

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.8 + phase: 0
(831 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
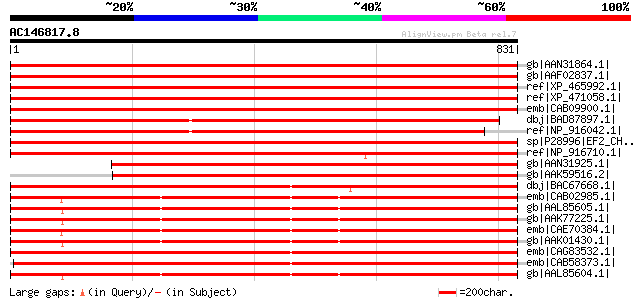
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN31864.1| putative elongation factor [Arabidopsis thaliana]... 1466 0.0
gb|AAF02837.1| elongation factor EF-2 [Arabidopsis thaliana] gi|... 1466 0.0
ref|XP_465992.1| putative elongation factor 2 [Oryza sativa (jap... 1459 0.0
ref|XP_471058.1| OSJNBa0020P07.3 [Oryza sativa (japonica cultiva... 1457 0.0
emb|CAB09900.1| elongation factor 2 [Beta vulgaris subsp. vulgar... 1455 0.0
dbj|BAD87897.1| putative Elongation factor 2 [Oryza sativa (japo... 1402 0.0
ref|NP_916042.1| putativeelongation factor 2 [Oryza sativa (japo... 1361 0.0
sp|P28996|EF2_CHLKE Elongation factor 2 (EF-2) gi|421771|pir||S3... 1298 0.0
ref|NP_916710.1| putative elongation factor 2 [Oryza sativa (jap... 1286 0.0
gb|AAN31925.1| putative elongation factor [Arabidopsis thaliana] 1174 0.0
gb|AAK59516.2| putative elongation factor [Arabidopsis thaliana]... 1172 0.0
dbj|BAC67668.1| elongation factor-2 [Cyanidioschyzon merolae] 1075 0.0
emb|CAB02985.1| Hypothetical protein F25H5.4 [Caenorhabditis ele... 1056 0.0
gb|AAL85605.1| elongation factor 2 [Aedes aegypti] 1055 0.0
gb|AAK77225.1| elongation factor 2 [Aedes aegypti] 1055 0.0
emb|CAE70384.1| Hypothetical protein CBG16945 [Caenorhabditis br... 1055 0.0
gb|AAK01430.1| elongation factor 2 [Aedes aegypti] 1054 0.0
emb|CAG83532.1| unnamed protein product [Yarrowia lipolytica CLI... 1053 0.0
emb|CAB58373.1| SPCP31B10.07 [Schizosaccharomyces pombe] gi|1907... 1053 0.0
gb|AAL85604.1| elongation factor 2 [Aedes aegypti] 1051 0.0
>gb|AAN31864.1| putative elongation factor [Arabidopsis thaliana]
gi|23397045|gb|AAN31808.1| putative elongation factor
[Arabidopsis thaliana] gi|27363422|gb|AAO11630.1|
At1g56070/T6H22_13 [Arabidopsis thaliana]
gi|13605865|gb|AAK32918.1| At1g56070/T6H22_13
[Arabidopsis thaliana] gi|30696056|ref|NP_849818.1|
elongation factor 2, putative / EF-2, putative
[Arabidopsis thaliana] gi|15450763|gb|AAK96653.1|
elongation factor EF-2 [Arabidopsis thaliana]
Length = 843
Score = 1466 bits (3795), Expect = 0.0
Identities = 711/831 (85%), Positives = 771/831 (92%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 13 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK+F G R+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYEMTDESLKSFTGARDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGV E KMM RLWGENFFD
Sbjct: 193 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVVESKMMERLWGENFFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+T+KW+ K+T +PTCKRGFVQFCYEPIKQII CMNDQKDKLWPML KLGV++K++EKEL
Sbjct: 253 ATRKWSGKNTGSPTCKRGFVQFCYEPIKQIIATCMNDQKDKLWPMLAKLGVSMKNDEKEL 312
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP AQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 313 MGKPLMKRVMQTWLPASTALLEMMIFHLPSPHTAQRYRVENLYEGPLDDQYANAIRNCDP 372
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
GPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTGMKVRIMGPNYIPGEKKDLY KSVQRT
Sbjct: 373 NGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGMKVRIMGPNYIPGEKKDLYTKSVQRT 432
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV VA
Sbjct: 433 VIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVA 492
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCT+ E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 493 VQCKVASDLPKLVEGLKRLAKSDPMVVCTMEESGEHIVAGAGELHLEICLKDLQDDFMGG 552
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETV ++S+ TVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRD+
Sbjct: 553 AEIIKSDPVVSFRETVCDRSTRTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDD 612
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KIL++EFGWDKDLAKK+W FGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 613 PKIRSKILAEEFGWDKDLAKKIWAFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 672
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEGP+A+EN+RG+CFEVCDVVLH+DAIHRGGGQ+IPTARRV YA+ +TAKPRLLEPVY+V
Sbjct: 673 KEGPLAEENMRGICFEVCDVVLHSDAIHRGGGQVIPTARRVIYASQITAKPRLLEPVYMV 732
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF F+ LRA T GQ
Sbjct: 733 EIQAPEGALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSSQLRAATSGQ 792
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHW+M+ SDPLEPGT A+ V +IRK+KGLKE + PLSEFED+L
Sbjct: 793 AFPQCVFDHWEMMSSDPLEPGTQASVLVADIRKRKGLKEAMTPLSEFEDKL 843
>gb|AAF02837.1| elongation factor EF-2 [Arabidopsis thaliana]
gi|25299536|pir||A96602 elongation factor EF-2
[imported] - Arabidopsis thaliana
Length = 846
Score = 1466 bits (3795), Expect = 0.0
Identities = 711/831 (85%), Positives = 771/831 (92%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 16 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 75
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK+F G R+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 76 TGISLYYEMTDESLKSFTGARDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 135
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T RVIE+ NV+MATY
Sbjct: 136 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY 195
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGV E KMM RLWGENFFD
Sbjct: 196 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVVESKMMERLWGENFFDP 255
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+T+KW+ K+T +PTCKRGFVQFCYEPIKQII CMNDQKDKLWPML KLGV++K++EKEL
Sbjct: 256 ATRKWSGKNTGSPTCKRGFVQFCYEPIKQIIATCMNDQKDKLWPMLAKLGVSMKNDEKEL 315
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP AQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 316 MGKPLMKRVMQTWLPASTALLEMMIFHLPSPHTAQRYRVENLYEGPLDDQYANAIRNCDP 375
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
GPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTGMKVRIMGPNYIPGEKKDLY KSVQRT
Sbjct: 376 NGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGMKVRIMGPNYIPGEKKDLYTKSVQRT 435
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV VA
Sbjct: 436 VIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVA 495
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCT+ E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 496 VQCKVASDLPKLVEGLKRLAKSDPMVVCTMEESGEHIVAGAGELHLEICLKDLQDDFMGG 555
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETV ++S+ TVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRD+
Sbjct: 556 AEIIKSDPVVSFRETVCDRSTRTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDD 615
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KIL++EFGWDKDLAKK+W FGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 616 PKIRSKILAEEFGWDKDLAKKIWAFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 675
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEGP+A+EN+RG+CFEVCDVVLH+DAIHRGGGQ+IPTARRV YA+ +TAKPRLLEPVY+V
Sbjct: 676 KEGPLAEENMRGICFEVCDVVLHSDAIHRGGGQVIPTARRVIYASQITAKPRLLEPVYMV 735
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF F+ LRA T GQ
Sbjct: 736 EIQAPEGALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSSQLRAATSGQ 795
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHW+M+ SDPLEPGT A+ V +IRK+KGLKE + PLSEFED+L
Sbjct: 796 AFPQCVFDHWEMMSSDPLEPGTQASVLVADIRKRKGLKEAMTPLSEFEDKL 846
>ref|XP_465992.1| putative elongation factor 2 [Oryza sativa (japonica
cultivar-group)] gi|49387779|dbj|BAD26337.1| putative
elongation factor 2 [Oryza sativa (japonica
cultivar-group)]
Length = 843
Score = 1459 bits (3776), Expect = 0.0
