

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.3 + phase: 0
(380 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
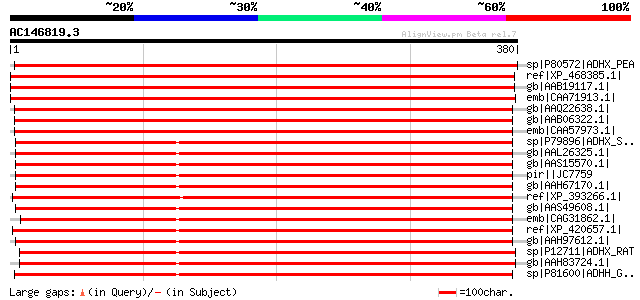
Score E
Sequences producing significant alignments: (bits) Value
sp|P80572|ADHX_PEA Alcohol dehydrogenase class III (Glutathione-... 737 0.0
ref|XP_468385.1| alcohol dehydrogenase class III [Oryza sativa (... 712 0.0
gb|AAB19117.1| class III ADH enzyme [Oryza sativa] gi|3023271|sp... 712 0.0
emb|CAA71913.1| glutothione-dependent formaldehyde dehydrogenase... 701 0.0
gb|AAQ22638.1| At5g43940/MRH10_4 [Arabidopsis thaliana] gi|21592... 699 0.0
gb|AAB06322.1| glutathione-dependent formaldehyde dehydrogenase 697 0.0
emb|CAA57973.1| class III ADH, glutathione-dependent formaldehyd... 697 0.0
sp|P79896|ADHX_SPAAU Alcohol dehydrogenase class III (Glutathion... 540 e-152
gb|AAL26325.1| alcohol dehydrogenase [Danio rerio] 538 e-152
gb|AAS15570.1| class III alcohol dehydrogenase [Oryzias latipes] 538 e-151
pir||JC7759 alcohol dehydrogenase (EC 1.1.1.1) 3 - zebra fish 538 e-151
gb|AAH67170.1| Alcohol dehydrogenase 5 [Danio rerio] gi|46195771... 537 e-151
ref|XP_393266.1| PREDICTED: similar to Alcohol dehydrogenase 5 [... 532 e-150
gb|AAS49608.1| alcohol dehydrogenase 5 [Xenopus laevis] 531 e-149
emb|CAG31862.1| hypothetical protein [Gallus gallus] 530 e-149
ref|XP_420657.1| PREDICTED: similar to Alcohol dehydrogenase cla... 530 e-149
gb|AAH97612.1| LOC445841 protein [Xenopus laevis] 530 e-149
sp|P12711|ADHX_RAT Alcohol dehydrogenase class III (Alcohol dehy... 530 e-149
gb|AAH83724.1| Unknown (protein for IMAGE:7191109) [Rattus norve... 529 e-149
sp|P81600|ADHH_GADMO Alcohol dehydrogenase class III H chain (Gl... 527 e-148
>sp|P80572|ADHX_PEA Alcohol dehydrogenase class III (Glutathione-dependent formaldehyde
dehydrogenase) (FDH) (FALDH) (GSH-FDH)
Length = 378
Score = 737 bits (1902), Expect = 0.0
Identities = 356/377 (94%), Positives = 367/377 (96%)
Query: 4 STEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEG 63
+T+GQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYT GKDPEG
Sbjct: 1 ATQGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTLGGKDPEG 60
Query: 64 LFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATG 123
LFPCILGHEAAGIVESVGEGVTDVKPGDHVIP YQAECGECKFCKS KTNLCGKVRAATG
Sbjct: 61 LFPCILGHEAAGIVESVGEGVTDVKPGDHVIPSYQAECGECKFCKSPKTNLCGKVRAATG 120
Query: 124 VGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
VGVMM DRK RFS+KGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDA LDKVCLLGCGVPT
Sbjct: 121 VGVMMADRKSRFSVKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAPLDKVCLLGCGVPT 180
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVT 243
GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNK+DTAKNFGVT
Sbjct: 181 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKYDTAKNFGVT 240
Query: 244 EFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASG 303
EFINPKDHEKPIQQVI DLTDGGVDYSFEC+GNVSVMRSALEC HKGWGTSV+VGVAASG
Sbjct: 241 EFINPKDHEKPIQQVIIDLTDGGVDYSFECLGNVSVMRSALECCHKGWGTSVIVGVAASG 300
Query: 304 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEA 363
QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEY+THNLTL EIN+A
Sbjct: 301 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYITHNLTLLEINKA 360
Query: 364 FDLMHEGKCLRCVLALH 380
FDL+HEG+CLRCVLA+H
Sbjct: 361 FDLLHEGQCLRCVLAVH 377
>ref|XP_468385.1| alcohol dehydrogenase class III [Oryza sativa (japonica
cultivar-group)] gi|47848172|dbj|BAD21999.1| alcohol
dehydrogenase class III [Oryza sativa (japonica
cultivar-group)] gi|47847883|dbj|BAD21676.1| alcohol
dehydrogenase class III [Oryza sativa (japonica
cultivar-group)]
Length = 381
Score = 712 bits (1839), Expect = 0.0
Identities = 343/378 (90%), Positives = 360/378 (94%)
Query: 1 MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD 60
MASST+GQVITCKAAVAWE N+P+TIEDV+VAPPQA EVR++ILFTALCHTD YTWSGKD
Sbjct: 1 MASSTQGQVITCKAAVAWEANRPMTIEDVQVAPPQAGEVRVKILFTALCHTDHYTWSGKD 60
Query: 61 PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRA 120
PEGLFPCILGHEAAGIVESVGEGVT+V+PGDHVIPCYQAEC ECKFCKSGKTNLCGKVRA
