

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146807.10 - phase: 0
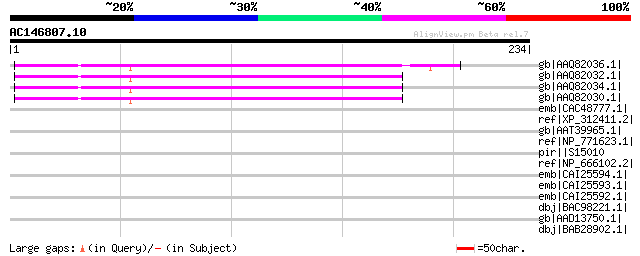
(234 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ82036.1| unknown [Pisum sativum] 150 4e-35
gb|AAQ82032.1| unknown [Pisum sativum] 149 6e-35
gb|AAQ82034.1| unknown [Pisum sativum] 149 8e-35
gb|AAQ82030.1| unknown [Pisum sativum] 148 1e-34
emb|CAC48777.1| putative cellulose synthase catalytic subunit pr... 37 0.59
ref|XP_312411.2| ENSANGP00000003008 [Anopheles gambiae str. PEST... 36 0.77
gb|AAT39965.1| hypothetical protein [Solanum demissum] 35 2.2
ref|NP_771623.1| hypothetical protein bll4983 [Bradyrhizobium ja... 35 2.2
pir||S15010 hypothetical protein B - Cryphonectria hypovirus 1 g... 34 3.8
ref|NP_666102.2| CDK5 regulatory subunit associated protein 2 [M... 33 6.5
emb|CAI25594.1| CDK5 regulatory subunit associated protein 2 [Mu... 33 6.5
emb|CAI25593.1| CDK5 regulatory subunit associated protein 2 [Mu... 33 6.5
emb|CAI25592.1| CDK5 regulatory subunit associated protein 2 [Mu... 33 6.5
dbj|BAC98221.1| mKIAA1633 protein [Mus musculus] 33 6.5
gb|AAD13750.1| unknown [Cryphonectria hypovirus 1] 33 6.5
dbj|BAB28902.1| unnamed protein product [Mus musculus] 33 6.5
>gb|AAQ82036.1| unknown [Pisum sativum]
Length = 546
Score = 150 bits (378), Expect = 4e-35
Identities = 85/209 (40%), Positives = 118/209 (55%), Gaps = 12/209 (5%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
AI IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP+S F D
Sbjct: 183 AISIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFMSLLPASGPFMDTQST 241
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + VK+++ CG+F NVPL+GT+G INYNP L++RQ GF
Sbjct: 242 LKWTQRVMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFI 301
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWNLM 180
M +P+ ++ + E AW + K LG + IA YT W
Sbjct: 302 MSRRPLEAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDW--- 358
Query: 181 LKKELVKV------RRELEEKDELLMRDS 203
+KK + + L+E+ L++ DS
Sbjct: 359 VKKRVETLLLPYDRMEPLQEQPPLILADS 387
>gb|AAQ82032.1| unknown [Pisum sativum]
Length = 562
Score = 149 bits (376), Expect = 6e-35
Identities = 78/177 (44%), Positives = 103/177 (58%), Gaps = 3/177 (1%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
A+ IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP S F D
Sbjct: 199 AVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQST 257
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF
Sbjct: 258 HKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFV 317
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQW 177
MK +P+ + + + + E AW + K LG + +A YT W
Sbjct: 318 MKGRPLEAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDW 374
>gb|AAQ82034.1| unknown [Pisum sativum]
Length = 562
Score = 149 bits (375), Expect = 8e-35
Identities = 78/177 (44%), Positives = 104/177 (58%), Gaps = 3/177 (1%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
A+ IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP S F D
Sbjct: 199 AVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQST 257
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF
Sbjct: 258 HKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFV 317
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQW 177
MK +P+ + ++ + + E AW + K LG + +A YT W
Sbjct: 318 MKGRPLEAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDW 374
>gb|AAQ82030.1| unknown [Pisum sativum]
Length = 562
Score = 148 bits (374), Expect = 1e-34
Identities = 78/177 (44%), Positives = 104/177 (58%), Gaps = 3/177 (1%)
Query: 3 AIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNSEN 60