Identities = 712/831 (85%), Positives = 773/831 (92%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K+NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 13 MDKKNNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISL+YEMSD LK +KGER+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLFYEMSDESLKLYKGERDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL ++ EEAY T RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVEGEEAYQTFSRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTL++FAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 193 EDTLLGDVQVYPEKGTVAFSAGLHGWAFTLSSFAKMYASKFGVDEFKMMERLWGENFFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWTNK+T + TCKRGFVQFCYEPIKQII CMNDQKDKLWPMLQKLGV +K++EKEL
Sbjct: 253 ATKKWTNKNTGSATCKRGFVQFCYEPIKQIINTCMNDQKDKLWPMLQKLGVVMKADEKEL 312
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GKALMKRVMQ+WLPAS+ALLEMMI+HLPSP+KAQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 313 MGKALMKRVMQTWLPASNALLEMMIYHLPSPSKAQRYRVENLYEGPLDDVYATAIRNCDP 372
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVFSG+V+TGMKVRIMGPNY+PG+KKDLYVKSVQRT
Sbjct: 373 EGPLMLYVSKMIPASDKGRFFAFGRVFSGRVATGMKVRIMGPNYVPGQKKDLYVKSVQRT 432
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQE+VEDVPCGNTVAMVGLDQFITKNATLTNEKE DA PIRAMKFSVSPVV VA
Sbjct: 433 VIWMGKKQESVEDVPCGNTVAMVGLDQFITKNATLTNEKESDACPIRAMKFSVSPVVRVA 492
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMV+CTI E+GEHIIA AGELHLEICLKDLQ+DFM G
Sbjct: 493 VQCKVASDLPKLVEGLKRLAKSDPMVLCTIEESGEHIIAGAGELHLEICLKDLQEDFMGG 552
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI S P+VSFRETVLEKS TVMSKSPNKHNRLYMEARP+EEGLAEAIDDGRIGPRD+
Sbjct: 553 AEIIVSPPVVSFRETVLEKSCRTVMSKSPNKHNRLYMEARPLEEGLAEAIDDGRIGPRDD 612
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KILS+EFGWDKDLAKK+WCFGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 613 PKVRSKILSEEFGWDKDLAKKIWCFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 672
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +A+EN+RG+CFEVCDVVLH DAIHRGGGQ+IPTARRV YA+ LTAKPRLLEPVYLV
Sbjct: 673 KEGALAEENMRGICFEVCDVVLHADAIHRGGGQVIPTARRVIYASQLTAKPRLLEPVYLV 732
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE+ALGGIY VLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F+ LRA T GQ
Sbjct: 733 EIQAPENALGGIYGVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFSSQLRAATSGQ 792
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHWDM+ SDPLE G+ A+ V +IRK+KGLKEQ+ PLS+FED+L
Sbjct: 793 AFPQCVFDHWDMMTSDPLEAGSQASTLVQDIRKRKGLKEQMTPLSDFEDKL 843
>ref|XP_471058.1| OSJNBa0020P07.3 [Oryza sativa (japonica cultivar-group)]
gi|38344860|emb|CAE01286.2| OSJNBa0020P07.3 [Oryza
sativa (japonica cultivar-group)]
Length = 843
Score = 1457 bits (3771), Expect = 0.0
Identities = 711/831 (85%), Positives = 771/831 (92%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K+NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 13 MDKKNNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISL+YEMSD LK +KGER+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLFYEMSDESLKLYKGERDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL ++ EEAY T RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVEGEEAYQTFSRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTL++FAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 193 EDTLLGDVQVYPEKGTVAFSAGLHGWAFTLSSFAKMYASKFGVDESKMMERLWGENFFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWTNK T + TCKRGFVQFCYEPIKQII CMNDQKDKLWPMLQKLGV +K++EK+L
Sbjct: 253 ATKKWTNKSTGSATCKRGFVQFCYEPIKQIINTCMNDQKDKLWPMLQKLGVVMKADEKDL 312
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GKALMKRVMQ+WLPAS+ALLEMMI+HLPSP+KAQKYRVENLYEGPLDD YA+AIRNCDP
Sbjct: 313 MGKALMKRVMQTWLPASNALLEMMIYHLPSPSKAQKYRVENLYEGPLDDVYATAIRNCDP 372
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVFSG+V+TGMKVRIMGPNY+PG+KKDLYVKSVQRT
Sbjct: 373 EGPLMLYVSKMIPASDKGRFFAFGRVFSGRVATGMKVRIMGPNYVPGQKKDLYVKSVQRT 432
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQE+VEDVPCGNTVAMVGLDQFITKNATLTNEKE DA PIRAMKFSVSPVV VA
Sbjct: 433 VIWMGKKQESVEDVPCGNTVAMVGLDQFITKNATLTNEKEADACPIRAMKFSVSPVVRVA 492
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMV+CTI E+GEHIIA AGELHLEICLKDLQ+DFM G
Sbjct: 493 VQCKVASDLPKLVEGLKRLAKSDPMVLCTIEESGEHIIAGAGELHLEICLKDLQEDFMGG 552
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI S P+VSFRETVLEKS TVMSKSPNKHNRLYMEARP+EEGLAEAIDDGRIGPRD+
Sbjct: 553 AEIIVSPPVVSFRETVLEKSCRTVMSKSPNKHNRLYMEARPLEEGLAEAIDDGRIGPRDD 612
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KILS+EFGWDKDLAKK+WCFGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 613 PKVRSKILSEEFGWDKDLAKKIWCFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 672
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +A+EN+RG+CFEVCDVVLH DAIHRGGGQ+IPTARRV YA+ LTAKPRLLEPVYLV
Sbjct: 673 KEGALAEENMRGICFEVCDVVLHADAIHRGGGQVIPTARRVIYASQLTAKPRLLEPVYLV 732
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE+ALGGIY VLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F+ LRA T GQ
Sbjct: 733 EIQAPENALGGIYGVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFSSQLRAATSGQ 792
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHWDM+ SDPLE + A V++IRK+KGLKEQ+ PLS+FED+L
Sbjct: 793 AFPQCVFDHWDMMTSDPLEVSSQANQLVLDIRKRKGLKEQMTPLSDFEDKL 843
>emb|CAB09900.1| elongation factor 2 [Beta vulgaris subsp. vulgaris]
gi|6015065|sp|O23755|EF2_BETVU Elongation factor 2
(EF-2) gi|7443464|pir||T14579 translation elongation
factor eEF-2 - beet
Length = 843
Score = 1455 bits (3766), Expect = 0.0
Identities = 706/831 (84%), Positives = 773/831 (92%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 13 MDCKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYY+M+D L+++KGER+GN YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYQMTDEALQSYKGERKGNDYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY+T Q+VIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYTTFQKVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTL+NFAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 193 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLSNFAKMYASKFGVDESKMMERLWGENFFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT K++ +CKRGFVQFCYEPIKQII CMNDQKDKL + KLG+ +K+EEK+L
Sbjct: 253 ATKKWTTKNSGNASCKRGFVQFCYEPIKQIIAACMNDQKDKLLAHVTKLGIQMKTEEKDL 312
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
G+ LMKRVMQ+WLPASSALLEMMI HLPSP AQ+YRVENLYEGP+DD YA+AIRNCDP
Sbjct: 313 MGRPLMKRVMQTWLPASSALLEMMIHHLPSPATAQRYRVENLYEGPMDDVYATAIRNCDP 372
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTGMKVRIMGPNY+PGEKKDLYVK+VQRT
Sbjct: 373 EGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGMKVRIMGPNYVPGEKKDLYVKNVQRT 432
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQETVEDVPCGNTVA+VGLDQ+ITKNATLTNEKE DAHPIRAMKFSVSPVV VA
Sbjct: 433 VIWMGKKQETVEDVPCGNTVALVGLDQYITKNATLTNEKESDAHPIRAMKFSVSPVVRVA 492
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVC+I E+GEHIIA AGELHLEICLKDLQDDFM G
Sbjct: 493 VQCKVASDLPKLVEGLKRLAKSDPMVVCSIEESGEHIIAGAGELHLEICLKDLQDDFMGG 552
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETVL++S TVMSKSPNKHNRLYMEARPMEEGLAEAID+GRIGPRD+
Sbjct: 553 AEIIKSDPVVSFRETVLDRSVRTVMSKSPNKHNRLYMEARPMEEGLAEAIDEGRIGPRDD 612
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PKN KIL++E+GWDKDLAKK+WCFGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 613 PKNRSKILAEEYGWDKDLAKKIWCFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 672
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +A+EN+RG+CFEVCDVVLHTDAIHRGGGQIIPTARRVFYA+ LTAKPRLLEPVYLV
Sbjct: 673 KEGALAEENMRGICFEVCDVVLHTDAIHRGGGQIIPTARRVFYASQLTAKPRLLEPVYLV 732
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE+ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF F+ +LRA T GQ
Sbjct: 733 EIQAPENALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSSTLRASTSGQ 792
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHW+M+PSDPLE G+ A+ V IRK+KGLKEQ+ PLSEFED+L