Sbjct: 61 PEGLFPCILGHEAAGIVESVGEGVTEVQPGDHVIPCYQAECRECKFCKSGKTNLCGKVRA 120
Query: 121 ATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCG 180
ATGVGVMMNDRK RFSI GKPIYHFMGTSTFSQYTVVHDVSVAKI+P A LDKVCLLGCG
Sbjct: 121 ATGVGVMMNDRKSRFSINGKPIYHFMGTSTFSQYTVVHDVSVAKINPQAPLDKVCLLGCG 180
Query: 181 VPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNF 240
V TGLGAVWNTAKVE GSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDS KFD AKNF
Sbjct: 181 VSTGLGAVWNTAKVEAGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSKKFDVAKNF 240
Query: 241 GVTEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVA 300
GVTEF+NPKDH+KPIQQVI DLTDGGVDYSFECIGNVSVMRSALEC HKGWGTSV+VGVA
Sbjct: 241 GVTEFVNPKDHDKPIQQVIVDLTDGGVDYSFECIGNVSVMRSALECCHKGWGTSVIVGVA 300
Query: 301 ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEI 360
ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYL KEIKVDEYVTH++ L +I
Sbjct: 301 ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLNKEIKVDEYVTHSMNLTDI 360
Query: 361 NEAFDLMHEGKCLRCVLA 378
N+AFDL+HEG CLRCVLA
Sbjct: 361 NKAFDLLHEGGCLRCVLA 378
>gb|AAB19117.1| class III ADH enzyme [Oryza sativa] gi|3023271|sp|P93436|ADHX_ORYSA
Alcohol dehydrogenase class III (Glutathione-dependent
formaldehyde dehydrogenase) (FDH) (FALDH) (GSH-FDH)
gi|7430933|pir||T04164 formaldehyde dehydrogenase
(glutathione) (EC 1.2.1.1) - rice
Length = 381
Score = 712 bits (1838), Expect = 0.0
Identities = 343/378 (90%), Positives = 359/378 (94%)
Query: 1 MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD 60
MASST+GQVITCKAAVAWE NKP+TIEDV+VAPPQA EVR++ILFTA CHTD YTWSGKD
Sbjct: 1 MASSTQGQVITCKAAVAWEANKPMTIEDVQVAPPQAGEVRVKILFTAFCHTDHYTWSGKD 60
Query: 61 PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRA 120
PEGLFPCILGHEAAGIVESVGEGVT+V+PGDHVIPCYQAEC ECKFCKSGKTNLCGKVRA
Sbjct: 61 PEGLFPCILGHEAAGIVESVGEGVTEVQPGDHVIPCYQAECRECKFCKSGKTNLCGKVRA 120
Query: 121 ATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCG 180
ATGVGVMMNDRK RFSI GKPIYHFMGTSTFSQYTVVHDVSVAKI+P A LDKVCLLGCG
Sbjct: 121 ATGVGVMMNDRKSRFSINGKPIYHFMGTSTFSQYTVVHDVSVAKINPQAPLDKVCLLGCG 180
Query: 181 VPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNF 240
V TGLGAVWNTAKVE GSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDS KFD AKNF
Sbjct: 181 VSTGLGAVWNTAKVEAGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSKKFDVAKNF 240
Query: 241 GVTEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVA 300
GVTEF+NPKDH+KPIQQVI DLTDGGVDYSFECIGNVSVMRSALEC HKGWGTSV+VGVA
Sbjct: 241 GVTEFVNPKDHDKPIQQVIVDLTDGGVDYSFECIGNVSVMRSALECCHKGWGTSVIVGVA 300
Query: 301 ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEI 360
ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYL KEIKVDEYVTH++ L +I
Sbjct: 301 ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLNKEIKVDEYVTHSMNLTDI 360
Query: 361 NEAFDLMHEGKCLRCVLA 378
N+AFDL+HEG CLRCVLA
Sbjct: 361 NKAFDLLHEGGCLRCVLA 378
>emb|CAA71913.1| glutothione-dependent formaldehyde dehydrogenase [Zea mays]
gi|6225010|sp|P93629|ADHX_MAIZE Alcohol dehydrogenase
class III (Glutathione-dependent formaldehyde
dehydrogenase) (FDH) (FALDH) (GSH-FDH)
gi|7430932|pir||T03289 formaldehyde dehydrogenase
(glutathione) (EC 1.2.1.1) - maize
Length = 381
Score = 701 bits (1810), Expect = 0.0
Identities = 330/379 (87%), Positives = 361/379 (95%)
Query: 1 MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD 60
MAS T+GQVITCKAAVA+EPNKPL IEDV+VAPPQA EVR++ILFTALCHTD YTWSGKD
Sbjct: 1 MASPTQGQVITCKAAVAYEPNKPLVIEDVQVAPPQAGEVRVKILFTALCHTDHYTWSGKD 60
Query: 61 PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRA 120
PEGLFPCILGHEAAGIVESVGEGVTDV+PGDHVIPCYQAEC ECKFCKSGKTNLCGKVR+
Sbjct: 61 PEGLFPCILGHEAAGIVESVGEGVTDVQPGDHVIPCYQAECKECKFCKSGKTNLCGKVRS 120
Query: 121 ATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCG 180
ATGVGVMMND K RFS+ GKPIYHFMGTSTFSQYTVVHDVSVAKI+P A LDKVCLLGCG
Sbjct: 121 ATGVGVMMNDMKSRFSVNGKPIYHFMGTSTFSQYTVVHDVSVAKINPQAPLDKVCLLGCG 180
Query: 181 VPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNF 240
VPTGLGAVWNTAKVE GS+VA+FGLGTVGLAVAEGAK+AGASR+IGIDID+ KFD AKNF
Sbjct: 181 VPTGLGAVWNTAKVESGSVVAVFGLGTVGLAVAEGAKAAGASRVIGIDIDNKKFDVAKNF 240
