

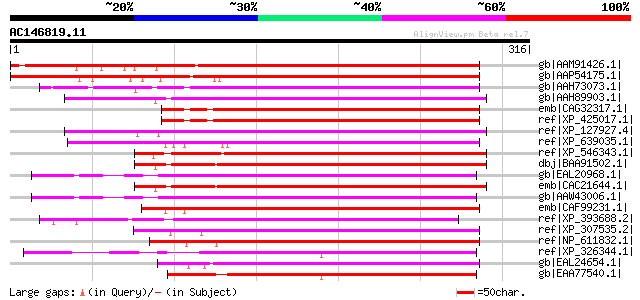
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.11 - phase: 0
(316 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91426.1| At2g20330/F11A3.12 [Arabidopsis thaliana] gi|4512... 376 e-103
gb|AAP54175.1| putative WD domain containing protein [Oryza sati... 346 4e-94
gb|AAH73073.1| MGC82743 protein [Xenopus laevis] 199 1e-49
gb|AAH89903.1| Hypothetical LOC294783 [Rattus norvegicus] gi|620... 193 5e-48
emb|CAG32317.1| hypothetical protein [Gallus gallus] 193 5e-48
ref|XP_425017.1| PREDICTED: similar to hypothetical protein FLJ1... 193 5e-48
ref|XP_127927.4| PREDICTED: similar to RGD1309487_predicted prot... 190 5e-47
ref|XP_639035.1| hypothetical protein DDB0218488 [Dictyostelium ... 187 3e-46
ref|XP_546343.1| PREDICTED: similar to hypothetical protein FLJ1... 186 8e-46
dbj|BAA91502.1| unnamed protein product [Homo sapiens] gi|192637... 185 2e-45
gb|EAL20968.1| hypothetical protein CNBD5690 [Cryptococcus neofo... 185 2e-45
emb|CAC21644.1| hypothetical protein [Homo sapiens] 185 2e-45
gb|AAW43006.1| transcription factor, putative [Cryptococcus neof... 183 5e-45
emb|CAF99231.1| unnamed protein product [Tetraodon nigroviridis] 180 6e-44
ref|XP_393688.2| PREDICTED: similar to CG5543-PA [Apis mellifera] 174 2e-42
ref|XP_307535.2| ENSANGP00000015367 [Anopheles gambiae str. PEST... 168 2e-40
ref|NP_611832.1| CG5543-PA [Drosophila melanogaster] gi|21064283... 167 4e-40
ref|XP_326344.1| hypothetical protein [Neurospora crassa] gi|289... 164 4e-39
gb|EAL24654.1| GA18958-PA [Drosophila pseudoobscura] 157 4e-37
gb|EAA77540.1| hypothetical protein FG07307.1 [Gibberella zeae P... 156 9e-37
>gb|AAM91426.1| At2g20330/F11A3.12 [Arabidopsis thaliana] gi|4512702|gb|AAD21755.1|
putative WD-40 repeat protein [Arabidopsis thaliana]
gi|13605895|gb|AAK32933.1| At2g20330/F11A3.12
[Arabidopsis thaliana] gi|16226753|gb|AAL16252.1|
At2g20330/F11A3.12 [Arabidopsis thaliana]
gi|25411973|pir||H84587 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana
gi|15225344|ref|NP_179623.1| transducin family protein /
WD-40 repeat family protein [Arabidopsis thaliana]
Length = 648
Score = 376 bits (966), Expect = e-103
Identities = 198/323 (61%), Positives = 232/323 (71%), Gaps = 41/323 (12%)
Query: 1 MEDSGEIYDGVRAEFPLSFGKQSKSQTPLETIHKATRRS-----------VPTSNPKTTS 49
MED E G+RA FP+SFGK SK E IH ATRR+ + T P +
Sbjct: 1 MEDELE---GLRAHFPVSFGKTSKVSASTEAIHSATRRTDVANDAGVSSDITTKKPDSDK 57
Query: 50 DDFPS-----KECLHSIRPP-KNPN--------PPSPPL---------QDDSPLVGPPPP 86
FPS + L S+R P +NPN PP PP +D+ ++GPPPP
Sbjct: 58 SGFPSLSSSSQTWLRSVRGPNRNPNSSGDVAMGPPPPPSSKRTQEEEEEDEEVMMGPPPP 117
Query: 87 PP---HHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRIPLSNEIVLKGH 143
P + DD+G+MIGPPPPPP + +D ED D+D D D NR++IPLSNEI LKGH
Sbjct: 118 PKGLGNINDSDDEGDMIGPPPPPPAARDSD-EDSDDDDDNDNEENRYQIPLSNEIQLKGH 176
Query: 144 TKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTA 203
TK+VS+LAVD G+RVLSGSYDY VRMYDFQGMNSRLQSFRQIEPSEGHQ+R++SWSPT+
Sbjct: 177 TKIVSSLAVDSAGARVLSGSYDYTVRMYDFQGMNSRLQSFRQIEPSEGHQVRSVSWSPTS 236
Query: 204 DRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILT 263
+FLCVTGSAQAKI+DRDGLTLGEF+KGDMYIRDLKNTKGHI GLTCGEWHP+TKET+LT
Sbjct: 237 GQFLCVTGSAQAKIFDRDGLTLGEFMKGDMYIRDLKNTKGHICGLTCGEWHPRTKETVLT 296