Sbjct: 793 AFPQCVFDHWEMMPSDPLEAGSQASTLVSVIRKRKGLKEQMTPLSEFEDKL 843
>dbj|BAD87897.1| putative Elongation factor 2 [Oryza sativa (japonica
cultivar-group)]
Length = 826
Score = 1402 bits (3629), Expect = 0.0
Identities = 686/804 (85%), Positives = 744/804 (92%), Gaps = 6/804 (0%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSV+AHVDHGKSTLTDSLVAAAGIIAQ+VAGDVRMTD+R DEAERGITIKS
Sbjct: 13 MDKKHNIRNMSVVAHVDHGKSTLTDSLVAAAGIIAQDVAGDVRMTDSRSDEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEMSD LK++KG+R+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYEMSDESLKSYKGDRDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL + EEAY T RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVGGEEAYQTFSRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
EDALLGDVQVYPEKGTV+FSAGLHGW+FTL+NFAKMYASKFGVDE KMM RLWGEN+FD
Sbjct: 193 EDALLGDVQVYPEKGTVAFSAGLHGWAFTLSNFAKMYASKFGVDESKMMERLWGENYFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT KHT + TCKRGF+QFCYEPI+QII CMNDQKDKL PMLQKLGV +K +L
Sbjct: 253 TTKKWTIKHTGSDTCKRGFIQFCYEPIRQIINTCMNDQKDKLLPMLQKLGVTMK----DL 308
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
+GKALMKRVMQ+WLPAS+ALLEMMI+HLPSP KAQ+YRVENLYEGPLDD YASAIRNCDP
Sbjct: 309 TGKALMKRVMQTWLPASNALLEMMIYHLPSPAKAQRYRVENLYEGPLDDIYASAIRNCDP 368
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRFYAFGRVFSG+V+TGMKVRIMGPNY PG+KKDLYVK+VQRT
Sbjct: 369 EGPLMLYVSKMIPASDKGRFYAFGRVFSGRVATGMKVRIMGPNYAPGQKKDLYVKNVQRT 428
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQE+VE VPCGNTVAMVGLDQFITKNATLTNEKEVDA PI+AMKFSVSPVV VA
Sbjct: 429 VIWMGKKQESVEGVPCGNTVAMVGLDQFITKNATLTNEKEVDACPIKAMKFSVSPVVRVA 488
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMV+CT+ E+GEHIIA AGELHLEICLKDLQ+DFM G
Sbjct: 489 VQCKVASDLPKLVEGLKRLAKSDPMVLCTVEESGEHIIAGAGELHLEICLKDLQEDFMGG 548
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEIT S P+VSFRETVLEKS TVMSKSPNKHNRLYMEARPMEEGL EAID+GRIGPRD+
Sbjct: 549 AEITVSPPVVSFRETVLEKSCRTVMSKSPNKHNRLYMEARPMEEGLPEAIDEGRIGPRDD 608
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KILS+EFGWDKDLAKK+WCFGPETTGPN++VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 609 PKVRSKILSEEFGWDKDLAKKIWCFGPETTGPNIVVDMCKGVQYLNEIKDSVVAGFQWAS 668
Query: 661 KEGPMADENL--RGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVY 718
KEG +A+EN+ RG+CFEVCDV+LH+DAIHRGGGQIIPTARRV YAA LTAKPRLLEPVY
Sbjct: 669 KEGALAEENMRGRGICFEVCDVILHSDAIHRGGGQIIPTARRVIYAAQLTAKPRLLEPVY 728
Query: 719 LVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTG 778
LVEIQAPE+ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F +LRA T
Sbjct: 729 LVEIQAPENALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFTSTLRAATS 788
Query: 779 GQAFPQLVFDHWDMVPSDPLEPGT 802
GQAFP VFDHW+M+ DPLEPGT
Sbjct: 789 GQAFPLFVFDHWEMLSVDPLEPGT 812
>ref|NP_916042.1| putativeelongation factor 2 [Oryza sativa (japonica
cultivar-group)]
Length = 795
Score = 1361 bits (3523), Expect = 0.0
Identities = 668/779 (85%), Positives = 724/779 (92%), Gaps = 6/779 (0%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSV+AHVDHGKSTLTDSLVAAAGIIAQ+VAGDVRMTD+R DEAERGITIKS
Sbjct: 13 MDKKHNIRNMSVVAHVDHGKSTLTDSLVAAAGIIAQDVAGDVRMTDSRSDEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEMSD LK++KG+R+GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYEMSDESLKSYKGDRDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL + EEAY T RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVGGEEAYQTFSRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
EDALLGDVQVYPEKGTV+FSAGLHGW+FTL+NFAKMYASKFGVDE KMM RLWGEN+FD
Sbjct: 193 EDALLGDVQVYPEKGTVAFSAGLHGWAFTLSNFAKMYASKFGVDESKMMERLWGENYFDP 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT KHT + TCKRGF+QFCYEPI+QII CMNDQKDKL PMLQKLGV +K +L
Sbjct: 253 TTKKWTIKHTGSDTCKRGFIQFCYEPIRQIINTCMNDQKDKLLPMLQKLGVTMK----DL 308
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
+GKALMKRVMQ+WLPAS+ALLEMMI+HLPSP KAQ+YRVENLYEGPLDD YASAIRNCDP
Sbjct: 309 TGKALMKRVMQTWLPASNALLEMMIYHLPSPAKAQRYRVENLYEGPLDDIYASAIRNCDP 368
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRFYAFGRVFSG+V+TGMKVRIMGPNY PG+KKDLYVK+VQRT
Sbjct: 369 EGPLMLYVSKMIPASDKGRFYAFGRVFSGRVATGMKVRIMGPNYAPGQKKDLYVKNVQRT 428
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQE+VE VPCGNTVAMVGLDQFITKNATLTNEKEVDA PI+AMKFSVSPVV VA
Sbjct: 429 VIWMGKKQESVEGVPCGNTVAMVGLDQFITKNATLTNEKEVDACPIKAMKFSVSPVVRVA 488
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMV+CT+ E+GEHIIA AGELHLEICLKDLQ+DFM G
Sbjct: 489 VQCKVASDLPKLVEGLKRLAKSDPMVLCTVEESGEHIIAGAGELHLEICLKDLQEDFMGG 548
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEIT S P+VSFRETVLEKS TVMSKSPNKHNRLYMEARPMEEGL EAID+GRIGPRD+
Sbjct: 549 AEITVSPPVVSFRETVLEKSCRTVMSKSPNKHNRLYMEARPMEEGLPEAIDEGRIGPRDD 608
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KILS+EFGWDKDLAKK+WCFGPETTGPN++VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 609 PKVRSKILSEEFGWDKDLAKKIWCFGPETTGPNIVVDMCKGVQYLNEIKDSVVAGFQWAS 668
Query: 661 KEGPMADENL--RGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVY 718
KEG +A+EN+ RG+CFEVCDV+LH+DAIHRGGGQIIPTARRV YAA LTAKPRLLEPVY
Sbjct: 669 KEGALAEENMRGRGICFEVCDVILHSDAIHRGGGQIIPTARRVIYAAQLTAKPRLLEPVY 728
Query: 719 LVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQT 777
LVEIQAPE+ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F +LRA T
Sbjct: 729 LVEIQAPENALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFTSTLRAAT 787
>sp|P28996|EF2_CHLKE Elongation factor 2 (EF-2) gi|421771|pir||S32819 translation
elongation factor eEF-2 - Chlorella kessleri
gi|167245|gb|AAA33028.1| elongation factor 2
gi|228693|prf||1808323A elongation factor 2
Length = 845
Score = 1298 bits (3358), Expect = 0.0
Identities = 637/833 (76%), Positives = 724/833 (86%), Gaps = 2/833 (0%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
M+ ++NIRNMSVIAHVDHGKSTLTDSLVAAAGIIA E AGD R+TDTR DE ERGITIKS
Sbjct: 13 MEYQNNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAFEQAGDQRLTDTRADEQERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYY+M+D LK F GER+GN +LINLIDSPGHVDFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYQMTDEQLKGFTGERQGNDFLINLIDSPGHVDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLT+NK+DRCFLEL LD EEAY +RVIE+ NV+MATY
Sbjct: 133 EGVCVQTETVLRQALGERIRPVLTINKIDRCFLELMLDPEEAYLAYRRVIENANVIMATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
D LGD Q +PE GTVSFSAGLHGW+FTLT FA MYA+KFG D ++MM +LWG+NFFD+
Sbjct: 193 ADEHLGDTQTHPEAGTVSFSAGLHGWAFTLTVFANMYAAKFGTDTKRMMEKLWGDNFFDA 252
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGV--NLKSEEK 298
+T+KWT KHT TCKRGF QF YEPIK +IE MND KDKL+ +L+KL V LK E++
Sbjct: 253 TTRKWTKKHTGADTCKRGFCQFIYEPIKTVIEAAMNDNKDKLFDLLKKLNVYSKLKPEDR 312
Query: 299 ELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNC 358
EL GK LMKRVMQ+WLPA ALLEMMI+HLPSP KAQKYRV+ LYEGPLDD YA+A+RNC
Sbjct: 313 ELMGKPLMKRVMQTWLPAHEALLEMMIWHLPSPAKAQKYRVDVLYEGPLDDTYATAVRNC 372
Query: 359 DPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQ 418
D +GPLM+YVSKMIPA+DKGRFYAFGRVFSG+++TG KVRIMGPNY+PG+KKDLYVK+VQ
Sbjct: 373 DADGPLMMYVSKMIPAADKGRFYAFGRVFSGRIATGRKVRIMGPNYVPGQKKDLYVKTVQ 432
Query: 419 RTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVS 478
RTV+ MG++QE VEDVPCGNTVA+VGLDQFITKNATLT+EK DAH I+AMKFSVSPVV
Sbjct: 433 RTVLCMGRRQEAVEDVPCGNTVALVGLDQFITKNATLTDEKCEDAHTIKAMKFSVSPVVR 492
Query: 479 VAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFM 538
VAV KVASDLPKLVEGLKRLAKSDPMV CTI ETGEHIIA AGELHLEICLKDLQDDFM
Sbjct: 493 VAVEPKVASDLPKLVEGLKRLAKSDPMVQCTIEETGEHIIAGAGELHLEICLKDLQDDFM 552
Query: 539 NGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPR 598
GAEI S+P+VSFRETV+ S H VMSKSPNKHNRLYM+ARPME+GLAEAID+G+IGPR
Sbjct: 553 GGAEIRVSEPVVSFRETVIGTSDHVVMSKSPNKHNRLYMQARPMEDGLAEAIDEGKIGPR 612
