PSP+ S S S SSSS S S +S + S S+ + P+ + S +S + S+P
Sbjct: 337 PSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSP 396
Query: 431 IRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQ 490
S +S + S + S ++ S S S +SS S S SSS S
Sbjct: 397 SPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSSSSSSSSSSSSSSSSSS 456
Query: 491 DEFDDTDFTCP 501
+ + P
Sbjct: 457 SSSSSSSSSSP 467
Score = 42.0 bits (97), Expect = 0.049
Identities = 45/163 (27%), Positives = 67/163 (40%), Gaps = 9/163 (5%)
Query: 291 SHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP 350
S SS SS S S S + +S + + SPSP+ S S + S++SS SS
Sbjct: 363 SSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSS 422
Query: 351 PHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPN 410
S S PSP+ S S S SSSS S S +S + S S + +
Sbjct: 423 SSSSSSS---------SSSSPSPSRSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSS 473
Query: 411 AEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTP 453
+ + S +S + +P S +S + S +Q + P
Sbjct: 474 SSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSPSIQPSSKP 516
Score = 41.6 bits (96), Expect = 0.064
Identities = 47/198 (23%), Positives = 73/198 (36%), Gaps = 5/198 (2%)
Query: 317 PAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFS 376
P + P PT S+ ++++SS P SS S S + + S + S
Sbjct: 314 PIRDVCEPLPTPSQD---INSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 370
Query: 377 PSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRC 436
S SPS SS S S +S + S S + ++ + S +S + S+ S
Sbjct: 371 SSSSPSPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 430
Query: 437 TSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDT 496
+S SR+ S ++ S S S +P SP S S SSS S +
Sbjct: 431 SSSPSPSRSSSSSSSSSSSSSSSSSSSSSSSSSSSSP--SPSSSSSSSSSSSSSSSSSSS 488
Query: 497 DFTCPFDVDDDDTTDPGS 514
P ++ S
Sbjct: 489 SSPSPSSSSSSSSSSSSS 506
Score = 39.3 bits (90), Expect = 0.32
Identities = 41/148 (27%), Positives = 60/148 (39%), Gaps = 7/148 (4%)
Query: 291 SHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL- 349
S SSPS S S S + +S P+ + S S + S S + S++SS SS
Sbjct: 370 SSSSSPSPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 429
Query: 350 ------PPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSES 403
P S S + + S + S S SPS SS S S +S + S S
Sbjct: 430 SSSSPSPSRSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSS 489
Query: 404 APVNIPNAEVTYSPGHTSRQNLPPSTPI 431
+P ++ + S +S P S P+
Sbjct: 490 SPSPSSSSSSSSSSSSSSPSIQPSSKPV 517
>ref|XP_524885.1| PREDICTED: hypothetical protein XP_524885 [Pan troglodytes]
Length = 328
Score = 52.8 bits (125), Expect = 3e-05
Identities = 70/274 (25%), Positives = 102/274 (36%), Gaps = 50/274 (18%)
Query: 218 TTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDR 277
+T+ +A+ L+ +T G L +++ S +PL T + G L +R
Sbjct: 49 STRLQAQKLRLAYTRSSHYGGSLPNVNQIGSGLAEFQSPLHSPLDSSRSTRHHG--LVER 106
Query: 278 LMRFPSL---PVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTL 334
+ R P P+ R P H S L S S Y SPP + + + + S HT
Sbjct: 107 VQRDPRRMVSPLRRYPRHISFTGDLGVSE--SSPYSPAYLSPPPESSWRRTSSDSALHTS 164
Query: 335 VSNASSQ----GYCPPSSLPP----------HPSEMSLLHKKNVNFDEYYPSPNFSPSPS 380
V N S Q G PPS LP HPS S L N+ P P S S
Sbjct: 165 VMNPSPQDTYPGPTPPSILPSRRGGLHSPLSHPSLQSSLSNPNLQASLSSPQPQLQGSHS 224
Query: 381 ---------------------PSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGH 419
PS S+P S S S +L + S P + P A SP H