Query: 241 GVTEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVA 300
GVTEF+NPK+H+KPIQQV+ DLTDGGVDYSFECIGNVS+MR+ALEC+ KGWGTSV+VGVA
Sbjct: 241 GVTEFVNPKEHDKPIQQVLVDLTDGGVDYSFECIGNVSIMRAALECSDKGWGTSVIVGVA 300
Query: 301 ASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEI 360
ASGQEISTRPFQLVTGRVWKGTAFGGFKSR+QVPWLV+KY+KKEIKVDEY+THN+ LA+I
Sbjct: 301 ASGQEISTRPFQLVTGRVWKGTAFGGFKSRTQVPWLVDKYMKKEIKVDEYITHNMNLADI 360
Query: 361 NEAFDLMHEGKCLRCVLAL 379
N+AF L+HEG CLRCVLA+
Sbjct: 361 NDAFHLLHEGGCLRCVLAM 379
>gb|AAQ22638.1| At5g43940/MRH10_4 [Arabidopsis thaliana] gi|21592856|gb|AAM64806.1|
alcohol dehydrogenase (EC 1.1.1.1) class III
[Arabidopsis thaliana] gi|9758553|dbj|BAB09054.1|
alcohol dehydrogenase (EC 1.1.1.1) class III
[Arabidopsis thaliana] gi|15240054|ref|NP_199207.1|
alcohol dehydrogenase class III / glutathione-dependent
formaldehyde dehydrogenase / GSH-FDH (ADHIII)
[Arabidopsis thaliana] gi|14517532|gb|AAK62656.1|
AT5g43940/MRH10_4 [Arabidopsis thaliana]
gi|20141281|sp|Q96533|ADHX_ARATH Alcohol dehydrogenase
class III (Glutathione-dependent formaldehyde
dehydrogenase) (FDH) (FALDH) (GSH-FDH)
Length = 379
Score = 699 bits (1803), Expect = 0.0
Identities = 332/374 (88%), Positives = 356/374 (94%)
Query: 4 STEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEG 63
+T+GQVITCKAAVA+EPNKPL IEDV+VAPPQA EVRI+IL+TALCHTDAYTWSGKDPEG
Sbjct: 2 ATQGQVITCKAAVAYEPNKPLVIEDVQVAPPQAGEVRIKILYTALCHTDAYTWSGKDPEG 61
Query: 64 LFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATG 123
LFPCILGHEAAGIVESVGEGVT+V+ GDHVIPCYQAEC ECKFCKSGKTNLCGKVR+ATG
Sbjct: 62 LFPCILGHEAAGIVESVGEGVTEVQAGDHVIPCYQAECRECKFCKSGKTNLCGKVRSATG 121
Query: 124 VGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
VG+MMNDRK RFS+ GKPIYHFMGTSTFSQYTVVHDVSVAKI P A LDKVCLLGCGVPT
Sbjct: 122 VGIMMNDRKSRFSVNGKPIYHFMGTSTFSQYTVVHDVSVAKIDPTAPLDKVCLLGCGVPT 181
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVT 243
GLGAVWNTAKVEPGS VAIFGLGTVGLAVAEGAK+AGASRIIGIDIDS K++TAK FGV
Sbjct: 182 GLGAVWNTAKVEPGSNVAIFGLGTVGLAVAEGAKTAGASRIIGIDIDSKKYETAKKFGVN 241
Query: 244 EFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASG 303
EF+NPKDH+KPIQ+VI DLTDGGVDYSFECIGNVSVMR+ALEC HKGWGTSV+VGVAASG
Sbjct: 242 EFVNPKDHDKPIQEVIVDLTDGGVDYSFECIGNVSVMRAALECCHKGWGTSVIVGVAASG 301
Query: 304 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEA 363
QEISTRPFQLVTGRVWKGTAFGGFKSR+QVPWLVEKY+ KEIKVDEY+THNLTL EIN+A
Sbjct: 302 QEISTRPFQLVTGRVWKGTAFGGFKSRTQVPWLVEKYMNKEIKVDEYITHNLTLGEINKA 361
Query: 364 FDLMHEGKCLRCVL 377
FDL+HEG CLRCVL
Sbjct: 362 FDLLHEGTCLRCVL 375
>gb|AAB06322.1| glutathione-dependent formaldehyde dehydrogenase
Length = 379
Score = 697 bits (1800), Expect = 0.0
Identities = 332/374 (88%), Positives = 355/374 (94%)
Query: 4 STEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEG 63
+T+GQVITCKAAVA+EPNKPL IEDV+VAPPQA EVRI+IL+TALCHTDAYTWSGKDPEG
Sbjct: 2 ATQGQVITCKAAVAYEPNKPLVIEDVQVAPPQAGEVRIKILYTALCHTDAYTWSGKDPEG 61
Query: 64 LFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATG 123
LFPCILGHEAAGIVESVGEGVT V+ GDHVIPCYQAEC ECKFCKSGKTNLCGKVR+ATG
Sbjct: 62 LFPCILGHEAAGIVESVGEGVTGVQAGDHVIPCYQAECRECKFCKSGKTNLCGKVRSATG 121
Query: 124 VGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
VG+MMNDRK RFS+ GKPIYHFMGTSTFSQYTVVHDVSVAKI P A LDKVCLLGCGVPT
Sbjct: 122 VGIMMNDRKSRFSVNGKPIYHFMGTSTFSQYTVVHDVSVAKIDPTAPLDKVCLLGCGVPT 181
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVT 243
GLGAVWNTAKVEPGS VAIFGLGTVGLAVAEGAK+AGASRIIGIDIDS K++TAK FGV
Sbjct: 182 GLGAVWNTAKVEPGSNVAIFGLGTVGLAVAEGAKTAGASRIIGIDIDSKKYETAKKFGVN 241
Query: 244 EFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASG 303
EF+NPKDH+KPIQ+VI DLTDGGVDYSFECIGNVSVMR+ALEC HKGWGTSV+VGVAASG
Sbjct: 242 EFVNPKDHDKPIQEVIVDLTDGGVDYSFECIGNVSVMRAALECCHKGWGTSVIVGVAASG 301
Query: 304 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEA 363
QEISTRPFQLVTGRVWKGTAFGGFKSR+QVPWLVEKY+ KEIKVDEY+THNLTL EIN+A
Sbjct: 302 QEISTRPFQLVTGRVWKGTAFGGFKSRTQVPWLVEKYMNKEIKVDEYITHNLTLGEINKA 361
Query: 364 FDLMHEGKCLRCVL 377
FDL+HEG CLRCVL
Sbjct: 362 FDLLHEGTCLRCVL 375
>emb|CAA57973.1| class III ADH, glutathione-dependent formaldehyde dehydrogenase.