Query: 599 DEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQI 658
D+PK KILS+EFGWDK+LAKK+ FGP+TTGPNM+ D KGVQYLNEIKDSVVA FQ
Sbjct: 613 DDPKVRSKILSEEFGWDKELAKKILAFGPDTTGPNMVTDITKGVQYLNEIKDSVVAAFQW 672
Query: 659 ASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVY 718
ASKEG +A+EN+RG+ FEVCDVVLH DAIHRGGGQIIPTARR YAA LTA+PRLLEPVY
Sbjct: 673 ASKEGVLAEENMRGIVFEVCDVVLHADAIHRGGGQIIPTARRSMYAAQLTAQPRLLEPVY 732
Query: 719 LVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTG 778
LVEIQ PE A+GG+YSVLNQKRG VF+E+QRP TP++N+KAYLPVIESF F +LRA T
Sbjct: 733 LVEIQCPEQAMGGVYSVLNQKRGMVFEELQRPGTPIFNLKAYLPVIESFGFTSTLRAATA 792
Query: 779 GQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
GQAFPQ VFDHW+ + SDP + G+ A V++IRK+KGLK + LSE+ED+L
Sbjct: 793 GQAFPQCVFDHWEAMGSDPTQVGSQANTLVMDIRKRKGLKPEPAALSEYEDKL 845
>ref|NP_916710.1| putative elongation factor 2 [Oryza sativa (japonica
cultivar-group)] gi|20160544|dbj|BAB89493.1| putative
elongation factor 2 [Oryza sativa (japonica
cultivar-group)] gi|18461242|dbj|BAB84439.1| putative
elongation factor 2 [Oryza sativa (japonica
cultivar-group)]
Length = 853
Score = 1286 bits (3327), Expect = 0.0
Identities = 624/841 (74%), Positives = 719/841 (85%), Gaps = 10/841 (1%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQ+VAGDVRMTDTR DEAERGITIKS
Sbjct: 13 MDKKDNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQDVAGDVRMTDTRADEAERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D L++F+G+R+GN YLINLIDSPGH+DFSSEVTAALRITDGALVVVDC+
Sbjct: 73 TGISLYYEMTDAALRSFEGKRDGNSYLINLIDSPGHIDFSSEVTAALRITDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQ+L ERIKPVLTVNKMDRCFLEL EEAY RVIESVNV MA Y
Sbjct: 133 EGVCVQTETVLRQSLAERIKPVLTVNKMDRCFLELQQSGEEAYQAFSRVIESVNVTMAPY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LGD V PEKGTV+FSAGLHGW+FTL+NFAKMY +KF VDE KMM RLWGEN+FD
Sbjct: 193 EDKNLGDCMVAPEKGTVAFSAGLHGWAFTLSNFAKMYKAKFKVDEAKMMERLWGENYFDH 252
Query: 241 STKKWTNK--HTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEK 298
+TKKWT TS+ TC+RGFVQFCYEPI++II CMND K+ LW ML KL + LK+EEK
Sbjct: 253 TTKKWTTTAPSTSSKTCQRGFVQFCYEPIRRIISACMNDDKENLWDMLTKLKITLKAEEK 312
Query: 299 ELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNC 358
EL+GK LMKRVMQ+WLPAS ALLEM++FHLPSP KAQ+YRV+ LY+GPLDDPYA+AIRNC
Sbjct: 313 ELTGKKLMKRVMQAWLPASDALLEMIVFHLPSPAKAQQYRVDTLYDGPLDDPYATAIRNC 372
Query: 359 DPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQ 418
DP+GPLM+YVSKMIPASDKGRF+AFGRVFSG V+TG KVRIMGPN++PGEKKDLYVK+VQ
Sbjct: 373 DPKGPLMVYVSKMIPASDKGRFFAFGRVFSGTVATGNKVRIMGPNFVPGEKKDLYVKTVQ 432
Query: 419 RTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVS 478
RTVIWMGKKQE+V+DVPCGNTVAMVGLDQFITKNATLT+EK VDAHPI+AMKFSVSPVV
Sbjct: 433 RTVIWMGKKQESVDDVPCGNTVAMVGLDQFITKNATLTDEKAVDAHPIKAMKFSVSPVVR 492
Query: 479 VAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFM 538
+V CK AS+LPKLVEGLKRLAKSDP+VVCTI E+GEH+IA G+LHLEIC+KDLQ+DFM
Sbjct: 493 KSVACKNASELPKLVEGLKRLAKSDPLVVCTIEESGEHVIAGVGQLHLEICIKDLQEDFM 552
Query: 539 NGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPM--------EEGLAEAI 590
GAEI PI+++RETV + S TVMSKSPNKHNRLYMEARP+ E L +AI
Sbjct: 553 GGAEIIVGPPIITYRETVTKNSCRTVMSKSPNKHNRLYMEARPLDKEDLQQDEPSLCKAI 612
Query: 591 DDGRIGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKD 650
DD RIGP+D+ K KILS+EFGWDKDLAKK+W FGPET GPN+LVD CKGVQYL+EIKD
Sbjct: 613 DDERIGPKDDIKERGKILSEEFGWDKDLAKKIWAFGPETKGPNLLVDMCKGVQYLSEIKD 672
Query: 651 SVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAK 710
SVVAGFQ ASKEG +A+EN+RGVCFE+CDV LH+D+IHRGGGQ+IPTARR YAA LTA
Sbjct: 673 SVVAGFQWASKEGALAEENMRGVCFELCDVTLHSDSIHRGGGQLIPTARRAMYAAQLTAS 732
Query: 711 PRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFN 770
PRL+EP+Y V+IQ P+ A+G +Y VLN + G + +E +R TPL N++ YLPV +SF F
Sbjct: 733 PRLMEPMYQVDIQVPKTAVGNVYGVLNSRNGELVEESERTGTPLSNLRFYLPVAKSFDFT 792
Query: 771 ESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDR 830
E LRA+T GQAFPQ +F HW + SDP + G+ AA + +IRK+KGLK+ + PLS++ED+
Sbjct: 793 EKLRAETSGQAFPQCIFHHWQTMRSDPFQEGSEAAKVITDIRKRKGLKDIITPLSDYEDK 852
Query: 831 L 831
L
Sbjct: 853 L 853
>gb|AAN31925.1| putative elongation factor [Arabidopsis thaliana]
Length = 665
Score = 1174 bits (3036), Expect = 0.0
Identities = 562/664 (84%), Positives = 615/664 (91%)
Query: 168 RVIESVNVVMATYEDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEK 227
RVIE+ NV+MATYED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGV E K
Sbjct: 2 RVIENANVIMATYEDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVVESK 61
Query: 228 MMNRLWGENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQ 287
MM RLWGENFFD +T+KW+ K+T +PTCKRGFVQFCYEPIKQII CMNDQKDKLWPML
Sbjct: 62 MMERLWGENFFDPATRKWSGKNTGSPTCKRGFVQFCYEPIKQIIATCMNDQKDKLWPMLA 121
Query: 288 KLGVNLKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPL 347
KLGV++K++EKEL GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP AQ+YRVENLYEGPL
Sbjct: 122 KLGVSMKNDEKELMGKPLMKRVMQTWLPASTALLEMMIFHLPSPHTAQRYRVENLYEGPL 181
Query: 348 DDPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPG 407
DD YA+AIRNCDP GPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTGMKVRIMGPNYIPG
Sbjct: 182 DDQYANAIRNCDPNGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGMKVRIMGPNYIPG 241
Query: 408 EKKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIR 467
EKKDLY KSVQRTVIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIR
Sbjct: 242 EKKDLYTKSVQRTVIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIR 301
Query: 468 AMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLE 527
AMKFSVSPVV VAV CKVASDLPKLVEGLKRLAKSDPMVVCT+ E+GEHI+A AGELHLE
Sbjct: 302 AMKFSVSPVVRVAVQCKVASDLPKLVEGLKRLAKSDPMVVCTMEESGEHIVAGAGELHLE 361
Query: 528 ICLKDLQDDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLA 587
ICLKDLQDDFM GAEI KSDP+VSFRETV ++S+ TVMSKSPNKHNRLYMEARPMEEGLA
Sbjct: 362 ICLKDLQDDFMGGAEIIKSDPVVSFRETVCDRSTRTVMSKSPNKHNRLYMEARPMEEGLA 421
Query: 588 EAIDDGRIGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNE 647
EAIDDGRIGPRD+PK KIL++EFGWDKDLAKK+W FGPETTGPNM+VD CKGVQYLNE
Sbjct: 422 EAIDDGRIGPRDDPKIRSKILAEEFGWDKDLAKKIWAFGPETTGPNMVVDMCKGVQYLNE 481
Query: 648 IKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAML 707
IKDSVVAGFQ ASKEGP+A+EN+RG+CFEVCDVVLH+DAIHRGGGQ+IPTARRV YA+ +
Sbjct: 482 IKDSVVAGFQWASKEGPLAEENMRGICFEVCDVVLHSDAIHRGGGQVIPTARRVIYASQI 541
Query: 708 TAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESF 767
TAKPRLLEPVY+VEIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF
Sbjct: 542 TAKPRLLEPVYMVEIQAPEGALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESF 601
Query: 768 QFNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEF 827
F+ LRA T GQAFPQ VFDHW+M+ SDPLEPGT A+ V +IRK+KGLKE + PLSEF
Sbjct: 602 GFSSQLRAATSGQAFPQCVFDHWEMMSSDPLEPGTQASVLVADIRKRKGLKEAMTPLSEF 661
Query: 828 EDRL 831
ED+L
Sbjct: 662 EDKL 665
>gb|AAK59516.2| putative elongation factor [Arabidopsis thaliana]
gi|29824421|gb|AAP04170.1| putative elongation factor
[Arabidopsis thaliana]
Length = 663
Score = 1172 bits (3031), Expect = 0.0
Identities = 561/663 (84%), Positives = 614/663 (91%)
Query: 169 VIESVNVVMATYEDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKM 228
VIE+ NV+MATYED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGV E KM
Sbjct: 1 VIENANVIMATYEDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVVESKM 60
Query: 229 MNRLWGENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQK 288
M RLWGENFFD +T+KW+ K+T +PTCKRGFVQFCYEPIKQII CMNDQKDKLWPML K
Sbjct: 61 MERLWGENFFDPATRKWSGKNTGSPTCKRGFVQFCYEPIKQIIATCMNDQKDKLWPMLAK 120
Query: 289 LGVNLKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLD 348
LGV++K++EKEL GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP AQ+YRVENLYEGPLD