Query: 264 SSEDGSLRIWDVNDFTTQKQVWK 286
SSEDGSLRIWDVN+F +Q QV K
Sbjct: 297 SSEDGSLRIWDVNNFLSQTQVIK 319
Score = 33.9 bits (76), Expect = 6.4
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 141 KGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWS 200
K HT ++++ G +LS S+D ++++D + M L+ F + P+ Q N+++S
Sbjct: 370 KAHTDDITSVKFSSDGRILLSRSFDGSLKVWDLRQMKEALKVFEGL-PNYYPQ-TNVAFS 427
Query: 201 PTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKET 260
P D + +TG++ K GL L + + + I + + C WHP+ +
Sbjct: 428 P--DEQIILTGTSVEKDSTTGGL-LCFYDRTKLEIVQKVGISPTSSVVQCA-WHPRLNQI 483
Query: 261 ILTSSE 266
TS +
Sbjct: 484 FATSGD 489
>gb|AAP54175.1| putative WD domain containing protein [Oryza sativa (japonica
cultivar-group)] gi|37535172|ref|NP_921888.1| putative
WD domain containing protein [Oryza sativa (japonica
cultivar-group)] gi|22758329|gb|AAN05533.1| putative WD
domain containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 646
Score = 346 bits (888), Expect = 4e-94
Identities = 191/321 (59%), Positives = 228/321 (70%), Gaps = 36/321 (11%)
Query: 1 MEDSGEI-YDGVRAEFPLSFGKQSKSQTPLETIHKATRRSVP---TSNPKTTS------- 49
M D GE+ + +RA FP+SFGK + H +T R P ++ P T+S
Sbjct: 1 MADGGEMDEEAMRAFFPMSFGKAPTRAGAAASAHASTLRKPPQNPSAKPSTSSAAAAAAA 60
Query: 50 -----DDFPSKECLHSIRPPKNPNPPSP--PLQDDSP----LVGPPPPPPHH----EADD 94
DD + + PP+ P P+ +DD ++GPP PPP E +D
Sbjct: 61 GDDDDDDDDDDDEGPMVGPPRPPPQPAGGGEGEDDEEGGGVMIGPPRPPPRSSSRGEGED 120
Query: 95 DDGEMIGPPPPPPGSNFNDSEDEDNDSDQ-----DEL---GNRF-RIPLSNEIVLKGHTK 145
DG MIGPP PPP + +D EDED+D D DE+ G R+ RIPLSNE+VL+GHTK
Sbjct: 121 ADGGMIGPPRPPPVKD-DDEEDEDDDDDDGDDSDDEMEDDGERYNRIPLSNEVVLRGHTK 179
Query: 146 VVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADR 205
VVSALAVDHTGSRVLSGSYDY V MYDFQGMNS+LQSFRQ+EP EGHQ+R+LSWSPT+DR
Sbjct: 180 VVSALAVDHTGSRVLSGSYDYTVCMYDFQGMNSKLQSFRQLEPFEGHQVRSLSWSPTSDR 239
Query: 206 FLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSS 265
FLCVTGSAQAKIYDRDGLTLGEF+KGDMYIRDLKNTKGHI+GLT GEW+PK+KETILTSS
Sbjct: 240 FLCVTGSAQAKIYDRDGLTLGEFIKGDMYIRDLKNTKGHISGLTGGEWNPKSKETILTSS 299
Query: 266 EDGSLRIWDVNDFTTQKQVWK 286
EDGS+R+WDV+DF +QKQV K
Sbjct: 300 EDGSIRLWDVSDFKSQKQVIK 320
>gb|AAH73073.1| MGC82743 protein [Xenopus laevis]
Length = 610
Score = 199 bits (505), Expect = 1e-49
Identities = 116/283 (40%), Positives = 165/283 (57%), Gaps = 23/283 (8%)
Query: 19 FGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQ--- 75
FGK++++ LE + + TRR+ + +T + KE S + P PS Q
Sbjct: 6 FGKKART-FDLEAMFEQTRRTAVERSKQTL--EAREKEEQLSNKSPVIKGAPSSSGQKKT 62
Query: 76 ------------DDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQ 123
D L+GPP PP + D E+IGPP PP + DS+DED++ +
Sbjct: 63 KASGSSSGSEDSSDDELIGPPLPP--NVTGDHGDELIGPPLPP---GYKDSDDEDDEEHE 117
Query: 124 DELGNRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSF 183
D+ IP S+EI L+ TK VSAL +D +G+R+++G YDY VR +DF GM++ LQ+F
Sbjct: 118 DDDNPVKDIPDSHEITLQHGTKTVSALGLDPSGARLVTGGYDYDVRFWDFAGMDASLQAF 177
Query: 184 RQIEPSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKG 243
R ++P E HQI++L +S T D L V G++QAK+ DRDG + E VKGD YI D+ NTKG
Sbjct: 178 RSLQPCECHQIKSLQYSNTGDVILVVAGNSQAKVLDRDGFPVMECVKGDQYIVDMANTKG 237
Query: 244 HITGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
H L G WHPK KE +T S DG++R WDV++ K ++K