[Arabidopsis thaliana] gi|2129562|pir||S71244 alcohol
dehydrogenase (EC 1.1.1.1) class III - Arabidopsis
thaliana
Length = 379
Score = 697 bits (1799), Expect = 0.0
Identities = 331/374 (88%), Positives = 356/374 (94%)
Query: 4 STEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEG 63
+T+GQVITCKAAVA+EPNKPL IEDV+VAPPQA EVRI+IL+TALCHTDAYTWSGKDPEG
Sbjct: 2 ATQGQVITCKAAVAYEPNKPLVIEDVQVAPPQAGEVRIKILYTALCHTDAYTWSGKDPEG 61
Query: 64 LFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATG 123
LFPCILGHEAAGIVESVGEGVT+V+ GDHVIPCYQAEC ECKFCKSGKTNLCGKVR+ATG
Sbjct: 62 LFPCILGHEAAGIVESVGEGVTEVQAGDHVIPCYQAECRECKFCKSGKTNLCGKVRSATG 121
Query: 124 VGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
VG+MMNDRK RFS+ GKPIYHFMGTSTFSQYTVVHDVSVAKI P A LDKVCLLGCGVPT
Sbjct: 122 VGIMMNDRKSRFSVNGKPIYHFMGTSTFSQYTVVHDVSVAKIDPTAPLDKVCLLGCGVPT 181
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVT 243
GLGAVWNTAKVEPGS VAIFGLGTVGLAVAEGAK+AGASRIIGIDIDS K++TAK FGV
Sbjct: 182 GLGAVWNTAKVEPGSNVAIFGLGTVGLAVAEGAKTAGASRIIGIDIDSKKYETAKKFGVN 241
Query: 244 EFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASG 303
EF+NPKDH+KPIQ+VI DLTDGGVDYSFECIGNVSVMR+ALEC HKGWGTSV+VGVAASG
Sbjct: 242 EFVNPKDHDKPIQEVIVDLTDGGVDYSFECIGNVSVMRAALECCHKGWGTSVIVGVAASG 301
Query: 304 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEA 363
QEISTRPFQLVTGRVWKGTAFGGFKSR+QVPWLVEKY+ KEIKVDEY+THNL+L EIN+A
Sbjct: 302 QEISTRPFQLVTGRVWKGTAFGGFKSRTQVPWLVEKYMNKEIKVDEYITHNLSLGEINKA 361
Query: 364 FDLMHEGKCLRCVL 377
FDL+HEG CLRCVL
Sbjct: 362 FDLLHEGTCLRCVL 375
>sp|P79896|ADHX_SPAAU Alcohol dehydrogenase class III (Glutathione-dependent formaldehyde
dehydrogenase) (FDH) (FALDH) gi|1814386|gb|AAB41888.1|
alcohol dehydrogenase class III [Sparus aurata]
Length = 376
Score = 540 bits (1391), Expect = e-152
Identities = 257/373 (68%), Positives = 303/373 (80%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWEP KPL+IE+VEVAPP A+EVRI++ T +CHTDAYT SG DPEGL
Sbjct: 3 TAGKVIKCKAAVAWEPGKPLSIEEVEVAPPNAHEVRIKLFATGVCHTDAYTLSGSDPEGL 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AG VESVGEGVT KPGD VIP Y +CGECKFCK+ KTNLC K+R G
Sbjct: 63 FPVILGHEGAGTVESVGEGVTKFKPGDTVIPLYVPQCGECKFCKNPKTNLCQKIRITQGQ 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G ++ D+ RF+ KGK ++HFMGTSTFS+YTVV D+S+AK++ A +DKVCLLGCG+ TG
Sbjct: 123 G-LLPDKTSRFTCKGKQVFHFMGTSTFSEYTVVADISLAKVNEKAPMDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GA NTAKVEPGS A+FGLG VGLAV G K AGA+RIIGID++ KF+TAK FG TE
Sbjct: 182 YGAALNTAKVEPGSTCAVFGLGAVGLAVIMGCKVAGATRIIGIDLNPAKFETAKEFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
F+NPKDH KPIQ+V+ ++TDGGVDYSFECIGNV +MR+ALE HKGWG SV++GVA +GQ
Sbjct: 242 FVNPKDHSKPIQEVLVEMTDGGVDYSFECIGNVQIMRAALEACHKGWGESVIIGVAGAGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EISTRPFQLVTGRVWKGTAFGG+KS VP LVE Y+ K++KVDE+VTH L +INE F
Sbjct: 302 EISTRPFQLVTGRVWKGTAFGGWKSVESVPKLVEDYMSKKLKVDEFVTHTLPFEKINEGF 361
Query: 365 DLMHEGKCLRCVL 377
+LMH GK +R VL
Sbjct: 362 ELMHAGKSIRTVL 374
>gb|AAL26325.1| alcohol dehydrogenase [Danio rerio]
Length = 376
Score = 538 bits (1387), Expect = e-152
Identities = 259/373 (69%), Positives = 301/373 (80%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWE KPLTIE+VEVAPP+A+EVR++I T +CHTDAYT SG DPEGL
Sbjct: 3 TTGKVIKCKAAVAWEAGKPLTIEEVEVAPPKAHEVRVKIHATGVCHTDAYTLSGSDPEGL 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AG VESVGEGVT KPGD VIP Y +CGECKFCK+ KTNLC K+R G
Sbjct: 63 FPVILGHEGAGTVESVGEGVTKFKPGDTVIPLYVPQCGECKFCKNPKTNLCQKIRVTQGQ 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G +M D RF+ KGK ++HFMGTSTFS+YTVV ++S+AK+ A LDKVCLLGCG+ TG
Sbjct: 123 G-LMPDNTSRFTCKGKQLFHFMGTSTFSEYTVVAEISLAKVDEHAPLDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GA NTAKVE GS A+FGLG VGLAV G KSAGA+RIIGID++ +KF+ AK FG TE
Sbjct: 182 YGAAINTAKVEAGSTCAVFGLGAVGLAVVMGCKSAGATRIIGIDVNPDKFEIAKKFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
F+NPKDH KPIQ+V+ +LTDGGVDYSFECIGNV +MR+ALE HKGWGTSV++GVA +GQ
Sbjct: 242 FVNPKDHSKPIQEVLVELTDGGVDYSFECIGNVGIMRAALEACHKGWGTSVIIGVAGAGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EISTRPFQLVTGR WKGTAFGG+KS VP LV Y+ K++ VDE+VTH L A+INEAF
Sbjct: 302 EISTRPFQLVTGRTWKGTAFGGWKSVESVPKLVNDYMNKKLMVDEFVTHTLPFAQINEAF 361