Sbjct: 121 LGVSMKNDEKELMGKPLMKRVMQTWLPASTALLEMMIFHLPSPHTAQRYRVENLYEGPLD 180
Query: 349 DPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGE 408
D YA+AIRNCDP GPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTGMKVRIMGPNYIPGE
Sbjct: 181 DQYANAIRNCDPNGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGMKVRIMGPNYIPGE 240
Query: 409 KKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRA 468
KKDLY KSVQRTVIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRA
Sbjct: 241 KKDLYTKSVQRTVIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRA 300
Query: 469 MKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEI 528
MKFSVSPVV VAV CKVASDLPKLVEGLKRLAKSDPMVVCT+ E+GEHI+A AGELHLEI
Sbjct: 301 MKFSVSPVVRVAVQCKVASDLPKLVEGLKRLAKSDPMVVCTMEESGEHIVAGAGELHLEI 360
Query: 529 CLKDLQDDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAE 588
CLKDLQDDFM GAEI KSDP+VSFRETV ++S+ TVMSKSPNKHNRLYMEARPMEEGLAE
Sbjct: 361 CLKDLQDDFMGGAEIIKSDPVVSFRETVCDRSTRTVMSKSPNKHNRLYMEARPMEEGLAE 420
Query: 589 AIDDGRIGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEI 648
AIDDGRIGPRD+PK KIL++EFGWDKDLAKK+W FGPETTGPNM+VD CKGVQYLNEI
Sbjct: 421 AIDDGRIGPRDDPKIRSKILAEEFGWDKDLAKKIWAFGPETTGPNMVVDMCKGVQYLNEI 480
Query: 649 KDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLT 708
KDSVVAGFQ ASKEGP+A+EN+RG+CFEVCDVVLH+DAIHRGGGQ+IPTARRV YA+ +T
Sbjct: 481 KDSVVAGFQWASKEGPLAEENMRGICFEVCDVVLHSDAIHRGGGQVIPTARRVIYASQIT 540
Query: 709 AKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQ 768
AKPRLLEPVY+VEIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF
Sbjct: 541 AKPRLLEPVYMVEIQAPEGALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFG 600
Query: 769 FNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFE 828
F+ LRA T GQAFPQ VFDHW+M+ SDPLEPGT A+ V +IRK+KGLKE + PLSEFE
Sbjct: 601 FSSQLRAATSGQAFPQCVFDHWEMMSSDPLEPGTQASVLVADIRKRKGLKEAMTPLSEFE 660
Query: 829 DRL 831
D+L
Sbjct: 661 DKL 663
>dbj|BAC67668.1| elongation factor-2 [Cyanidioschyzon merolae]
Length = 846
Score = 1075 bits (2779), Expect = 0.0
Identities = 540/837 (64%), Positives = 649/837 (77%), Gaps = 9/837 (1%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD+ IRNMSVIAHVDHGKSTLTDSLVAAAGIIA E AGD R+TDTR DE ER ITIKS
Sbjct: 13 MDIPEQIRNMSVIAHVDHGKSTLTDSLVAAAGIIAIEAAGDTRLTDTRPDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISL++ DL+ K + +L+NLIDSPGHVDFSSEVTAALR+TDGALVVVDCV
Sbjct: 73 TGISLFFHYPP-DLELPKDSGDSRDFLVNLIDSPGHVDFSSEVTAALRVTDGALVVVDCV 131
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQAL ERIKPVLT+NK+DR FLEL L+AEE Y T RVIE+ NV++ATY
Sbjct: 132 EGVCVQTETVLRQALAERIKPVLTINKLDRAFLELQLEAEEMYQTFSRVIENANVILATY 191
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
+DA LGDVQV P KGTV+FSAGLHGW+FTLT FA+MYA KFGVD EKM RLWGEN+F+
Sbjct: 192 QDAALGDVQVSPAKGTVAFSAGLHGWAFTLTRFARMYAKKFGVDVEKMTQRLWGENYFNR 251
Query: 241 STKKWTNKHTSTP--TCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEK 298
TKKWT K T +R F +F +P+K+IIELCM+DQ + L +L L V+L +++K
Sbjct: 252 KTKKWTTKSTDAEGEQLERAFCEFVIKPVKKIIELCMSDQVEALEKLLSGLDVSLTNDDK 311
Query: 299 ELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNC 358
+L K LMKRV+Q WLPA ALLEM++ HLPSP KAQKYR E LYEGP+DD A+A+RNC
Sbjct: 312 QLRQKPLMKRVLQKWLPADQALLEMIVTHLPSPVKAQKYRTELLYEGPMDDVAATAMRNC 371
Query: 359 DPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQ 418
DP+GPLMLYVSKM+PASDKGRF AFGRVFSG + TGMKVRI GPNY PGEKKDL VK++Q
Sbjct: 372 DPKGPLMLYVSKMVPASDKGRFVAFGRVFSGTIRTGMKVRIYGPNYEPGEKKDLAVKNIQ 431
Query: 419 RTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVS 478
RT++ MG++ E V+ VP GNTV +VG+DQF+ K+ T+T+E+ A PI+ MK+SVSPVV
Sbjct: 432 RTLLMMGRRTEAVDSVPAGNTVGLVGVDQFLVKSGTITDEES--AFPIKNMKYSVSPVVR 489
Query: 479 VAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFM 538
VAV K SDLPKLVEGLKRLAKSDP+V T+ E+GEHIIA AGELHLEICLKDLQ+DFM
Sbjct: 490 VAVEPKNPSDLPKLVEGLKRLAKSDPLVEVTMEESGEHIIAGAGELHLEICLKDLQEDFM 549
Query: 539 NGAEITKSDPIVSFRETVL----EKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGR 594
NGAEI +P+VS+RETV ++ +SKSPNKHNRLY+ A P+ EG+A+AID+G+
Sbjct: 550 NGAEIRVGNPVVSYRETVEGVPDPLNTAVCLSKSPNKHNRLYIYADPLPEGVAQAIDEGK 609
Query: 595 IGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVA 654
I PRDEPK KIL DE+ D+D A+++WCF P+TTGPN+ +D K VQ+LNEIKDS VA
Sbjct: 610 ITPRDEPKARAKILKDEYNMDEDAARRIWCFAPDTTGPNLFMDRTKAVQFLNEIKDSCVA 669
Query: 655 GFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLL 714
Q A KEG + +E +R + F + DV LH DAIHRGGGQIIPT RR Y A L AKPRL
Sbjct: 670 AMQWACKEGVLCEEPMRNIGFNLVDVTLHADAIHRGGGQIIPTCRRCLYGAQLLAKPRLF 729
Query: 715 EPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLR 774
EP++LV+I PE A+G IY + ++KRG V +E QR TPL+ +KAYLPV+ESF F LR
Sbjct: 730 EPMFLVDITCPEQAVGSIYGLFSRKRGMVTEEQQRAGTPLWILKAYLPVVESFGFTAELR 789
Query: 775 AQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
+ T GQAFPQ++F HW++VP PLE G A R +KGLKE + +S F D+L
Sbjct: 790 SATSGQAFPQMMFSHWELVPGSPLETGNLAYDFCKATRLRKGLKESVPDISNFYDKL 846
>emb|CAB02985.1| Hypothetical protein F25H5.4 [Caenorhabditis elegans]
gi|3123205|sp|P29691|EF2_CAEEL Elongation factor 2
(EF-2) gi|17506493|ref|NP_492457.1| translation
Elongation FacTor (94.8 kD) (eft-2) [Caenorhabditis
elegans]
Length = 852
Score = 1056 bits (2730), Expect = 0.0
Identities = 528/845 (62%), Positives = 645/845 (75%), Gaps = 19/845 (2%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE ER ITIKS
Sbjct: 13 MDRKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGSKAGETRFTDTRKDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREG------------NKYLINLIDSPGHVDFSSEVTAALR 108
T ISL++E+ DL+ KGE + N +LINLIDSPGHVDFSSEVTAALR
Sbjct: 73 TAISLFFELEKKDLEFVKGENQFETVEVDGKKEKYNGFLINLIDSPGHVDFSSEVTAALR 132
Query: 109 ITDGALVVVDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQR 168
+TDGALVVVDCV GVCVQTETVLRQA+ ERIKPVL +NKMDR LEL L AEE + T QR
Sbjct: 133 VTDGALVVVDCVSGVCVQTETVLRQAIAERIKPVLFMNKMDRALLELQLGAEELFQTFQR 192
Query: 169 VIESVNVVMATY--EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEE 226
++E++NV++ATY +D +G + V P G V F +GLHGW+FTL FA+MYA KFGV +
Sbjct: 193 IVENINVIIATYGDDDGPMGPIMVDPSIGNVGFGSGLHGWAFTLKQFAEMYAGKFGVQVD 252
Query: 227 KMMNRLWGENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPML 286
K+M LWG+ FFD TKKW++ T T KRGF QF +PI + + MN +KDK ++
Sbjct: 253 KLMKNLWGDRFFDLKTKKWSS--TQTDESKRGFCQFVLDPIFMVFDAVMNIKKDKTAALV 310
Query: 287 QKLGVNLKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGP 346
+KLG+ L ++EK+L GK LMK M+ WLPA +L+M+ FHLPSP AQKYR+E LYEGP
Sbjct: 311 EKLGIKLANDEKDLEGKPLMKVFMRKWLPAGDTMLQMIAFHLPSPVTAQKYRMEMLYEGP 370
Query: 347 LDDPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIP 406
DD A AI+ CDP GPLM+Y+SKM+P SDKGRFYAFGRVFSGKV+TGMK RI GPNY+P
Sbjct: 371 HDDEAAVAIKTCDPNGPLMMYISKMVPTSDKGRFYAFGRVFSGKVATGMKARIQGPNYVP 430
Query: 407 GEKKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPI 466
G+K+DLY K++QRT++ MG+ E +ED+P GN +VG+DQ++ K T+T K DAH +
Sbjct: 431 GKKEDLYEKTIQRTILMMGRFIEPIEDIPSGNIAGLVGVDQYLVKGGTITTYK--DAHNM 488
Query: 467 RAMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHL 526
R MKFSVSPVV VAV K +DLPKLVEGLKRLAKSDPMV C E+GEHIIA AGELHL
Sbjct: 489 RVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKSDPMVQCIFEESGEHIIAGAGELHL 548
Query: 527 EICLKDLQDDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGL 586
EICLKDL++D + KSDP+VS+RETV +S+ +SKSPNKHNRL+ A+PM +GL
Sbjct: 549 EICLKDLEEDHAC-IPLKKSDPVVSYRETVQSESNQICLSKSPNKHNRLHCTAQPMPDGL 607
Query: 587 AEAIDDGRIGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLN 646
A+ I+ G + RDE K KIL++++ +D A+K+WCFGP+ TGPN+L+D KGVQYLN
Sbjct: 608 ADDIEGGTVNARDEFKARAKILAEKYEYDVTEARKIWCFGPDGTGPNLLMDVTKGVQYLN 667
Query: 647 EIKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAM 706