Sbjct: 238 HTAMLNGGCWHPKIKEEFMTCSNDGTVRTWDVSNEKKHKGIFK 280
>gb|AAH89903.1| Hypothetical LOC294783 [Rattus norvegicus]
gi|62078549|ref|NP_001013931.1| hypothetical protein
LOC294783 [Rattus norvegicus]
Length = 655
Score = 193 bits (491), Expect = 5e-48
Identities = 103/262 (39%), Positives = 151/262 (57%), Gaps = 8/262 (3%)
Query: 34 KATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPP-----P 88
K R+ + P +S + S R + + D L+GPP PP P
Sbjct: 71 KELRKQLEDIEPTPSSSSAVRERSKSSSRDTSSSDSDHSSGSSDDELIGPPLPPEMVGGP 130
Query: 89 HHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRIPLSNEIVLKGHTKVVS 148
+ D+D ++GP PPP D +D+D + + +E RIP S+EI LK TK VS
Sbjct: 131 VNTVDED---ILGPLPPPLCEEGEDDDDDDLEDEGEEDNPIHRIPDSHEITLKHGTKTVS 187
Query: 149 ALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLC 208
AL +D +G+R+++G YDY V+ +DF GM++ ++FR ++P E HQI++L +S T D L
Sbjct: 188 ALGLDPSGARLVTGGYDYDVKFWDFAGMDASFKAFRSLQPCECHQIKSLQYSNTGDMILV 247
Query: 209 VTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDG 268
V+GS+QAK+ DRDG + E +KGD YI D+ NTKGH L G WHPK K +T S D
Sbjct: 248 VSGSSQAKVIDRDGFEVMECIKGDQYIVDMANTKGHTAMLHTGSWHPKIKGEFMTCSNDA 307
Query: 269 SLRIWDVNDFTTQKQVWKIFTL 290
++R+W+V + QK V+K T+
Sbjct: 308 TVRLWEVENPKKQKSVFKPRTM 329
>emb|CAG32317.1| hypothetical protein [Gallus gallus]
Length = 387
Score = 193 bits (491), Expect = 5e-48
Identities = 99/195 (50%), Positives = 132/195 (66%), Gaps = 7/195 (3%)
Query: 93 DDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFR-IPLSNEIVLKGHTKVVSALA 151
DDDD E IGPP PP F DS+D+D + DE N + IP S+EI L+ TK VSAL
Sbjct: 127 DDDDEEFIGPPLPP---GFKDSDDDDTE---DEDNNPIKKIPDSHEITLQHGTKTVSALG 180
Query: 152 VDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTG 211
+D +G+R+++G YDY V+ +DF GM++ LQ+FR ++P E HQI++L +S T D L V+G
Sbjct: 181 LDPSGARLITGGYDYDVKFWDFAGMDASLQAFRSLQPCECHQIKSLQYSSTGDVILVVSG 240
Query: 212 SAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLR 271
++QAK+ DRDG + E +KGD YI D+ NTKGH L G WHPK KE LT S DG++R
Sbjct: 241 NSQAKVLDRDGFPVMECIKGDQYIVDMTNTKGHTAMLNSGCWHPKIKEEFLTCSNDGTVR 300
Query: 272 IWDVNDFTTQKQVWK 286
WDVN+ K V+K
Sbjct: 301 TWDVNNEKKHKSVFK 315
>ref|XP_425017.1| PREDICTED: similar to hypothetical protein FLJ10233 [Gallus gallus]
Length = 644
Score = 193 bits (491), Expect = 5e-48
Identities = 99/195 (50%), Positives = 132/195 (66%), Gaps = 7/195 (3%)
Query: 93 DDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFR-IPLSNEIVLKGHTKVVSALA 151
DDDD E IGPP PP F DS+D+D + DE N + IP S+EI L+ TK VSAL
Sbjct: 127 DDDDEEFIGPPLPP---GFKDSDDDDTE---DEDNNPIKKIPDSHEITLQHGTKTVSALG 180
Query: 152 VDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTG 211
+D +G+R+++G YDY V+ +DF GM++ LQ+FR ++P E HQI++L +S T D L V+G
Sbjct: 181 LDPSGARLITGGYDYDVKFWDFAGMDASLQAFRSLQPCECHQIKSLQYSSTGDVILVVSG 240
Query: 212 SAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGSLR 271
++QAK+ DRDG + E +KGD YI D+ NTKGH L G WHPK KE LT S DG++R
Sbjct: 241 NSQAKVLDRDGFPVMECIKGDQYIVDMTNTKGHTAMLNSGCWHPKIKEEFLTCSNDGTVR 300
Query: 272 IWDVNDFTTQKQVWK 286
WDVN+ K V+K
Sbjct: 301 TWDVNNEKKHKSVFK 315
>ref|XP_127927.4| PREDICTED: similar to RGD1309487_predicted protein [Mus musculus]
Length = 722
Score = 190 bits (482), Expect = 5e-47
Identities = 102/261 (39%), Positives = 148/261 (56%), Gaps = 4/261 (1%)