Query: 365 DLMHEGKCLRCVL 377
DLMH GK +R VL
Sbjct: 362 DLMHAGKSIRAVL 374
>gb|AAS15570.1| class III alcohol dehydrogenase [Oryzias latipes]
Length = 379
Score = 538 bits (1386), Expect = e-151
Identities = 257/373 (68%), Positives = 301/373 (79%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWEP KPL+IE+VEVAPP+A+EVRI++ T +CHTDAYT SG DPEGL
Sbjct: 3 TAGKVIKCKAAVAWEPGKPLSIEEVEVAPPKAHEVRIKLFATGVCHTDAYTLSGSDPEGL 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AG VESVGEGVT KPGD VIP Y +CGECKFCK+ KTNLC K+R G
Sbjct: 63 FPVILGHEGAGTVESVGEGVTSFKPGDTVIPLYVPQCGECKFCKNPKTNLCQKIRVTQGQ 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G ++ D+ RF+ KGK I+HFMGTSTFS+YTVV D+S+AK++ A LDKVCLLGCG+ TG
Sbjct: 123 G-LLPDKTSRFTCKGKQIFHFMGTSTFSEYTVVADISLAKVNEKAPLDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GA NTAKVEPGS A+FGLG VGLA G K AGA+RIIG+DI+ KF+ AK FG TE
Sbjct: 182 YGAALNTAKVEPGSTCAVFGLGAVGLAAIMGCKVAGANRIIGVDINPEKFERAKEFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
F+NPKDH KPIQ V+ ++TDGGVDYSFECIGNV +MR+ALE HKGWG SV++GVA +GQ
Sbjct: 242 FVNPKDHSKPIQGVLVEMTDGGVDYSFECIGNVQIMRAALEACHKGWGQSVIIGVAGAGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EISTRPFQLVTGRVWKGTAFGG+KS VP LVE Y+ K++KVDE+VTH L +INE F
Sbjct: 302 EISTRPFQLVTGRVWKGTAFGGWKSVESVPRLVEDYMNKKLKVDEFVTHTLPFDQINEGF 361
Query: 365 DLMHEGKCLRCVL 377
+LMH GK +R VL
Sbjct: 362 ELMHAGKSIRTVL 374
>pir||JC7759 alcohol dehydrogenase (EC 1.1.1.1) 3 - zebra fish
Length = 376
Score = 538 bits (1385), Expect = e-151
Identities = 258/373 (69%), Positives = 301/373 (80%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWE KPLTIE+VEVAPP+A+EVR++I T +CHTDAYT SG DPEGL
Sbjct: 3 TTGKVIKCKAAVAWEAGKPLTIEEVEVAPPKAHEVRVKIHATGVCHTDAYTLSGSDPEGL 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AG VESVGEGVT KPGD VIP Y +CGECKFCK+ KTNLC K+R G
Sbjct: 63 FPVILGHEGAGTVESVGEGVTKFKPGDTVIPLYVPQCGECKFCKNPKTNLCQKIRVTQGQ 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G +M D RF+ KGK ++HFMGTSTFS+YTVV ++S+AK+ A LDKVCLLGCG+ TG
Sbjct: 123 G-LMPDNTSRFTCKGKQLFHFMGTSTFSEYTVVAEISLAKVDEHAPLDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GA NTAKVE GS A+FGLG VGLAV G KSAGA+RIIGID++ +KF+ AK FG TE
Sbjct: 182 YGAAINTAKVEAGSTCAVFGLGAVGLAVVMGCKSAGATRIIGIDVNPDKFEIAKKFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
F+NPKDH KPIQ+V+ +LTDGGVDYSFECIGNV +MR+A+E HKGWGTSV++GVA +GQ
Sbjct: 242 FVNPKDHSKPIQEVLVELTDGGVDYSFECIGNVGIMRAAIEACHKGWGTSVIIGVAGAGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EISTRPFQLVTGR WKGTAFGG+KS VP LV Y+ K++ VDE+VTH L A+INEAF
Sbjct: 302 EISTRPFQLVTGRTWKGTAFGGWKSVESVPKLVNDYMNKKLMVDEFVTHTLPFAQINEAF 361
Query: 365 DLMHEGKCLRCVL 377
DLMH GK +R VL
Sbjct: 362 DLMHAGKSIRAVL 374
>gb|AAH67170.1| Alcohol dehydrogenase 5 [Danio rerio] gi|46195771|ref|NP_571924.2|
alcohol dehydrogenase 5 [Danio rerio]
Length = 376
Score = 537 bits (1383), Expect = e-151
Identities = 258/373 (69%), Positives = 301/373 (80%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWE KPL+IE+VEVAPP+A+EVR++I T +CHTDAYT SG DPEGL
Sbjct: 3 TTGKVIKCKAAVAWEAGKPLSIEEVEVAPPKAHEVRVKIHATGVCHTDAYTLSGSDPEGL 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AG VESVGEGVT KPGD VIP Y +CGECKFCK+ KTNLC K+R G
Sbjct: 63 FPVILGHEGAGTVESVGEGVTKFKPGDTVIPLYVPQCGECKFCKNPKTNLCQKIRVTQGQ 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G +M D RF+ KGK ++HFMGTSTFS+YTVV ++S+AK+ A LDKVCLLGCG+ TG
Sbjct: 123 G-LMPDNTSRFTCKGKQLFHFMGTSTFSEYTVVAEISLAKVDEHAPLDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GA NTAKVE GS A+FGLG VGLAV G KSAGA+RIIGID++ +KF+ AK FG TE
Sbjct: 182 YGAAINTAKVEAGSTCAVFGLGAVGLAVVMGCKSAGATRIIGIDVNPDKFEIAKKFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
F+NPKDH KPIQ+V+ +LTDGGVDYSFECIGNV +MR+ALE HKGWGTSV++GVA +GQ
Sbjct: 242 FVNPKDHSKPIQEVLVELTDGGVDYSFECIGNVGIMRAALEACHKGWGTSVIIGVAGAGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EISTRPFQLVTGR WKGTAFGG+KS VP LV Y+ K++ VDE+VTH L A+INEAF
Sbjct: 302 EISTRPFQLVTGRTWKGTAFGGWKSVESVPKLVNDYMNKKLMVDEFVTHTLPFAQINEAF 361
Query: 365 DLMHEGKCLRCVL 377
DLMH GK +R VL
Sbjct: 362 DLMHAGKSIRAVL 374
>ref|XP_393266.1| PREDICTED: similar to Alcohol dehydrogenase 5 [Apis mellifera]
Length = 377
Score = 532 bits (1371), Expect = e-150