Sbjct: 225 HPSLPASSLARHALPTTSLGHPSLSAPALSSSSSTSSPVLGAPSYPASTPGA----SPHH 280
Query: 420 TSRQNLPPSTPIRISRCTSETERSRNLMQSCTTP 453
+ P +P+ + ++ RS+ + +P
Sbjct: 281 ----HRVPLSPLSLLAGPADARRSQQQLPKQFSP 310
>emb|CAG31044.1| hypothetical protein [Gallus gallus]
Length = 888
Score = 52.0 bits (123), Expect = 5e-05
Identities = 62/220 (28%), Positives = 84/220 (38%), Gaps = 33/220 (15%)
Query: 273 PLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESH 332
P R R PSLP R PS P RR S S R SPP YSPSP
Sbjct: 558 PPPPRRRRSPSLPRRRSPS--------PPPRRRSPSPR--RYSPPIQRRYSPSPPPKRR- 606
Query: 333 TLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSL-- 390
+S PP P + ++ + P P SP+ SP S
Sbjct: 607 -------------TASPPPPPKRRASPSPQSKRRVSHSPPPKQRSSPAAKRRSPSISSKH 653
Query: 391 --GSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQ 448
GS S++ + S P N + SP R + S+P + R S + + R
Sbjct: 654 RKGSPPSRSNRETRSPPQNKRD-----SPSPRPRASHTSSSPPPLRRGASASPQRRQSPS 708
Query: 449 SCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRS 488
T P + S E +K +P S+ ++S SRS+S S
Sbjct: 709 PSTRPIRRVSRTPEPKKTKASTPSPRSARRVSSSRSASGS 748
>ref|XP_317690.2| ENSANGP00000010305 [Anopheles gambiae str. PEST]
gi|55237903|gb|EAA12308.2| ENSANGP00000010305 [Anopheles
gambiae str. PEST]
Length = 1877
Score = 50.8 bits (120), Expect = 1e-04
Identities = 66/215 (30%), Positives = 92/215 (42%), Gaps = 35/215 (16%)
Query: 242 LSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPF 301
+S M+ S+ +S +P SP SP + PS PV S SPSS +
Sbjct: 1635 MSPMHPTSASNFASPGYSPTSPSY------SPTSPSYS--PSSPVFSPTSPSYSPSSPSY 1686
Query: 302 SRRH-SWSYENCRASPPAVNY-------------YSPSPTHSESHTLVSNASSQGYCP-- 345
S S+S + SP + N+ YSP+ H + + SS Y P
Sbjct: 1687 SPTSPSYSPTSPSYSPTSPNFTPVTPSYSPSSPNYSPTSPHYSPASPSYSPSSPKYSPTS 1746
Query: 346 PSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSP-SPSPSSSSPIYSLGSLASKTLLRSES- 403
PS P PS + H + SPN+SP SPS S SSP +S GS ++ + S S
Sbjct: 1747 PSYSPTSPSYIGSPHYTPASPQYGQASPNYSPTSPSYSPSSPQHSPGSSSNHYSIGSSSY 1806
Query: 404 ----APVNIPNAEVTYSPGHTSRQNLPPSTPIRIS 434
AP + PN +YSP S P++P+ S
Sbjct: 1807 SQTPAPYS-PNMS-SYSP---SSPKYSPTSPVYTS 1836
Score = 49.7 bits (117), Expect = 2e-04
Identities = 73/243 (30%), Positives = 98/243 (40%), Gaps = 36/243 (14%)
Query: 248 PCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSS------LPF 301
P S + + +SP + GSP + P P S SPSS P
Sbjct: 1525 PSFSPSAQSDASGMSPSSWSPIRGSPSSPGPSMSPYPPSSPSMSPSYSPSSPNYAPNSPG 1584
Query: 302 SRRHSWSYENCRASPPAVNY------YSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSE 355
S+S + SPP+ Y YSPS + S T + +S Y P S P HP+
Sbjct: 1585 GASPSYSPTSPGYSPPSPQYSPGSSNYSPSSPNIHSATQNYSPTSPSYSPMS--PMHPTS 1642
Query: 356 MSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEV 413
S N Y P SP++SP SPS S SSP++S S + S S+P P +
Sbjct: 1643 AS-----NFASPGYSPTSPSYSPTSPSYSPSSPVFSPTSPS-----YSPSSPSYSPTSP- 1691
Query: 414 TYSPGHTSRQNLPPS-TPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIA 472
+YSP S P+ TP+ S S S T+P +S S S K +
Sbjct: 1692 SYSPTSPSYSPTSPNFTPVTPSYSPSSPN------YSPTSPH--YSPASPSYSPSSPKYS 1743
Query: 473 PNS 475
P S
Sbjct: 1744 PTS 1746
Score = 40.4 bits (93), Expect = 0.14
Identities = 56/208 (26%), Positives = 80/208 (37%), Gaps = 19/208 (9%)
Query: 294 SSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSES-HTLVSNASSQGYCP------- 345
++PS P+ + + Y S PA P+ S S + S S + P
Sbjct: 1493 ATPSMTPWQQSATPGYGGWSPSGPASGMTPGGPSFSPSAQSDASGMSPSSWSPIRGSPSS 1552