Query: 34 KATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQD--DSPLVGPPPPPPH-- 89
K R+ + P +S + S R + + S D D L+GPP PP
Sbjct: 136 KELRKQIEDMEPAPSSSSAARERSQSSCRDTSSSDSESDDSSDSSDDELIGPPLPPKMVG 195
Query: 90 HEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRIPLSNEIVLKGHTKVVSA 149
D +GP PPP D +D++ D + +E RIP S+EI L+ TK VSA
Sbjct: 196 ESVTTVDEGTLGPLPPPLCEEGEDDDDDELDDEGEEDNPVQRIPDSHEITLRHGTKTVSA 255
Query: 150 LAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCV 209
L +D +G+R+++G YDY V+ +DF GM++ ++FR ++P E HQI++L +S T D L V
Sbjct: 256 LGLDPSGARLVTGGYDYDVKFWDFAGMDASFKAFRSLQPCECHQIKSLQYSNTGDMILVV 315
Query: 210 TGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSSEDGS 269
+GS+QAK+ DRDG + E +KGD YI D+ NTKGH L G WHPK K +T S D +
Sbjct: 316 SGSSQAKVIDRDGFEVMECIKGDQYIVDMANTKGHTAMLHTGSWHPKIKGEFMTCSNDAT 375
Query: 270 LRIWDVNDFTTQKQVWKIFTL 290
+R+W+V + QK V+K T+
Sbjct: 376 VRLWEVENPKKQKSVFKPRTM 396
>ref|XP_639035.1| hypothetical protein DDB0218488 [Dictyostelium discoideum]
gi|60467713|gb|EAL65732.1| hypothetical protein
DDB0218488 [Dictyostelium discoideum]
Length = 772
Score = 187 bits (475), Expect = 3e-46
Identities = 104/280 (37%), Positives = 159/280 (56%), Gaps = 29/280 (10%)
Query: 36 TRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEAD-- 93
T +V P TT++ P K +++ P K P + + +GP P + D
Sbjct: 202 TTTNVGPQKPTTTTNVGPQKPTTNNVGPQKPSTTNVGPQKPTTNTIGPQRPTIKDDDDSD 261
Query: 94 -------DDDGE---------MIGPPPP----PPGSNFNDSEDEDNDSDQDELGN--RFR 131
DDD ++GP P S+ +D +D D+DSD DE R+R
Sbjct: 262 SDSDSDSDDDNSNKIKNEQQGIVGPTRPIMKKDDSSDSDDDDDSDDDSDDDEKEEIERWR 321
Query: 132 -----IPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQI 186
+P+S+E+ +KGH + VS++++D +GSR+LSGSYD V+ +DF GM+S +SFR+I
Sbjct: 322 QLAISLPISHEVTIKGHARTVSSISIDPSGSRMLSGSYDCKVKFWDFNGMDSSFRSFREI 381
Query: 187 EPSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHIT 246
EPSEG Q+R++ +S D F + + Q +IYDRDG E V GD +I+DL +TKGH++
Sbjct: 382 EPSEGTQVRSIQFSNLGDIFSALPYTTQIRIYDRDGHFKNESVSGDRFIQDLYHTKGHVS 441
Query: 247 GLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
LT WHP ++ +++SS DG++R+WDVN + K V K
Sbjct: 442 TLTAQCWHPTERDELISSSIDGTIRVWDVNQLSKNKSVIK 481
>ref|XP_546343.1| PREDICTED: similar to hypothetical protein FLJ10233 [Canis
familiaris]
Length = 723
Score = 186 bits (472), Expect = 8e-46
Identities = 95/220 (43%), Positives = 140/220 (63%), Gaps = 11/220 (5%)
Query: 77 DSPLVGPPPP------PPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRF 130
D L+GPP P P +H +D ++GP PPP + +D+D DS+++E +
Sbjct: 151 DDELIGPPLPLKMMEEPSNHIEED----ILGPLPPPLYEEVEEDDDDDGDSEEEENPVQ- 205
Query: 131 RIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSE 190
+IP S+EI LK TK VSAL +D +G+R+++G YDY V+ +DF GM++ ++FR ++P E
Sbjct: 206 KIPDSHEITLKHGTKTVSALGLDPSGARLVTGGYDYDVKFWDFAGMDASFKAFRSLQPCE 265
Query: 191 GHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTC 250
HQI++L +S T D L V+GS+QAK+ DRDG + E +KGD YI D+ NTKGH L
Sbjct: 266 CHQIKSLQYSNTGDMILVVSGSSQAKVIDRDGFEVMECIKGDQYIVDMANTKGHTAMLHT 325
Query: 251 GEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWKIFTL 290
G WHPK K +T S D ++R W+V + Q+ V+K T+
Sbjct: 326 GSWHPKIKGEFMTCSNDATVRTWEVENPRKQRSVFKPRTM 365
>dbj|BAA91502.1| unnamed protein product [Homo sapiens] gi|19263773|gb|AAH25315.1|