EIKDSVVAGFQ A++EG ++DEN+RGV F V DV LH DAIHRGGGQIIPTARRVFYA++
Sbjct: 668 EIKDSVVAGFQWATREGVLSDENMRGVRFNVHDVTLHADAIHRGGGQIIPTARRVFYASV 727
Query: 707 LTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIES 766
LTA+PRLLEPVYLVEIQ PE A+GGIY VLN++RGHVF+E Q TP++ VKAYLPV ES
Sbjct: 728 LTAEPRLLEPVYLVEIQCPEAAVGGIYGVLNRRRGHVFEESQVTGTPMFVVKAYLPVNES 787
Query: 767 FQFNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSE 826
F F LR+ TGGQAFPQ VFDHW ++P DPLE GT V++ RK+KGLKE + L
Sbjct: 788 FGFTADLRSNTGGQAFPQCVFDHWQVLPGDPLEAGTKPNQIVLDTRKRKGLKEGVPALDN 847
Query: 827 FEDRL 831
+ D++
Sbjct: 848 YLDKM 852
>gb|AAL85605.1| elongation factor 2 [Aedes aegypti]
Length = 844
Score = 1055 bits (2729), Expect = 0.0
Identities = 530/837 (63%), Positives = 647/837 (76%), Gaps = 11/837 (1%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE ER ITIKS
Sbjct: 13 MDRKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGAKAGETRFTDTRKDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNK----YLINLIDSPGHVDFSSEVTAALRITDGALVV 116
T IS+Y+E+ D DL + +K +LINLIDSPGHVDFSSEVTAALR+TDGALVV
Sbjct: 73 TAISMYFELEDQDLVFITNPDQRDKDCKGFLINLIDSPGHVDFSSEVTAALRVTDGALVV 132
Query: 117 VDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVV 176
VDCV GVCVQTETVLRQA+ ERIKPVL +NKMDR LEL LDAE+ Y T QR++E+VNV+
Sbjct: 133 VDCVSGVCVQTETVLRQAIAERIKPVLFMNKMDRALLELQLDAEDLYQTFQRIVENVNVI 192
Query: 177 MATYED--ALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWG 234
+ATY D +G+V+V P KG+V F +GLHGW+FTL FA+MYA+ F +D K+MNRLWG
Sbjct: 193 IATYNDDGGPMGEVRVDPSKGSVGFGSGLHGWAFTLKQFAEMYAAMFKIDVVKLMNRLWG 252
Query: 235 ENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLK 294
ENFF+ TKKW T KR FV + +PI ++ + MN + D++ +L+K+ V LK
Sbjct: 253 ENFFNPKTKKWAK--TKDDDNKRSFVMYVLDPIYKVFDAIMNYKTDEIPKLLEKIKVTLK 310
Query: 295 SEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASA 354
E+K+ GK L+K VM+SWLPA ALL+M+ HLPSP AQKYR+E LYEGP DD A A
Sbjct: 311 HEDKDKDGKNLLKVVMRSWLPAGEALLQMIAIHLPSPVVAQKYRMEMLYEGPHDDEAAVA 370
Query: 355 IRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYV 414
++NCDPEGPLM+YVSKM+P SDKGRFYAFGRVF+GKV+TG K RIMGPNY PG+K+DLY
Sbjct: 371 VKNCDPEGPLMMYVSKMVPTSDKGRFYAFGRVFAGKVATGQKCRIMGPNYTPGKKEDLYE 430
Query: 415 KSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVS 474
K++QRT++ MG+ E +EDVPCGN +VG+DQF+ K T++ K DAH ++ MKFSVS
Sbjct: 431 KAIQRTILMMGRYVEAIEDVPCGNICGLVGVDQFLVKTGTISTFK--DAHNMKVMKFSVS 488
Query: 475 PVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQ 534
PVV VAV K +DLPKLVEGLKRLAKSDPMV C I E+GEHIIA AGELHLEICLKDL+
Sbjct: 489 PVVRVAVEPKNPADLPKLVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELHLEICLKDLE 548
Query: 535 DDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGR 594
+D + KSDP+VS+RETV ++S +SKSPNKHNRL+M+A PM +GLAE ID+G
Sbjct: 549 EDHAC-IPLKKSDPVVSYRETVSDESDQMCLSKSPNKHNRLFMKAVPMPDGLAEDIDNGD 607
Query: 595 IGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVA 654
+ RD+ K + L++++ +D A+K+WCFGP+ TGPN++VD KGVQYLNEIKDSVVA
Sbjct: 608 VNSRDDFKVRARYLAEKYDYDVTEARKIWCFGPDGTGPNIVVDCTKGVQYLNEIKDSVVA 667
Query: 655 GFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLL 714
GFQ ASKEG +A+EN+R V F + DV LH DAIHRGGGQIIPTARRV YA+ +TA PR++
Sbjct: 668 GFQWASKEGVLAEENMRAVRFNIYDVTLHADAIHRGGGQIIPTARRVLYASYITAAPRIM 727
Query: 715 EPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLR 774
EPVYL EIQ PE A GGIY VLN++RGHVF+E Q TP++ VKAYLPV ESF F LR
Sbjct: 728 EPVYLCEIQCPEVAAGGIYDVLNRRRGHVFEEAQVVGTPMFVVKAYLPVNESFGFTADLR 787
Query: 775 AQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
+ TGGQAFPQ VFDHW ++P DP EPGT + V +IRK+KGLKE L LS++ D+L
Sbjct: 788 SNTGGQAFPQCVFDHWQILPGDPAEPGTKPYSVVQDIRKRKGLKEGLPDLSQYLDKL 844
>gb|AAK77225.1| elongation factor 2 [Aedes aegypti]
Length = 844
Score = 1055 bits (2728), Expect = 0.0
Identities = 529/837 (63%), Positives = 648/837 (77%), Gaps = 11/837 (1%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE ER ITIKS
Sbjct: 13 MDRKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGAKAGETRFTDTRKDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNK----YLINLIDSPGHVDFSSEVTAALRITDGALVV 116
T IS+Y+E+ D DL + +K +LINLIDSPGHVDFSSEVTAALR+TDGALVV
Sbjct: 73 TAISMYFELEDQDLVFITNPDQRDKDCKGFLINLIDSPGHVDFSSEVTAALRVTDGALVV 132
Query: 117 VDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVV 176
VDCV GVCVQTE+VLRQA+ ERIKPVL +NKMDR LEL LDAE+ Y T QR++E+VNV+
Sbjct: 133 VDCVSGVCVQTESVLRQAIAERIKPVLFMNKMDRALLELQLDAEDLYQTFQRIVENVNVI 192
Query: 177 MATYED--ALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWG 234
+ATY D +G+V+V P KG+V F +GLHGW+FTL FA+MYA+ F +D K+MNRLWG
Sbjct: 193 IATYNDDGGPMGEVRVDPSKGSVGFGSGLHGWAFTLKQFAEMYAAMFKIDVVKLMNRLWG 252
Query: 235 ENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLK 294
ENFF+ TKKW T KR FV + +PI ++ + MN + D++ +L+K+ V LK
Sbjct: 253 ENFFNPKTKKWAK--TKDDDNKRSFVMYVLDPIYKVFDAIMNYKTDEIPKLLEKIKVTLK 310
Query: 295 SEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASA 354
E+K+ GK L+K VM+SWLPA ALL+M+ HLPSP AQKYR+E LYEGP DD A A
Sbjct: 311 HEDKDKDGKNLLKVVMRSWLPAGEALLQMIAIHLPSPVVAQKYRMEMLYEGPHDDEAAVA 370
Query: 355 IRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYV 414
++NCDPEGPLM+YVSKM+P SDKGRFYAFGRVF+GKV+TG K RIMGPNY PG+K+DLY
Sbjct: 371 VKNCDPEGPLMMYVSKMVPTSDKGRFYAFGRVFAGKVATGQKCRIMGPNYTPGKKEDLYE 430
Query: 415 KSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVS 474
K++QRT++ MG+ E +EDVPCGN +VG+DQF+ K T++ K DAH ++ MKFSVS
Sbjct: 431 KAIQRTILMMGRYVEAIEDVPCGNICGLVGVDQFLVKTGTISTFK--DAHNMKVMKFSVS 488
Query: 475 PVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQ 534
PVV VAV K +DLPKLVEGLKRLAKSDPMV C I E+GEHIIA AGELHLEICLKDL+
Sbjct: 489 PVVRVAVEPKNPADLPKLVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELHLEICLKDLE 548
Query: 535 DDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGR 594
+D + KSDP+VS+RETV ++S +SKSPNKHNRL+M+A PM +GLAE ID+G
Sbjct: 549 EDHAC-IPLKKSDPVVSYRETVSDESDQMCLSKSPNKHNRLFMKAVPMPDGLAEDIDNGD 607
Query: 595 IGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVA 654
+ RD+ K + L++++ +D A+K+WCFGP+ TGPN++VD KGVQYLNEIKDSVVA
Sbjct: 608 VNSRDDFKVRARYLAEKYDYDVTEARKIWCFGPDGTGPNIVVDCTKGVQYLNEIKDSVVA 667
Query: 655 GFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLL 714
GFQ ASKEG +A+EN+R V F + DV LH DAIHRGGGQIIPTARRV YA+ +TA PR++
Sbjct: 668 GFQWASKEGVLAEENMRAVRFNIYDVTLHADAIHRGGGQIIPTARRVLYASYITAAPRIM 727
Query: 715 EPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLR 774
EPVYL EIQ PE A+GGIY VLN++RGHVF+E Q TP++ VKAYLPV ESF F LR
Sbjct: 728 EPVYLCEIQCPEVAVGGIYGVLNRRRGHVFEEAQVAGTPMFVVKAYLPVNESFGFTADLR 787
Query: 775 AQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
+ TGGQAFPQ VFDHW ++P DP EPGT + V +IRK+KGLKE L LS++ D+L
Sbjct: 788 SNTGGQAFPQCVFDHWQILPGDPAEPGTKPYSVVQDIRKRKGLKEGLPDLSQYLDKL 844
>emb|CAE70384.1| Hypothetical protein CBG16945 [Caenorhabditis briggsae]
Length = 852
Score = 1055 bits (2727), Expect = 0.0
Identities = 527/845 (62%), Positives = 644/845 (75%), Gaps = 19/845 (2%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE ER ITIKS
Sbjct: 13 MDRKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGAKAGETRFTDTRKDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREG------------NKYLINLIDSPGHVDFSSEVTAALR 108
T ISL++E+ DL KGE++ N +LINLIDSPGHVDFSSEVTAALR
Sbjct: 73 TAISLFFELDKKDLDFVKGEQQFETVEVDGKKEKYNGFLINLIDSPGHVDFSSEVTAALR 132
Query: 109 ITDGALVVVDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQR 168
+TDGALVVVDCV GVCVQTETVLRQA+ ERIKPVL +NKMDR LEL L AEE + T QR
Sbjct: 133 VTDGALVVVDCVSGVCVQTETVLRQAIAERIKPVLFMNKMDRALLELQLGAEELFQTFQR 192
Query: 169 VIESVNVVMATY--EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEE 226
++E++NV++ATY +D +G + V P G V F +GLHGW+FTL F++MYA KFGV +
Sbjct: 193 IVENINVIIATYGDDDGPMGPIMVDPSVGNVGFGSGLHGWAFTLKQFSEMYADKFGVQVD 252