A+ IF NPVPTLLADTY+AIH R KG G I CC+PLL RWF+S LP S F D
Sbjct: 199 AVSIFIGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQST 257
Query: 61 WSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFP 120
++QR+M+LT ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF
Sbjct: 258 HKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFV 317
Query: 121 MKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQW 177
MK +P+ + ++ + + E AW + K LG + +A YT W
Sbjct: 318 MKGRPLEAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDW 374
>emb|CAC48777.1| putative cellulose synthase catalytic subunit protein
[Sinorhizobium meliloti 1021]
gi|16264125|ref|NP_436917.1| putative cellulose synthase
catalytic subunit protein [Sinorhizobium meliloti 1021]
gi|25501792|pir||A95889 probable cellulose synthase
catalytic subunit protein [imported] - Sinorhizobium
meliloti (strain 1021) magaplasmid pSymB
Length = 664
Score = 36.6 bits (83), Expect = 0.59
Identities = 26/92 (28%), Positives = 41/92 (44%), Gaps = 7/92 (7%)
Query: 34 YILCCVPLLYRWFISHLPSSFHDNSENWSYSQRIMALTPNEVVWITPTAQVKEIITGCGD 93
+I+ +P++Y WF LP F D ++ S ++A + W+TPT + + T G
Sbjct: 380 FIVLLIPIVYLWF-GALPLYFTDVADYVSNQVPLLAAYFLLMFWLTPTRYLPLVSTAVGT 438
Query: 94 FLNVPLLGTRGGINYNPELAMRQFGFPMKAKP 125
F +L T +R FG P K P
Sbjct: 439 FSTFRMLPT------VLSSLVRPFGKPFKVTP 464
>ref|XP_312411.2| ENSANGP00000003008 [Anopheles gambiae str. PEST]
gi|55242238|gb|EAA07494.2| ENSANGP00000003008 [Anopheles
gambiae str. PEST]
Length = 1333
Score = 36.2 bits (82), Expect = 0.77
Identities = 19/52 (36%), Positives = 27/52 (51%)
Query: 182 KKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESEDHPTTSY 233
K +L K+R+ELEEK ELLM + +CN Y + + SE +Y
Sbjct: 250 KSKLRKLRQELEEKSELLMEVKEELDHKCNQYEKLRTESQDWYSESRRAAAY 301
>gb|AAT39965.1| hypothetical protein [Solanum demissum]
Length = 607
Score = 34.7 bits (78), Expect = 2.2
Identities = 48/193 (24%), Positives = 65/193 (32%), Gaps = 33/193 (17%)
Query: 13 VPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHL---------------------- 50
VP +LAD Y A+ G Y C LL W I H+
Sbjct: 214 VPMILADIYRAL-TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIGGHEE 272
Query: 51 ---PSSFHDNSENWSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGIN 107
SF E W + + LT +++VW E+I + L+G RG
Sbjct: 273 RIKDHSFPKGIEAWK--KYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRGVQP 330
Query: 108 YNPELAMRQFGFPMKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVR- 166
Y P MRQ G P + ++ P + E F W C S H V
Sbjct: 331 YMPLRVMRQLGRRQFLPPNEDIREFMYEFHPEIPLRKSEIF-KIWGG-CMLSSPHDRVED 388
Query: 167 --SGIAHEAYTQW 177
G +AY +W
Sbjct: 389 RTKGEVDQAYLEW 401
>ref|NP_771623.1| hypothetical protein bll4983 [Bradyrhizobium japonicum USDA 110]
gi|27353248|dbj|BAC50248.1| bll4983 [Bradyrhizobium
japonicum USDA 110]
Length = 518
Score = 34.7 bits (78), Expect = 2.2
Identities = 35/139 (25%), Positives = 51/139 (36%), Gaps = 13/139 (9%)
Query: 45 WFISHLPSSFHDNSENWSYSQRIMALTPNEVVWITPTA-QVKEIITGCGDFLNVPLLGTR 103
W + + HDN + + + ++ W A Q+K I TG GD +N+ + T
Sbjct: 288 WGLFQASVAAHDNHVGYYGASEVTGHPEDKWGWAVQLALQIKNIPTGAGDVINLSGVYTD 347
Query: 104 GGINYN-PELAMRQFGFPMKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKH 162
G YN ELA A ++ S Y S G +A G + + K
Sbjct: 348 GASRYNFQELA---------ATSYSMFGSSNAAYQSIGFAGVSDAVFGPGGGL--ELTKT 396
Query: 163 LGVRSGIAHEAYTQWNLML 181
G R H WN L
Sbjct: 397 YGFRGAYTHNWSPYWNTAL 415
>pir||S15010 hypothetical protein B - Cryphonectria hypovirus 1
gi|9626820|ref|NP_041091.1| ORF B [Cryphonectria
hypovirus 1] gi|331159|gb|AAA67458.1| ORF B