Hypothetical protein LOC55100 [Homo sapiens]
gi|16307120|gb|AAH09648.1| Hypothetical protein LOC55100
[Homo sapiens] gi|8922301|ref|NP_060504.1| hypothetical
protein LOC55100 [Homo sapiens]
Length = 654
Score = 185 bits (469), Expect = 2e-45
Identities = 94/219 (42%), Positives = 140/219 (63%), Gaps = 9/219 (4%)
Query: 77 DSPLVGPPPPP-----PHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFR 131
D L+GPP PP P + ++D ++GP PPP ++E+E+ + +++E +
Sbjct: 114 DDELIGPPLPPKMVGKPVNFMEED---ILGPLPPPLNEEEEEAEEEEEEEEEEE-NPVHK 169
Query: 132 IPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEG 191
IP S+EI LK TK VSAL +D +G+R+++G YDY V+ +DF GM++ ++FR ++P E
Sbjct: 170 IPDSHEITLKHGTKTVSALGLDPSGARLVTGGYDYDVKFWDFAGMDASFKAFRSLQPCEC 229
Query: 192 HQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCG 251
HQI++L +S T D L V+GS+QAK+ DRDG + E +KGD YI D+ NTKGH L G
Sbjct: 230 HQIKSLQYSNTGDMILVVSGSSQAKVIDRDGFEVMECIKGDQYIVDMANTKGHTAMLHTG 289
Query: 252 EWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWKIFTL 290
WHPK K +T S D ++R W+V + QK V+K T+
Sbjct: 290 SWHPKIKGEFMTCSNDATVRTWEVENPKKQKSVFKPRTM 328
>gb|EAL20968.1| hypothetical protein CNBD5690 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 550
Score = 185 bits (469), Expect = 2e-45
Identities = 99/272 (36%), Positives = 156/272 (56%), Gaps = 25/272 (9%)
Query: 14 EFPLSFGKQSKS-QTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSP 72
+ P+SFGK+SK+ + L+ + T+R+ P ++ P +E P P
Sbjct: 5 DLPMSFGKKSKAGNSNLKAKVENTKRA---DIPTPVKEEKPEEEV------------PGP 49
Query: 73 PLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRI 132
+ EA+++D E IGP PP E++++D +Q+ R+
Sbjct: 50 SISTSGRKT---------EAEEEDDEEIGPIPPSATGKRKAEENDESDDEQEAEEEVDRL 100
Query: 133 PLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGH 192
P+S+EI+LK HTKVVSALAVD +G+R+ +GS+DY +++DF GM+ RL+ F+ E + +
Sbjct: 101 PISHEIILKDHTKVVSALAVDPSGARIATGSHDYDTKLWDFGGMDHRLKPFKSFEANGNY 160
Query: 193 QIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGE 252
+ +LS+SP + L ++G+ K+++R+G EF KGD+Y+RD+KNT GH + G
Sbjct: 161 YVHDLSYSPDGQQLLVISGTVHPKVFNRNGEDEMEFNKGDVYLRDMKNTNGHTAEINAGA 220
Query: 253 WHPKTKETILTSSEDGSLRIWDVNDFTTQKQV 284
WHP K LT S D +LRIWD ++ QKQV
Sbjct: 221 WHPTDKSIFLTCSNDSTLRIWDTSNKRKQKQV 252
>emb|CAC21644.1| hypothetical protein [Homo sapiens]
Length = 674
Score = 185 bits (469), Expect = 2e-45
Identities = 94/219 (42%), Positives = 140/219 (63%), Gaps = 9/219 (4%)
Query: 77 DSPLVGPPPPP-----PHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFR 131
D L+GPP PP P + ++D ++GP PPP ++E+E+ + +++E +
Sbjct: 134 DDELIGPPLPPKMVGKPVNFMEED---ILGPLPPPLNEEEEEAEEEEEEEEEEE-NPVHK 189
Query: 132 IPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEG 191
IP S+EI LK TK VSAL +D +G+R+++G YDY V+ +DF GM++ ++FR ++P E
Sbjct: 190 IPDSHEITLKHGTKTVSALGLDPSGARLVTGGYDYDVKFWDFAGMDASFKAFRSLQPCEC 249
Query: 192 HQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCG 251
HQI++L +S T D L V+GS+QAK+ DRDG + E +KGD YI D+ NTKGH L G
Sbjct: 250 HQIKSLQYSNTGDMILVVSGSSQAKVIDRDGFEVMECIKGDQYIVDMANTKGHTAMLHTG 309
Query: 252 EWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWKIFTL 290
WHPK K +T S D ++R W+V + QK V+K T+
Sbjct: 310 SWHPKIKGEFMTCSNDATVRTWEVENPKKQKSVFKPRTM 348
>gb|AAW43006.1| transcription factor, putative [Cryptococcus neoformans var.