Query: 346 --PSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGS--LASKTLLRS 401
PS P PS S+ + + Y P+ SPS S +SP YS S + + S
Sbjct: 1553 PGPSMSPYPPSSPSMSPSYSPSSPNYAPNSPGGASPSYSPTSPGYSPPSPQYSPGSSNYS 1612
Query: 402 ESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGK 461
S+P NI +A YSP S + P P S S + S T+P S
Sbjct: 1613 PSSP-NIHSATQNYSPTSPSYSPMSPMHPTSASNFASPGYSPTSPSYSPTSP----SYSP 1667
Query: 462 ESQKYSGGKIAPNSSPQISFSRSSSRSY 489
S +S +P+ SP +S SY
Sbjct: 1668 SSPVFS--PTSPSYSPSSPSYSPTSPSY 1693
Score = 38.9 bits (89), Expect = 0.41
Identities = 46/132 (34%), Positives = 57/132 (42%), Gaps = 22/132 (16%)
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL 349
PS+ SP P S ++ + N SP + +Y SP HS + SN S G S
Sbjct: 1754 PSYIGSPHYTPASPQYGQASPN--YSPTSPSYSPSSPQHSPGSS--SNHYSIGSSSYSQT 1809
Query: 350 PPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASKTLLR--SESAP 405
P S N Y P SP +SP SP +SSSP YS S A + S S+P
Sbjct: 1810 PAPYSP---------NMSSYSPSSPKYSPTSPVYTSSSPTYSPSSPAYSPTPQGYSPSSP 1860
Query: 406 VNIPNAEVTYSP 417
A TYSP
Sbjct: 1861 -----ASPTYSP 1867
>gb|AAO72634.1| p67-like superoxide-generating NADPH oxidase [Dictyostelium
discoideum] gi|29028302|gb|AAO62420.1| p67-like
superoxide-generating NADPH oxidase [Dictyostelium
discoideum] gi|66805627|ref|XP_636535.1| hypothetical
protein DDB0191152 [Dictyostelium discoideum]
gi|60464893|gb|EAL63008.1| hypothetical protein
DDB0191152 [Dictyostelium discoideum]
Length = 604
Score = 50.8 bits (120), Expect = 1e-04
Identities = 42/147 (28%), Positives = 66/147 (44%), Gaps = 20/147 (13%)
Query: 355 EMSLLHKKNVNFDEYYPSPNFSPSPSP---SSSSPIYSLGSLASKTLLRSESAPVNIPNA 411
+M + +N + PSP+ SPSPSP ++S+ YS S +S + S S+
Sbjct: 370 KMICMEINEINVKDIIPSPSPSPSPSPDKTNNSTSSYSSSSSSSSSSSSSSSSSSYDNKP 429
Query: 412 EVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKM--------FSIGKES 463
+ ++ P T+R LPP+T T+ T S N ++ T P+K FS S
Sbjct: 430 KSSFIPKTTTRPILPPTT-------TTTTSTSNNFNRNATLPKKFGSTPSSPSFSSPSSS 482
Query: 464 QKYSGG--KIAPNSSPQISFSRSSSRS 488
GG I SP IS + +++
Sbjct: 483 SSGGGGGPPIPTRGSPSISLLKQQNQT 509
Score = 34.7 bits (78), Expect = 7.8
Identities = 37/141 (26%), Positives = 59/141 (41%), Gaps = 17/141 (12%)
Query: 271 GSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSE 330
GS L P + + P +P ++ S + + ++S P+ S P++
Sbjct: 164 GSQLNFSTRPIPLSLLFKPPKVSDAPQ-----KQRSATTSSIQSSSPSTPMSSSPPSY-- 216
Query: 331 SHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSL 390
++ SS PPSS P S SL + P P+F SP PSSSS S
Sbjct: 217 ---ILKGPSS----PPSSSSPSSSSPSLSSSSSPKLPPT-PKPSFGSSPPPSSSSSSSSS 268
Query: 391 GSLASKTL--LRSESAPVNIP 409
S +S ++ L +++ P P
Sbjct: 269 SSSSSSSISPLTNKTLPPKPP 289
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.127 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 930,988,276
Number of Sequences: 2540612
Number of extensions: 42254259
Number of successful extensions: 170084
Number of sequences better than 10.0: 1077
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 1107
Number of HSP's that attempted gapping in prelim test: 163757
Number of HSP's gapped (non-prelim): 4137
length of query: 535
length of database: 863,360,394
effective HSP length: 133
effective length of query: 402
effective length of database: 525,458,998
effective search space: 211234517196
effective search space used: 211234517196
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146842.6