Length = 3165
Score = 33.9 bits (76), Expect = 3.8
Identities = 25/77 (32%), Positives = 37/77 (47%), Gaps = 12/77 (15%)
Query: 134 FFYYSNAPTG---QREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWNLMLKKELVKVRR 190
F+ + PTG +E FI W++ RK H+ G++HEA ELV+ R
Sbjct: 239 FWNEKDIPTGLKLSKEGFIKLWAQKSRKWQDHMARSIGLSHEAAV--------ELVRATR 290
Query: 191 ELEEKDELL-MRDSKRA 206
E K L+ M ++K A
Sbjct: 291 VNEAKPHLVPMEEAKEA 307
>ref|NP_666102.2| CDK5 regulatory subunit associated protein 2 [Mus musculus]
Length = 1822
Score = 33.1 bits (74), Expect = 6.5
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 178 NLMLKKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESED 227
N+ LK E+ ++REL+EKD+LL++ SK A GS+ + ED
Sbjct: 102 NIELKVEVESLKRELQEKDQLLVKASKAVES----LAERGGSEVQRVKED 147
>emb|CAI25594.1| CDK5 regulatory subunit associated protein 2 [Mus musculus]
Length = 1778
Score = 33.1 bits (74), Expect = 6.5
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 178 NLMLKKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESED 227
N+ LK E+ ++REL+EKD+LL++ SK A GS+ + ED
Sbjct: 102 NIELKVEVESLKRELQEKDQLLVKASKAVES----LAERGGSEVQRVKED 147
>emb|CAI25593.1| CDK5 regulatory subunit associated protein 2 [Mus musculus]
Length = 1788
Score = 33.1 bits (74), Expect = 6.5
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 178 NLMLKKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESED 227
N+ LK E+ ++REL+EKD+LL++ SK A GS+ + ED
Sbjct: 102 NIELKVEVESLKRELQEKDQLLVKASKAVES----LAERGGSEVQRVKED 147
>emb|CAI25592.1| CDK5 regulatory subunit associated protein 2 [Mus musculus]
Length = 1820
Score = 33.1 bits (74), Expect = 6.5
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 178 NLMLKKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESED 227
N+ LK E+ ++REL+EKD+LL++ SK A GS+ + ED
Sbjct: 102 NIELKVEVESLKRELQEKDQLLVKASKAVES----LAERGGSEVQRVKED 147
>dbj|BAC98221.1| mKIAA1633 protein [Mus musculus]
Length = 1855
Score = 33.1 bits (74), Expect = 6.5
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 178 NLMLKKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESED 227
N+ LK E+ ++REL+EKD+LL++ SK A GS+ + ED
Sbjct: 135 NIELKVEVESLKRELQEKDQLLVKASKAVES----LAERGGSEVQRVKED 180
>gb|AAD13750.1| unknown [Cryphonectria hypovirus 1]
Length = 3164
Score = 33.1 bits (74), Expect = 6.5
Identities = 27/89 (30%), Positives = 43/89 (47%), Gaps = 15/89 (16%)
Query: 125 PINLATSPE---FFYYSNAPTG---QREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWN 178
PI+L T+ + F+ + PTG +E FI W++ RK H+ G++H A
Sbjct: 227 PISLLTNRQHARFWNDKDVPTGLKLSKEGFIKLWAQKSRKWQDHMARAIGLSHAA----- 281
Query: 179 LMLKKELVKVRRELEEKDELL-MRDSKRA 206
ELV+ + E K L+ M ++K A
Sbjct: 282 ---TAELVRATKVNEAKPHLVPMEEAKEA 307
>dbj|BAB28902.1| unnamed protein product [Mus musculus]
Length = 294
Score = 33.1 bits (74), Expect = 6.5
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 178 NLMLKKELVKVRRELEEKDELLMRDSKRARGRCNFYARYCGSDSESESED 227
N+ LK E+ ++REL+EKD+LL++ SK A GS+ + ED
Sbjct: 102 NIELKVEVESLKRELQEKDQLLVKASKAVES----LAERGGSEVQRVKED 147
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 440,462,854
Number of Sequences: 2540612
Number of extensions: 19188987
Number of successful extensions: 45480
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 45462
Number of HSP's gapped (non-prelim): 20
length of query: 234
length of database: 863,360,394
effective HSP length: 124
effective length of query: 110
effective length of database: 548,324,506
effective search space: 60315695660
effective search space used: 60315695660
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146807.10