neoformans JEC21] gi|58266314|ref|XP_570313.1|
transcription factor, putative [Cryptococcus neoformans
var. neoformans JEC21]
Length = 550
Score = 183 bits (465), Expect = 5e-45
Identities = 99/272 (36%), Positives = 155/272 (56%), Gaps = 25/272 (9%)
Query: 14 EFPLSFGKQSKS-QTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSP 72
+ P+SFGK+SK+ + L+ + T+R+ P ++ P +E P P
Sbjct: 5 DLPMSFGKKSKAGNSNLKAKVENTKRA---DIPTPVKEEKPEEEV------------PGP 49
Query: 73 PLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRI 132
+ EA+++D E IGP PP E++ +D +Q+ R+
Sbjct: 50 SISTSGRKT---------EAEEEDDEEIGPIPPSATGKRKAEENDASDDEQEAEEEVDRL 100
Query: 133 PLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGH 192
P+S+EI+LK HTKVVSALAVD +G+R+ +GS+DY +++DF GM+ RL+ F+ E + +
Sbjct: 101 PISHEIILKDHTKVVSALAVDPSGARIATGSHDYDTKLWDFGGMDHRLKPFKSFEANGNY 160
Query: 193 QIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGE 252
+ +LS+SP + L ++G+ K+++R+G EF KGD+Y+RD+KNT GH + G
Sbjct: 161 YVHDLSYSPDGQQLLVISGTVHPKVFNRNGEDEMEFNKGDVYLRDMKNTNGHTAEINAGA 220
Query: 253 WHPKTKETILTSSEDGSLRIWDVNDFTTQKQV 284
WHP K LT S D +LRIWD ++ QKQV
Sbjct: 221 WHPTDKSIFLTCSNDSTLRIWDTSNKRKQKQV 252
>emb|CAF99231.1| unnamed protein product [Tetraodon nigroviridis]
Length = 642
Score = 180 bits (456), Expect = 6e-44
Identities = 93/219 (42%), Positives = 133/219 (60%), Gaps = 13/219 (5%)
Query: 81 VGPPPPPPHHEAD-----DDDGEMIGPPPP--------PPGSNFNDSEDEDNDSDQDELG 127
+ P P P ++ D D +++GPP P G +D ++E++ SD +E
Sbjct: 78 ISPCPRKPQSDSSGSSDSDSDSDLVGPPLPRQYSDEHMKQGDTGSDDDEEEDASDAEEDN 137
Query: 128 NRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIE 187
+IP S+EI L+ K VSALA+D +G+R+++G YDY V+ +DF GM+ LQ+FR ++
Sbjct: 138 PVRKIPDSHEITLQHGNKTVSALALDPSGARLVTGGYDYDVKFWDFAGMDQALQAFRSLQ 197
Query: 188 PSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITG 247
P E HQI++L +S T D L +G+AQAK+ DRDG + E +KGD YI D+ NTKGH
Sbjct: 198 PCECHQIKSLQYSITGDVVLVASGNAQAKVLDRDGFNVMECIKGDQYIVDMVNTKGHTAM 257
Query: 248 LTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
L G WHPK KE +T S DG++R WDV K V+K
Sbjct: 258 LNGGCWHPKIKEEFMTCSNDGTVRTWDVKSEKQHKSVFK 296
>ref|XP_393688.2| PREDICTED: similar to CG5543-PA [Apis mellifera]
Length = 664
Score = 174 bits (442), Expect = 2e-42
Identities = 104/263 (39%), Positives = 148/263 (55%), Gaps = 15/263 (5%)
Query: 19 FGKQSKS---QTPLETIHKATRRS----VPTSNPKTTSDDFPSKECLHSIRPPKNPNPPS 71
FGK++KS Q LE I K S V + + S P+KE + I N +
Sbjct: 69 FGKKAKSFDVQEMLENITKTINNSKVSNVNIKSIEKESKTIPAKEVIKDIDIINNESTND 128
Query: 72 PPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSD-QDELGNRF 130
++D +GPP PP + + + N N+S++++ SD ++EL +
Sbjct: 129 N--EEDDDFIGPPIPPELQNSIN-----LEQSTKIKDQNDNESDEDEEQSDIEEELQLKD 181
Query: 131 RIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSE 190
+IP S+E+ + TK V+A+A D +G+R+ SGS DY V +DF GM+S ++SFR ++P E
Sbjct: 182 KIPCSHEVTMTHGTKAVTAIAADPSGARLASGSIDYDVCFWDFAGMDSSMRSFRTLQPCE 241
Query: 191 GHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTC 250
H I+ L +S T D L ++GSAQAK+ DRDG E VKGD YI D+ TKGH GL
Sbjct: 242 NHPIKCLQYSMTGDVILVISGSAQAKVLDRDGFEKCETVKGDQYITDMARTKGHTAGLNS 301
Query: 251 GEWHPKTKETILTSSEDGSLRIW 273
G WHP TKE LT S+D + RIW
Sbjct: 302 GCWHPFTKEEYLTCSQDSTCRIW 324
>ref|XP_307535.2| ENSANGP00000015367 [Anopheles gambiae str. PEST]