Query: 227 KMMNRLWGENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPML 286
K+M LWG+ FFD TKKW+N T T KRGF QF +PI + + MN +KDK ++
Sbjct: 253 KLMKNLWGDRFFDLKTKKWSN--TQTDDAKRGFNQFVLDPIFMVFDAIMNLKKDKTAALV 310
Query: 287 QKLGVNLKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGP 346
+KLG+ L ++EK+L GK LMK M+ WLPA +L+M+ FHLPSP AQKYR+E LYEGP
Sbjct: 311 EKLGIKLANDEKDLEGKPLMKAFMRRWLPAGDTMLQMITFHLPSPVTAQKYRMEMLYEGP 370
Query: 347 LDDPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIP 406
DD A AI+ CDP GPLM+YVSKM+P SDKGRFYAFGRVFSGKV+TGMK RI GPNY+P
Sbjct: 371 HDDEAAVAIKTCDPNGPLMMYVSKMVPTSDKGRFYAFGRVFSGKVATGMKARIQGPNYVP 430
Query: 407 GEKKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPI 466
G+K+DLY K++QRT++ MG+ E +ED+P GN +VG+DQ++ K T+T K DAH +
Sbjct: 431 GKKEDLYEKTIQRTILMMGRYIEPIEDIPSGNIAGLVGVDQYLVKGGTITTYK--DAHNM 488
Query: 467 RAMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHL 526
R MKFSVSPVV VAV K +DLPKLVEGLKRLAKSDPMV C E+GEHIIA AGELHL
Sbjct: 489 RVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKSDPMVQCIFEESGEHIIAGAGELHL 548
Query: 527 EICLKDLQDDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGL 586
EICLKDL++D + KSDP+VS+RETV +S+ +SKSPNKHNRL+ A+PM +GL
Sbjct: 549 EICLKDLEEDHAC-IPLKKSDPVVSYRETVQAESNQICLSKSPNKHNRLHCTAQPMPDGL 607
Query: 587 AEAIDDGRIGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLN 646
A+ I+ G + RDE K KIL++++ +D A+K+WCFGP+ TGPN+L D KGVQYLN
Sbjct: 608 ADDIEGGTVNARDEFKARAKILAEKYEYDVTEARKIWCFGPDGTGPNLLFDVTKGVQYLN 667
Query: 647 EIKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAM 706
EIKDSVVAGFQ A++EG ++DEN+RGV F + DV LH DAIHRGGGQIIPTARRVFYA++
Sbjct: 668 EIKDSVVAGFQWATREGVLSDENMRGVRFNIHDVTLHADAIHRGGGQIIPTARRVFYASI 727
Query: 707 LTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIES 766
LTA+PR+LEPVYLVEIQ PE A+GGIY VLN++RGHVF+E Q TP++ VKAYLPV ES
Sbjct: 728 LTAEPRILEPVYLVEIQCPEAAVGGIYGVLNRRRGHVFEESQVTGTPMFVVKAYLPVNES 787
Query: 767 FQFNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSE 826
F F LR+ TGGQAFPQ VFDHW ++P DPLE GT V++ RK+KGLKE + L
Sbjct: 788 FGFTADLRSNTGGQAFPQCVFDHWQVLPGDPLEAGTKPNQIVLDTRKRKGLKEGIPALDN 847
Query: 827 FEDRL 831
+ D++
Sbjct: 848 YLDKM 852
>gb|AAK01430.1| elongation factor 2 [Aedes aegypti]
Length = 844
Score = 1054 bits (2726), Expect = 0.0
Identities = 529/837 (63%), Positives = 647/837 (77%), Gaps = 11/837 (1%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE ER ITIKS
Sbjct: 13 MDRKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGAKAGETRFTDTRKDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNK----YLINLIDSPGHVDFSSEVTAALRITDGALVV 116
T IS+Y+E+ D DL + +K +LINLIDSPGHVDFSSEVTAALR+TDGALVV
Sbjct: 73 TAISMYFELEDQDLVFITNPDQRDKDCKGFLINLIDSPGHVDFSSEVTAALRVTDGALVV 132
Query: 117 VDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVV 176
VDCV GVCVQTETVLRQA+ ERIKPVL +NKMDR LEL LDAE+ Y T QR++E+VNV+
Sbjct: 133 VDCVSGVCVQTETVLRQAIAERIKPVLFMNKMDRALLELQLDAEDLYQTFQRIVENVNVI 192
Query: 177 MATYED--ALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWG 234
+ATY D +G+V+V P KG+V F +GLHGW+FTL FA+MYA+ F +D K+MNRLWG
Sbjct: 193 IATYNDDGGPMGEVRVDPSKGSVGFGSGLHGWAFTLKQFAEMYAAMFKIDVVKLMNRLWG 252
Query: 235 ENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLK 294
ENFF+ KKW T KR FV + +PI ++ + MN + D++ +L+K+ V LK
Sbjct: 253 ENFFNPKIKKWAK--TKDDDNKRSFVMYVLDPIYKVFDAIMNYKTDEIPKLLEKIKVTLK 310
Query: 295 SEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASA 354
E+K+ GK L+K VM+SWLPA ALL+M+ HLPSP AQKYR+E LYEGP DD A A
Sbjct: 311 HEDKDKDGKNLLKVVMRSWLPAGEALLQMIAIHLPSPVVAQKYRMEMLYEGPHDDEAAVA 370
Query: 355 IRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYV 414
++NCDPEGPLM+YVSKM+P SDKGRFYAFGRVF+GKV+TG K RIMGPNY PG+K+DLY
Sbjct: 371 VKNCDPEGPLMMYVSKMVPTSDKGRFYAFGRVFAGKVATGQKCRIMGPNYTPGKKEDLYE 430
Query: 415 KSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVS 474
K++QRT++ MG+ E +EDVPCGN +VG+DQF+ K T++ K DAH ++ MKFSVS
Sbjct: 431 KAIQRTILMMGRYVEAIEDVPCGNICGLVGVDQFLVKTGTISTFK--DAHNMKVMKFSVS 488
Query: 475 PVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQ 534
PVV VAV K +DLPKLVEGLKRLAKSDPMV C I E+GEHIIA AGELHLEICLKDL+
Sbjct: 489 PVVRVAVEPKNPADLPKLVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELHLEICLKDLE 548
Query: 535 DDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGR 594
+D + KSDP+VS+RETV ++S +SKSPNKHNRL+M+A PM +GLAE ID+G
Sbjct: 549 EDHAC-IPLKKSDPVVSYRETVSDESDQMCLSKSPNKHNRLFMKAVPMPDGLAEDIDNGD 607
Query: 595 IGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVA 654
+ RD+ K + L++++ +D A+K+WCFGP+ TGPN++VD KGVQYLNEIKDSVVA
Sbjct: 608 VNSRDDFKVRARYLAEKYDYDVTEARKIWCFGPDGTGPNIVVDCTKGVQYLNEIKDSVVA 667
Query: 655 GFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLL 714
GFQ ASKEG +A+EN+R V F + DV LH DAIHRGGGQIIPTARRV YA+ +TA PR++
Sbjct: 668 GFQWASKEGVLAEENMRAVRFNIYDVTLHADAIHRGGGQIIPTARRVLYASYITAAPRIM 727
Query: 715 EPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLR 774
EPVYL EIQ PE A+GGIY VLN++RGHVF+E Q TP++ VKAYLPV ESF F LR
Sbjct: 728 EPVYLCEIQCPEVAVGGIYGVLNRRRGHVFEEAQVAGTPMFVVKAYLPVNESFGFTADLR 787
Query: 775 AQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
+ TGGQAFPQ VFDHW ++P DP EPGT + V +IRK+KGLKE L LS++ D+L
Sbjct: 788 SNTGGQAFPQCVFDHWQILPGDPAEPGTKPYSVVQDIRKRKGLKEGLPDLSQYLDKL 844
>emb|CAG83532.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50542892|ref|XP_499612.1| hypothetical protein
[Yarrowia lipolytica]
Length = 842
Score = 1053 bits (2723), Expect = 0.0
Identities = 519/833 (62%), Positives = 642/833 (76%), Gaps = 5/833 (0%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD N+RNMSVIAHVDHGKSTLTDSLV AGII+ AG+ R TDTR+DE ERGITIKS
Sbjct: 13 MDKVSNVRNMSVIAHVDHGKSTLTDSLVQKAGIISAAKAGEARFTDTRKDEQERGITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
T ISLY +M D D+K K + GN++L+NLIDSPGHVDFSSEVTAALR+TDGALVVVDC+
Sbjct: 73 TAISLYAQMDDEDVKEIKQKTVGNEFLVNLIDSPGHVDFSSEVTAALRVTDGALVVVDCI 132
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERIKPV +NK+DR LEL + E+ Y++ QR +ESVNV++ATY
Sbjct: 133 EGVCVQTETVLRQALGERIKPVCVINKVDRALLELQITKEDLYTSFQRTVESVNVIIATY 192
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
D LGD QVYPE+GTV+F++GLHGW+FT+ FA YA KFGVD EKMM RLWG+++F+
Sbjct: 193 VDKALGDCQVYPERGTVAFASGLHGWAFTVRQFAVRYAKKFGVDREKMMQRLWGDSYFNP 252
Query: 241 STKKWTNKHTST--PTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEK 298
TKKWTNK T R F F +PI +I MN +KD++ +L+KL +NLK++EK
Sbjct: 253 KTKKWTNKDTDADGKPLDRAFNMFVLDPIFRIFSAIMNFKKDEIPALLEKLEINLKTDEK 312
Query: 299 ELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNC 358
EL GKAL+K VM+ +LPA+ ALLEM++ HLPSP AQ YR + LYEGP+DDP+ I+NC
Sbjct: 313 ELEGKALLKVVMRKFLPAADALLEMIVIHLPSPITAQNYRADTLYEGPIDDPFGQGIKNC 372
Query: 359 DPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQ 418
DP LMLYVSKM+P SDKGRFYAFGRVF+G V +G KVRI GP+YIPG+KKDL+VK++Q
Sbjct: 373 DPNADLMLYVSKMVPTSDKGRFYAFGRVFAGTVKSGQKVRIQGPDYIPGQKKDLFVKAIQ 432
Query: 419 RTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVS 478
R V+ MG+ E +EDVP GN V +VG+DQF+ K+ TLT + AH ++ MKFSVSPVV
Sbjct: 433 RCVLMMGRFVEPIEDVPAGNIVGLVGVDQFLLKSGTLTTNEA--AHNLKVMKFSVSPVVQ 490
Query: 479 VAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFM 538
VAV K A+DLPKLVEGLKRL+KSDP V+ ISE+GEHI+A GELHLEICL DL+ D
Sbjct: 491 VAVEVKNANDLPKLVEGLKRLSKSDPCVLTYISESGEHIVACTGELHLEICLLDLEQDHA 550
Query: 539 NGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPR 598
G + KS P+VS+RETV +SS T +SKSPNKHNRLY+ A P++E ++ AI+ G+I PR
Sbjct: 551 -GVPLKKSPPVVSYRETVSAESSMTALSKSPNKHNRLYVVAVPLDEEVSLAIESGKISPR 609
Query: 599 DEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQI 658
D+ K ++L+D++GWD A+K+WCFGP+ TG N++VDT K VQYL EIKDSVVAGF