Identities = 254/375 (67%), Positives = 305/375 (80%), Gaps = 1/375 (0%)
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPE 62
S+T G++I CKAAVAW+ +PL++E++EVAPP+A+EVRI+I+F ALCHTDAYT G DPE
Sbjct: 2 SNTAGKIIKCKAAVAWKEKEPLSLEEIEVAPPKAHEVRIKIIFVALCHTDAYTLDGLDPE 61
Query: 63 GLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAAT 122
G+FPCILGHE +GIVESVGE VT+ +PGDHVIP Y +CG+CKFCKS KTNLC K+R
Sbjct: 62 GIFPCILGHEGSGIVESVGEEVTEFQPGDHVIPLYVPQCGDCKFCKSPKTNLCSKIRTTQ 121
Query: 123 GVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVP 182
G GVM D RF+ KG+ + HFMG STFS+YTVV D+S+AKI P A LDKV LLGCGV
Sbjct: 122 GKGVMP-DGTSRFTCKGQTLAHFMGCSTFSEYTVVADISLAKIDPTAPLDKVSLLGCGVS 180
Query: 183 TGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGV 242
TG GA NTAKVE GS AI+GLG VGLAVA G K AGA RIIGID++ NK + AK FG
Sbjct: 181 TGYGAALNTAKVESGSTCAIWGLGAVGLAVAFGCKKAGAKRIIGIDVNPNKSEVAKKFGC 240
Query: 243 TEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAAS 302
TEFINPKD+ KPIQ+V+ +LTDGG+DY+FEC+GNV+ MR+ALE HKGWGTSV+VGVAA+
Sbjct: 241 TEFINPKDYNKPIQEVLIELTDGGLDYTFECVGNVNTMRAALESCHKGWGTSVIVGVAAA 300
Query: 303 GQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINE 362
GQEISTRPFQLVTGRVWKGTAFGG+KS+ VP LV++Y+ K + +DE+VTH L +INE
Sbjct: 301 GQEISTRPFQLVTGRVWKGTAFGGWKSKESVPKLVKEYMSKSLILDEFVTHTLPFEKINE 360
Query: 363 AFDLMHEGKCLRCVL 377
FDL+H G CLR VL
Sbjct: 361 GFDLLHSGNCLRAVL 375
>gb|AAS49608.1| alcohol dehydrogenase 5 [Xenopus laevis]
Length = 376
Score = 531 bits (1367), Expect = e-149
Identities = 264/373 (70%), Positives = 300/373 (79%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWE KPL++E+VEVAPP+A+EVRI+I+ TA+CHTDAYT SG DPEG
Sbjct: 3 TAGKVIKCKAAVAWEAGKPLSMEEVEVAPPKAHEVRIKIVSTAVCHTDAYTLSGADPEGC 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AGIVESVGEGVT VKPGD VIP Y +CGECKFC + KTNLC K+R G
Sbjct: 63 FPVILGHEGAGIVESVGEGVTRVKPGDKVIPLYIPQCGECKFCLNPKTNLCQKIRITQGK 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G M D RF+ KG+ I+HFMGTSTFS+YTVV D+SVAKI A LDKVCLLGCG+ TG
Sbjct: 123 G-FMPDGTSRFTCKGQQIFHFMGTSTFSEYTVVADISVAKIEDSAPLDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GAV NTAKVEPGS A+FGLG VGLAV G K AGA+RIIGID++ +KF A FG TE
Sbjct: 182 YGAVINTAKVEPGSTCAVFGLGGVGLAVIMGCKVAGATRIIGIDLNKDKFAKATEFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
+NP D +KPIQ V+ D+TDGGVDYSFECIGNV VMRSALE HKGWGTSV+VGVAASGQ
Sbjct: 242 CLNPADFDKPIQDVLIDMTDGGVDYSFECIGNVRVMRSALEACHKGWGTSVIVGVAASGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EI+TRPFQLVTGRVWKGTAFGG+KS VP LV +Y+ K+IKVDE+VTH L INEAF
Sbjct: 302 EIATRPFQLVTGRVWKGTAFGGWKSVDSVPKLVSEYMTKKIKVDEFVTHTLPFDSINEAF 361
Query: 365 DLMHEGKCLRCVL 377
+LMH GK +R VL
Sbjct: 362 ELMHAGKSIRSVL 374
>emb|CAG31862.1| hypothetical protein [Gallus gallus]
Length = 374
Score = 530 bits (1366), Expect = e-149
Identities = 263/369 (71%), Positives = 298/369 (80%), Gaps = 1/369 (0%)
Query: 9 VITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCI 68
VI CKAAVAWE KPL+IE+VEVAPP+A+EVRI+I+ TALCHTDAYT SG DPEG FP I
Sbjct: 5 VIKCKAAVAWEAGKPLSIEEVEVAPPKAHEVRIKIVATALCHTDAYTLSGADPEGCFPVI 64
Query: 69 LGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGVMM 128
LGHE AGIVESVGEGVT VKPGD VIP Y +CGECK+CK+ KTNLC K+R G G +M
Sbjct: 65 LGHEGAGIVESVGEGVTKVKPGDTVIPLYIPQCGECKYCKNPKTNLCQKIRVTQGKG-LM 123
Query: 129 NDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGAV 188
D RF+ KGK IYHFMGTSTFS+YTVV D+SVAKI P A DKVCLLGCGV TG GA
Sbjct: 124 PDGTIRFTCKGKQIYHFMGTSTFSEYTVVADISVAKIDPAAPFDKVCLLGCGVSTGYGAA 183
Query: 189 WNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFINP 248
NTAKVEPGS A+FGLG VGLA G K+AGASRIIGIDI+ N + AK FG E I+P
Sbjct: 184 VNTAKVEPGSTCAVFGLGGVGLATVMGCKAAGASRIIGIDINKNTYAKAKEFGAAECISP 243
Query: 249 KDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEIST 308
+D EKPIQ+V+ ++TDGGVDYSFECIGNV VMR+ALE HKGWG SV+VGVAA+GQEIST
Sbjct: 244 QDFEKPIQEVLVEMTDGGVDYSFECIGNVGVMRAALEACHKGWGVSVIVGVAAAGQEIST 303
Query: 309 RPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMH 368
RPFQLVTGR WKGTAFGG+KS VP LV Y+ K+IKVDE+VTH L +INEAFDL+H
Sbjct: 304 RPFQLVTGRTWKGTAFGGWKSVDSVPKLVNDYMAKKIKVDEFVTHTLPFDKINEAFDLLH 363
Query: 369 EGKCLRCVL 377
+GK +R VL
Sbjct: 364 KGKSIRTVL 372