gi|55246598|gb|EAA03327.2| ENSANGP00000015367 [Anopheles
gambiae str. PEST]
Length = 647
Score = 168 bits (425), Expect = 2e-40
Identities = 94/221 (42%), Positives = 126/221 (56%), Gaps = 10/221 (4%)
Query: 76 DDSPLVGPPPPPPHHEADDDD-----GEMIGPPPPPPGSNFNDSE-----DEDNDSDQDE 125
D+ ++ P + DDDD EMIGP P G DS E++DS+ DE
Sbjct: 102 DEKAIIDPDDERKGKDDDDDDEEEEEDEMIGPVPTAAGGGATDSRRKKVHKEEDDSEFDE 161
Query: 126 LGNRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQ 185
++P S+E+ ++ +K V ALA D +G R+ SGS DY + +DF GM+ ++SFR
Sbjct: 162 DAPINKLPHSHEVEMRHGSKAVIALASDPSGVRLASGSMDYNLNFWDFSGMDKSMKSFRS 221
Query: 186 IEPSEGHQIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHI 245
++P E H IRNLS+S + D L V+GS+QAK+ DRDG E VKGD YI D+ TKGHI
Sbjct: 222 MQPCENHPIRNLSYSFSGDMMLVVSGSSQAKVLDRDGFEKMECVKGDQYIADMSKTKGHI 281
Query: 246 TGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
GLT G W P +E LTS D +LR W Q+ V K
Sbjct: 282 AGLTSGCWSPVRREEFLTSGMDSTLRTWVFAKTKEQRAVIK 322
>ref|NP_611832.1| CG5543-PA [Drosophila melanogaster] gi|21064283|gb|AAM29371.1|
LD24649p [Drosophila melanogaster]
gi|7291643|gb|AAF47065.1| CG5543-PA [Drosophila
melanogaster] gi|16198109|gb|AAL13853.1| LD31556p
[Drosophila melanogaster]
Length = 655
Score = 167 bits (423), Expect = 4e-40
Identities = 90/214 (42%), Positives = 129/214 (60%), Gaps = 13/214 (6%)
Query: 86 PPPHHEADDDDGEMIGPPPPP----------PGSNFNDSEDEDNDSDQD---ELGNRFRI 132
P + D+++ ++IGP PP S DS+D+D SD+D E RI
Sbjct: 111 PKEDDQKDEEEEDVIGPLPPAVTTDKEKATKESSKDEDSDDDDYSSDEDSDDEQSLAKRI 170
Query: 133 PLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGH 192
P ++E+ ++ ++ V ALA D +G+R++SGS DY + +DF GM+S ++SFRQ++P E H
Sbjct: 171 PYTHEVQMQHGSRAVLALAGDPSGARLVSGSIDYDMCFWDFAGMDSSMRSFRQLQPCENH 230
Query: 193 QIRNLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGE 252
IR+L +S T D L ++G+AQAK+ DRDG E KGD YI D+ TKGH+ LT G
Sbjct: 231 PIRSLQYSVTGDMILVISGNAQAKVLDRDGFEKLECCKGDQYISDMSRTKGHVAQLTSGC 290
Query: 253 WHPKTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
WHP +E LT++ DG+LRIW Q QV K
Sbjct: 291 WHPFNREQFLTAALDGTLRIWQGLKSKEQLQVIK 324
>ref|XP_326344.1| hypothetical protein [Neurospora crassa] gi|28918215|gb|EAA27893.1|
hypothetical protein [Neurospora crassa]
Length = 585
Score = 164 bits (414), Expect = 4e-39
Identities = 99/287 (34%), Positives = 145/287 (50%), Gaps = 62/287 (21%)
Query: 9 DGVRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPN 68
D +RA PLSFGK S+ I +A R+ + P P
Sbjct: 20 DPMRAYVPLSFGKTSREANIAAQIERARRQ----------------------VETPAAPK 57
Query: 69 PPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDELGN 128
DDS +++DDDD D+D+DSD +
Sbjct: 58 KGDRKKDDDSD----------NDSDDDD-------------------DDDDDSDDESEDE 88
Query: 129 RFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMN-SRLQSFRQIE 187
R P+S+E+VLK H + V+++++D G R+L+GS D ++++DF M + L++FR ++
Sbjct: 89 SDRFPVSHELVLKTHERAVTSVSLDPAGGRLLTGSLDGTIKLHDFSAMTPTTLRAFRSVD 148
Query: 188 P---------SEGHQIRNLSWSPTA-DRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRD 237
P ++ H I+ + PT+ FLCVT QAKI RDG L EFVKGDMY+RD
Sbjct: 149 PWESNKSSATTDSHPIQRAEFCPTSGSHFLCVTAHPQAKIMSRDGDILTEFVKGDMYLRD 208
Query: 238 LKNTKGHITGLTCGEWHPKTKETILTSSEDGSLRIWDVNDFTTQKQV 284