Sbjct: 610 DDFKARARVLADDYGWDVTEARKIWCFGPDGTGANVVVDTTKAVQYLAEIKDSVVAGFNW 669
Query: 659 ASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVY 718
A+KEGP+ +EN+R V + DV LH DAIHRG GQI+PT R V YAAML A+PR+ EPV+
Sbjct: 670 ATKEGPIFNENMRSVRVNIMDVTLHADAIHRGTGQIMPTMRSVTYAAMLLAEPRIQEPVF 729
Query: 719 LVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTG 778
LVEIQ PE+A+GGIYSVLN+KRG V E QRP TPL+ +KAYLPV ESF F LR TG
Sbjct: 730 LVEIQCPENAVGGIYSVLNKKRGQVVSEEQRPGTPLFTIKAYLPVNESFGFTGELRQATG 789
Query: 779 GQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
GQAFPQ+VFDHW+ + PL+P + A V E RK++G+KE + E+ D+L
Sbjct: 790 GQAFPQMVFDHWEAMSGSPLDPSSKPGAIVCETRKRRGMKENVPGYEEYYDKL 842
>emb|CAB58373.1| SPCP31B10.07 [Schizosaccharomyces pombe]
gi|19075363|ref|NP_587863.1| hypothetical protein
SPCP31B10.07 [Schizosaccharomyces pombe 972h-]
gi|12643989|sp|O14460|EF2_SCHPO Elongation factor 2
(EF-2)
Length = 842
Score = 1053 bits (2722), Expect = 0.0
Identities = 520/828 (62%), Positives = 635/828 (75%), Gaps = 5/828 (0%)
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
N+RNMSVIAHVDHGKSTLTDSLV AGII+ AGD R DTR DE ERG+TIKST ISL
Sbjct: 18 NVRNMSVIAHVDHGKSTLTDSLVQKAGIISAAKAGDARFMDTRADEQERGVTIKSTAISL 77
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
+ EM+D D+K+ K +G +L+NLIDSPGHVDFSSEVTAALR+TDGALVVVD +EGVCV
Sbjct: 78 FAEMTDDDMKDMKEPADGTDFLVNLIDSPGHVDFSSEVTAALRVTDGALVVVDTIEGVCV 137
Query: 126 QTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYEDALL 185
QTETVLRQALGERI+PV+ VNK+DR LEL + EE Y RV+ESVNVV++TY D +L
Sbjct: 138 QTETVLRQALGERIRPVVVVNKVDRALLELQISQEELYQNFARVVESVNVVISTYYDKVL 197
Query: 186 GDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDSSTKKW 245
GD QV+P+KGTV+F++GLHGW+FT+ FA YA KFG+D KMM RLWGEN+F+ TKKW
Sbjct: 198 GDCQVFPDKGTVAFASGLHGWAFTVRQFANRYAKKFGIDRNKMMQRLWGENYFNPKTKKW 257
Query: 246 TNKHTSTP--TCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKELSGK 303
+ T + +R F F +PI +I + MN +KD+++ +L KL V +K +EKEL GK
Sbjct: 258 SKSATDANGNSNQRAFNMFILDPIYRIFDAVMNSRKDEVFTLLSKLEVTIKPDEKELEGK 317
Query: 304 ALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDPEGP 363
AL+K VM+ +LPA+ AL+EM++ HLPSP AQ+YR E LYEGP+DD A IRNCD P
Sbjct: 318 ALLKVVMRKFLPAADALMEMIVLHLPSPKTAQQYRAETLYEGPMDDECAVGIRNCDANAP 377
Query: 364 LMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRTVIW 423
LM+YVSKM+P SD+GRFYAFGRVFSG V +G+KVRI GPNY+PG+K DL++K++QRTV+
Sbjct: 378 LMIYVSKMVPTSDRGRFYAFGRVFSGTVRSGLKVRIQGPNYVPGKKDDLFIKAIQRTVLM 437
Query: 424 MGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVAVTC 483
MG + E +ED P GN + +VG+DQF+ K+ TLT + AH ++ MKFSVSPVV VAV
Sbjct: 438 MGSRIEPIEDCPAGNIIGLVGVDQFLVKSGTLTTSEV--AHNMKVMKFSVSPVVQVAVEV 495
Query: 484 KVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNGAEI 543
K +DLPKLVEGLKRL+KSDP V+CT SE+GEHI+A AGELHLEICLKDLQ+D G +
Sbjct: 496 KNGNDLPKLVEGLKRLSKSDPCVLCTTSESGEHIVAGAGELHLEICLKDLQEDHA-GIPL 554
Query: 544 TKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDEPKN 603
S P+VS+RE+V E SS T +SKSPNKHNR++M A PM E L+ AI+ G + PRD+ K
Sbjct: 555 KISPPVVSYRESVSEPSSMTALSKSPNKHNRIFMTAEPMSEELSVAIETGHVNPRDDFKV 614
Query: 604 HLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIASKEG 663
+I++DEFGWD A+K+WCFGP+TTG N++VD K V YLNEIKDSVVA F ASKEG
Sbjct: 615 RARIMADEFGWDVTDARKIWCFGPDTTGANVVVDQTKAVAYLNEIKDSVVAAFAWASKEG 674
Query: 664 PMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLVEIQ 723
PM +ENLR F + DVVLH DAIHRGGGQIIPTARRV YA+ L A P + EPV+LVEIQ
Sbjct: 675 PMFEENLRSCRFNILDVVLHADAIHRGGGQIIPTARRVVYASTLLASPIIQEPVFLVEIQ 734
Query: 724 APEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFP 783
E+A+GGIYSVLN+KRGHVF E QR TPLYN+KAYLPV ESF F LR T GQAFP
Sbjct: 735 VSENAMGGIYSVLNKKRGHVFSEEQRVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFP 794
Query: 784 QLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
QLVFDHW + DPL+P + V E RK+KGLKE + +E+ DRL
Sbjct: 795 QLVFDHWSPMSGDPLDPTSKPGQIVCEARKRKGLKENVPDYTEYYDRL 842
>gb|AAL85604.1| elongation factor 2 [Aedes aegypti]
Length = 844
Score = 1051 bits (2718), Expect = 0.0
Identities = 529/837 (63%), Positives = 645/837 (76%), Gaps = 11/837 (1%)
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE ER ITIKS
Sbjct: 13 MDRKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGAKAGETRFTDTRKDEQERCITIKS 72
Query: 61 TGISLYYEMSDGDLKNFKGEREGNK----YLINLIDSPGHVDFSSEVTAALRITDGALVV 116
T IS+Y+E+ D DL + +K +LINLIDSPGHVDFSSEVTAALR+TDGALVV
Sbjct: 73 TAISMYFELEDQDLVFITNPDQRDKDCKGFLINLIDSPGHVDFSSEVTAALRVTDGALVV 132
Query: 117 VDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVV 176
VDCV GVCVQTETVLRQA+ ERIKPVL +NKMDR LEL LDAE+ Y T QR++E+VNV+
Sbjct: 133 VDCVSGVCVQTETVLRQAIAERIKPVLFMNKMDRALLELQLDAEDLYQTFQRIVENVNVI 192
Query: 177 MATYED--ALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWG 234
+ATY D +G+V+V P KG+V F +GLHGW+FTL FA+MYA+ F +D K+MNRLWG
Sbjct: 193 IATYNDDGGPMGEVRVDPSKGSVGFGSGLHGWAFTLKQFAEMYAAMFKIDVVKLMNRLWG 252
Query: 235 ENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLK 294
ENFF+ TKKW T KR FV + +PI ++ + MN + D++ +L+K+ V LK
Sbjct: 253 ENFFNPKTKKWAK--TKDDDNKRSFVMYVLDPIYKVFDAIMNYKTDEIPKLLEKIKVTLK 310
Query: 295 SEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASA 354
E+K+ GK L+K VM+SWLPA ALL+M+ HLPSP AQKYR+E LYEGP DD A
Sbjct: 311 HEDKDKDGKNLLKVVMRSWLPAGEALLQMIAIHLPSPVVAQKYRMEMLYEGPHDDEAAVC 370
Query: 355 IRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYV 414
+NCDPEGPLM+YVSKM+P SDKGRFYAFGRVF+GKV+TG K RIMGPNY PG+K+DLY
Sbjct: 371 SQNCDPEGPLMMYVSKMVPTSDKGRFYAFGRVFAGKVATGQKCRIMGPNYTPGKKEDLYE 430
Query: 415 KSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVS 474
K++QRT++ MG+ E +EDVPCGN +VG+DQF+ K T++ K DAH ++ MKFSVS
Sbjct: 431 KAIQRTILMMGRYVEAIEDVPCGNICGLVGVDQFLVKTGTISTFK--DAHNMKVMKFSVS 488
Query: 475 PVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQ 534
PVV VAV K +DLPKLVEGLKRLAKSDPMV C I E+GEHIIA AGELHLEICLKDL
Sbjct: 489 PVVRVAVEPKNPADLPKLVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELHLEICLKDLG 548
Query: 535 DDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGR 594
+D + KSDP+VS+RETV ++S +SKSPNKHNRL+M+A PM +GLAE ID+G
Sbjct: 549 EDHAC-IPLKKSDPVVSYRETVSDESDQMCLSKSPNKHNRLFMKAVPMPDGLAEDIDNGD 607
Query: 595 IGPRDEPKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVA 654
+ RD+ K + L++++ +D A+K+WCFGP+ TGPN++VD KGVQYLNEIKDSVVA
Sbjct: 608 VNSRDDFKVRARYLAEKYDYDVTEARKIWCFGPDGTGPNIVVDCTKGVQYLNEIKDSVVA 667
Query: 655 GFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLL 714
GFQ ASKEG +A+EN+R V F + DV LH DAIHRGGGQIIPTARRV YA+ +TA PR++
Sbjct: 668 GFQWASKEGVLAEENMRAVRFNIYDVTLHADAIHRGGGQIIPTARRVLYASYITAAPRIM 727
Query: 715 EPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLR 774
EPVYL EIQ PE A+GGIY VLN++RGHVF+E Q TP++ VKAYLPV ESF F LR
Sbjct: 728 EPVYLCEIQCPEVAVGGIYGVLNRRRGHVFEEAQVAGTPMFVVKAYLPVNESFGFTADLR 787
Query: 775 AQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
+ TGGQAFPQ VFDHW ++P DP EPGT + V +IRK+KGLKE L LS++ D+L
Sbjct: 788 SNTGGQAFPQCVFDHWQILPGDPAEPGTKPYSVVQDIRKRKGLKEGLPDLSQYLDKL 844
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,402,444,557
Number of Sequences: 2540612
Number of extensions: 59917898
Number of successful extensions: 164440
Number of sequences better than 10.0: 4528
Number of HSP's better than 10.0 without gapping: 2729
Number of HSP's successfully gapped in prelim test: 1799
Number of HSP's that attempted gapping in prelim test: 151909
Number of HSP's gapped (non-prelim): 6969
length of query: 831
length of database: 863,360,394
effective HSP length: 137
effective length of query: 694
effective length of database: 515,296,550
effective search space: 357615805700
effective search space used: 357615805700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146817.8