>ref|XP_420657.1| PREDICTED: similar to Alcohol dehydrogenase class III
(Glutathione-dependent formaldehyde dehydrogenase) (FDH)
[Gallus gallus]
Length = 484
Score = 530 bits (1366), Expect = e-149
Identities = 263/375 (70%), Positives = 300/375 (79%), Gaps = 1/375 (0%)
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPE 62
S+ VI CKAAVAWE KPL+IE+VEVAPP+A+EVRI+I+ TALCHTDAYT SG DPE
Sbjct: 109 SAMASGVIKCKAAVAWEAGKPLSIEEVEVAPPKAHEVRIKIVATALCHTDAYTLSGADPE 168
Query: 63 GLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAAT 122
G FP ILGHE AGIVESVGEGVT VKPGD VIP Y +CGECK+CK+ KTNLC K+R
Sbjct: 169 GCFPVILGHEGAGIVESVGEGVTKVKPGDTVIPLYIPQCGECKYCKNPKTNLCQKIRVTQ 228
Query: 123 GVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVP 182
G G +M D RF+ KGK IYHFMGTSTFS+YTVV D+SVAKI P A DKVCLLGCG+
Sbjct: 229 GKG-LMPDGTIRFTCKGKQIYHFMGTSTFSEYTVVADISVAKIDPAAPFDKVCLLGCGIS 287
Query: 183 TGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGV 242
TG GA NTAKVEPGS A+FGLG VGLA G K+AGASRIIGIDI+ N + AK FG
Sbjct: 288 TGYGAAVNTAKVEPGSTCAVFGLGGVGLATVMGCKAAGASRIIGIDINKNTYAKAKEFGA 347
Query: 243 TEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAAS 302
E I+P+D EKPIQ+V+ ++TDGGVDYSFECIGNV VMR+ALE HKGWG SV+VGVAA+
Sbjct: 348 AECISPQDFEKPIQEVLVEMTDGGVDYSFECIGNVGVMRAALEACHKGWGVSVIVGVAAA 407
Query: 303 GQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINE 362
GQEISTRPFQLVTGR WKGTAFGG+KS VP LV Y+ K+IKVDE+VTH L +INE
Sbjct: 408 GQEISTRPFQLVTGRTWKGTAFGGWKSVDSVPKLVNDYMAKKIKVDEFVTHTLPFDKINE 467
Query: 363 AFDLMHEGKCLRCVL 377
AFDL+H+GK +R VL
Sbjct: 468 AFDLLHKGKSIRTVL 482
>gb|AAH97612.1| LOC445841 protein [Xenopus laevis]
Length = 376
Score = 530 bits (1365), Expect = e-149
Identities = 264/373 (70%), Positives = 299/373 (79%), Gaps = 1/373 (0%)
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
T G+VI CKAAVAWE KPL++E+VEVAPP+A+EVRI+I+ TA+CHTDAYT SG DPEG
Sbjct: 3 TAGKVIKCKAAVAWEAGKPLSMEEVEVAPPKAHEVRIKIVSTAVCHTDAYTLSGADPEGC 62
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHE AGIVESVGEGVT VKPGD VIP Y +CGECKFC + KTNLC K+R G
Sbjct: 63 FPVILGHEGAGIVESVGEGVTRVKPGDKVIPLYIPQCGECKFCLNPKTNLCQKIRITQGK 122
Query: 125 GVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
G M D RF+ KG+ I+HFMGTSTFS+YTVV D+SVAKI A LDKVCLLGCG+ TG
Sbjct: 123 G-FMPDGTSRFTCKGQQIFHFMGTSTFSEYTVVADISVAKIEDSAPLDKVCLLGCGISTG 181
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GAV NTAKVEPGS A+FGLG VGLAV G K AGA+RIIGID++ +KF A FG TE
Sbjct: 182 YGAVINTAKVEPGSTCAVFGLGGVGLAVIMGCKVAGATRIIGIDLNKDKFAKATEFGATE 241
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
+NP D KPIQ V+ D+TDGGVDYSFECIGNV VMRSALE HKGWGTSV+VGVAASGQ
Sbjct: 242 CLNPADFNKPIQDVLIDMTDGGVDYSFECIGNVRVMRSALEACHKGWGTSVIVGVAASGQ 301
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
EI+TRPFQLVTGRVWKGTAFGG+KS VP LV +Y+ K+IKVDE+VTH L INEAF
Sbjct: 302 EIATRPFQLVTGRVWKGTAFGGWKSVDSVPKLVSEYMAKKIKVDEFVTHTLPFDSINEAF 361
Query: 365 DLMHEGKCLRCVL 377
+LMH GK +R VL
Sbjct: 362 ELMHAGKSIRSVL 374
>sp|P12711|ADHX_RAT Alcohol dehydrogenase class III (Alcohol dehydrogenase 2)
(Glutathione-dependent formaldehyde dehydrogenase) (FDH)
(FALDH) (Alcohol dehydrogenase-B2)
Length = 373
Score = 530 bits (1364), Expect = e-149
Identities = 264/372 (70%), Positives = 299/372 (79%), Gaps = 1/372 (0%)
Query: 8 QVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPC 67
QVI CKAAVAWE KPL+IE++EVAPPQA+EVRI+I+ TA+CHTDAYT SG DPEG FP
Sbjct: 3 QVIRCKAAVAWEAGKPLSIEEIEVAPPQAHEVRIKIIATAVCHTDAYTLSGADPEGCFPV 62
Query: 68 ILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGVM 127
ILGHE AGIVESVGEGVT +K GD VIP Y +CGECKFC + KTNLC K+R G G +
Sbjct: 63 ILGHEGAGIVESVGEGVTKLKAGDTVIPLYIPQCGECKFCLNPKTNLCQKIRVTQGKG-L 121
Query: 128 MNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGA 187
M D RF+ KGKPI HFMGTSTFS+YTVV D+SVAKI P A LDKVCLLGCG+ TG GA
Sbjct: 122 MPDGTSRFTCKGKPILHFMGTSTFSEYTVVADISVAKIDPSAPLDKVCLLGCGISTGYGA 181
Query: 188 VWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFIN 247
NTAKVEPGS A+FGLG VGLAV G K AGASRIIGIDI+ +KF AK FG TE IN
Sbjct: 182 AVNTAKVEPGSTCAVFGLGGVGLAVIMGCKVAGASRIIGIDINKDKFAKAKEFGATECIN 241
Query: 248 PKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEIS 307