+ NTKGH+ +T G WHP +T+ D +LRIWDVN+ +QK+V
Sbjct: 209 MHNTKGHVGEITTGTWHPSDPNLCVTAGSDSTLRIWDVNNKRSQKEV 255
Score = 33.5 bits (75), Expect = 8.3
Identities = 31/124 (25%), Positives = 53/124 (42%), Gaps = 26/124 (20%)
Query: 156 GSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWSPTADRFLCVTGSAQA 215
G V++ D +++++D + L S+ + N+ +SP D +TGSA
Sbjct: 331 GRMVVTRGGDGLIKLWDTRKFKEPLVKVEHPSTSDRFPMTNIRYSP--DSRSIITGSAS- 387
Query: 216 KIYDRDGLTLGEFVKGDMYIRDLKNTKG-HITGLTCG------EWHPKTKETILTSSEDG 268
G ++I + N + H+T +T G EWHPK + I+T S +
Sbjct: 388 ---------------GHLHILNPGNLRPEHVTPITPGIPIIVVEWHPKLNQ-IITGSANA 431
Query: 269 SLRI 272
RI
Sbjct: 432 ETRI 435
>gb|EAL24654.1| GA18958-PA [Drosophila pseudoobscura]
Length = 643
Score = 157 bits (397), Expect = 4e-37
Identities = 88/211 (41%), Positives = 126/211 (59%), Gaps = 16/211 (7%)
Query: 91 EADDDDGEMIGPPPPPP------------GSNFNDSEDE---DNDSDQDELGNRFRIPLS 135
E + + ++IGP PP + DS+D+ D DSD DE RIP +
Sbjct: 103 EVAEQEEDVIGPLPPAAVEDSKQGCKDEATNKVEDSDDDCSSDEDSD-DEQSLSKRIPYT 161
Query: 136 NEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIR 195
+E+ ++ ++ V ALA D +G+R++SGS DY + +DF GM+S ++SFRQ++P E H IR
Sbjct: 162 HEVQMQHGSRAVLALAGDPSGARLVSGSIDYDMCFWDFVGMDSSMRSFRQLQPCENHPIR 221
Query: 196 NLSWSPTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHP 255
+L +S T + L ++G+AQAK+ DRDG E KGD YI D+ TKGH+ LT G WHP
Sbjct: 222 SLQYSVTGEMILVISGNAQAKVLDRDGFEKLECCKGDQYISDMSRTKGHVAQLTSGCWHP 281
Query: 256 KTKETILTSSEDGSLRIWDVNDFTTQKQVWK 286
+E LT++ DG+LRIW Q QV K
Sbjct: 282 FNREQFLTAALDGTLRIWQGLKAKEQCQVIK 312
>gb|EAA77540.1| hypothetical protein FG07307.1 [Gibberella zeae PH-1]
gi|46125859|ref|XP_387483.1| hypothetical protein
FG07307.1 [Gibberella zeae PH-1]
Length = 639
Score = 156 bits (394), Expect = 9e-37
Identities = 82/199 (41%), Positives = 121/199 (60%), Gaps = 17/199 (8%)
Query: 97 GEMIGPPPPPPGSNFNDSEDEDNDSDQDELGNRFRIPLSNEIVLKGHTKVVSALAVDHTG 156
G+ GP S+ +DS+D D+ D+DE P+S+E+VLK H + V+ +++D G
Sbjct: 58 GQAQGPKKKGNSSDDSDSDDSDDSDDEDEF------PVSHELVLKTHERAVTTVSLDPAG 111
Query: 157 SRVLSGSYDYMVRMYDFQGMN-SRLQSFRQIEP---------SEGHQIRNLSWSPTADR- 205
R+ + S D V+++DF M + L++FR ++P SE H + + ++P A
Sbjct: 112 GRLATASTDCTVKLHDFASMTPTTLRAFRSVDPYEQKASSASSEAHPVHRIEFNPHAGGV 171
Query: 206 FLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKETILTSS 265
FLCV+ QAKI RDG + EFVKGDMY+RD+ NTKGHI+ +T G WHP K +T+
Sbjct: 172 FLCVSAHPQAKIMSRDGDIVTEFVKGDMYLRDMHNTKGHISEITTGTWHPTDKNLCITAG 231
Query: 266 EDGSLRIWDVNDFTTQKQV 284
D +LRIWDVN+ +QK V
Sbjct: 232 TDSTLRIWDVNNKRSQKDV 250
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 683,850,278
Number of Sequences: 2540612
Number of extensions: 36612091
Number of successful extensions: 346648
Number of sequences better than 10.0: 5824
Number of HSP's better than 10.0 without gapping: 1481
Number of HSP's successfully gapped in prelim test: 4688
Number of HSP's that attempted gapping in prelim test: 258463
Number of HSP's gapped (non-prelim): 33986
length of query: 316
length of database: 863,360,394
effective HSP length: 128
effective length of query: 188
effective length of database: 538,162,058
effective search space: 101174466904
effective search space used: 101174466904
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146819.11