P+D K IQ+V+ ++TDGGVD+SFECIGNV VMRSALE HKGWG SVVVGVAASG+EIS
Sbjct: 242 PQDFSKSIQEVLIEMTDGGVDFSFECIGNVKVMRSALEAAHKGWGVSVVVGVAASGEEIS 301
Query: 308 TRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLM 367
TRPFQLVTGR WKGTAFGG+KS VP LV +Y+ K+IKVDE+VT NL+ +IN+AFDLM
Sbjct: 302 TRPFQLVTGRTWKGTAFGGWKSVESVPKLVSEYMSKKIKVDEFVTGNLSFDQINKAFDLM 361
Query: 368 HEGKCLRCVLAL 379
H G +R VL L
Sbjct: 362 HSGNSIRTVLKL 373
>gb|AAH83724.1| Unknown (protein for IMAGE:7191109) [Rattus norvegicus]
Length = 379
Score = 529 bits (1362), Expect = e-149
Identities = 263/372 (70%), Positives = 299/372 (79%), Gaps = 1/372 (0%)
Query: 8 QVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPC 67
QVI CKAAVAWE KPL+IE++EVAPPQA+EVRI+I+ TA+CHTDAYT SG DPEG FP
Sbjct: 9 QVIRCKAAVAWEAGKPLSIEEIEVAPPQAHEVRIKIIATAVCHTDAYTLSGADPEGCFPV 68
Query: 68 ILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGVM 127
ILGHE AGIVESVGEGVT +K GD VIP Y +CGECKFC + KTNLC K+R G G +
Sbjct: 69 ILGHEGAGIVESVGEGVTKLKAGDTVIPLYIPQCGECKFCLNPKTNLCQKIRVTQGKG-L 127
Query: 128 MNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGA 187
M D RF+ KGKPI HFMGTSTFS+YTVV D+SVAKI P A LDKVCLLGCG+ TG GA
Sbjct: 128 MPDGTSRFTCKGKPILHFMGTSTFSEYTVVADISVAKIDPSAPLDKVCLLGCGISTGYGA 187
Query: 188 VWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFIN 247
NTAKVEPGS A+FGLG VGLAV G K AGASRIIGIDI+ +KF AK FG TE IN
Sbjct: 188 AVNTAKVEPGSTCAVFGLGGVGLAVIMGCKVAGASRIIGIDINKDKFAKAKEFGATECIN 247
Query: 248 PKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEIS 307
P+D K IQ+V+ ++TDGGVD+SFECIGNV VMRSALE HKGWG SVVVGVAASG+EIS
Sbjct: 248 PQDFSKSIQEVLIEMTDGGVDFSFECIGNVKVMRSALEAAHKGWGVSVVVGVAASGEEIS 307
Query: 308 TRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLM 367
TRPFQLVTGR WKGTAFGG+KS VP LV +Y+ K+IKVDE+VT NL+ +IN+AFDLM
Sbjct: 308 TRPFQLVTGRTWKGTAFGGWKSVESVPKLVSEYMSKKIKVDEFVTGNLSFDQINKAFDLM 367
Query: 368 HEGKCLRCVLAL 379
H G +R VL +
Sbjct: 368 HSGNSIRTVLKM 379
>sp|P81600|ADHH_GADMO Alcohol dehydrogenase class III H chain (Glutathione-dependent
formaldehyde dehydrogenase) (FDH)
Length = 375
Score = 527 bits (1357), Expect = e-148
Identities = 251/374 (67%), Positives = 295/374 (78%), Gaps = 1/374 (0%)
Query: 4 STEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEG 63
+T GQVI CKAAVAWE KPL++E+VEVAPP+A EVRI+++ T +CHTDAYT SG DPEG
Sbjct: 1 ATAGQVIKCKAAVAWEAGKPLSLEEVEVAPPRAGEVRIKVVATGVCHTDAYTLSGSDPEG 60
Query: 64 LFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATG 123
FP ILGHE AG+VESVGEGVT K GD VIP Y +CGECKFCK+ KTNLC K+R G
Sbjct: 61 AFPVILGHEGAGLVESVGEGVTKFKAGDTVIPLYVPQCGECKFCKNPKTNLCQKIRVTQG 120
Query: 124 VGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
G +M D RF+ KGK ++HFMGTSTFS+YTVV D+S+A + P A LDKVCLLGCG+ T
Sbjct: 121 RG-LMPDNTSRFTCKGKQLFHFMGTSTFSEYTVVADISLANVDPKAPLDKVCLLGCGIST 179
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVT 243
G GA NTAKVEPGS A+FGLG VGLA G K AGA+RIIG+DI+ KF A FG T
Sbjct: 180 GYGAALNTAKVEPGSTCAVFGLGAVGLAAIMGCKVAGATRIIGVDINPEKFGKAAEFGAT 239
Query: 244 EFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASG 303
E +NPKDH +P+Q+V+ ++TDGGVDYSFECIGNV +MRSALE HKGWG SV++GVA +G
Sbjct: 240 ECLNPKDHARPVQEVLVEMTDGGVDYSFECIGNVEIMRSALEACHKGWGESVIIGVAGAG 299
Query: 304 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEA 363
QEI+TRPFQLVTGRVWK TAFGG+KS VP LVE Y+ K++KVDE+VTH L INE
Sbjct: 300 QEIATRPFQLVTGRVWKATAFGGWKSVESVPKLVEDYMNKKLKVDEFVTHTLPFDSINEG 359
Query: 364 FDLMHEGKCLRCVL 377
FDLMH GK +RCVL
Sbjct: 360 FDLMHAGKSIRCVL 373
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 656,756,713
Number of Sequences: 2540612
Number of extensions: 27874704
Number of successful extensions: 76813
Number of sequences better than 10.0: 3507
Number of HSP's better than 10.0 without gapping: 2761
Number of HSP's successfully gapped in prelim test: 746
Number of HSP's that attempted gapping in prelim test: 69993
Number of HSP's gapped (non-prelim): 4159
length of query: 380
length of database: 863,360,394
effective HSP length: 130
effective length of query: 250
effective length of database: 533,080,834
effective search space: 133270208500
effective search space used: 133270208500
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146819.3


