

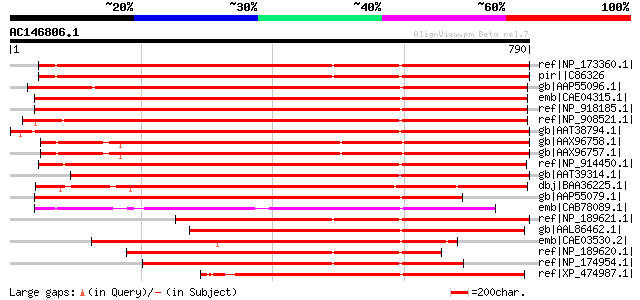
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146806.1 - phase: 0
(790 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_173360.1| hAT dimerisation domain-containing protein [Ara... 632 e-179
pir||C86326 hypothetical protein T29M8.13 [imported] - Arabidops... 632 e-179
gb|AAP55096.1| putative transposase [Oryza sativa (japonica cult... 618 e-175
emb|CAE04315.1| OSJNBb0016D16.6 [Oryza sativa (japonica cultivar... 616 e-175
ref|NP_918185.1| putative transposase [Oryza sativa (japonica cu... 614 e-174
ref|NP_908521.1| putative transposable element Tip100 protein [O... 613 e-174
gb|AAT38794.1| putative hAT family dimerisation domain containin... 603 e-171
gb|AAX96758.1| hAT family dimerisation domain, putative [Oryza s... 598 e-169
gb|AAX96757.1| hAT family dimerisation domain, putative [Oryza s... 597 e-169
ref|NP_914450.1| putative transposable element [Oryza sativa (ja... 585 e-165
gb|AAT39314.1| putative transposase [Solanum demissum] 571 e-161
dbj|BAA36225.1| transposase [Ipomoea purpurea] 553 e-156
gb|AAP55079.1| putative transposase [Oryza sativa (japonica cult... 517 e-145
emb|CAB78089.1| putative protein [Arabidopsis thaliana] gi|45388... 508 e-142
ref|NP_189621.1| hAT dimerisation domain-containing protein [Ara... 499 e-139
gb|AAL86462.1| putative transposase protein, 5'-partial [Oryza s... 465 e-129
emb|CAE03530.2| OSJNBa0061C06.15 [Oryza sativa (japonica cultiva... 427 e-118
ref|NP_189620.1| hypothetical protein [Arabidopsis thaliana] 421 e-116
ref|NP_174954.1| hypothetical protein [Arabidopsis thaliana] 409 e-112
ref|XP_474987.1| OSJNBa0065B15.7 [Oryza sativa (japonica cultiva... 394 e-108
>ref|NP_173360.1| hAT dimerisation domain-containing protein [Arabidopsis thaliana]
Length = 769
Score = 632 bits (1630), Expect = e-179
Identities = 345/756 (45%), Positives = 491/756 (64%), Gaps = 15/756 (1%)
Query: 44 LETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKNWFNLY 103
L +DP R I SYHP+ +DEVR+ YL IR P + RRFN WF+LY
Sbjct: 20 LPSDPAKRKSILSYHPNQRDEVRREYL-IRGPCQPRGHKFKQIAIGKVLRRFNPKWFDLY 78
Query: 104 -DWLEYSESKNLAFCLPCFLFKN-VSNYGG-DHFVGDGFGDWKNPRKLANHA--TSNNSH 158
DWLEYS K AFCL C+LF++ N GG D F+ GF W +L H N+ H
Sbjct: 79 GDWLEYSVEKEKAFCLYCYLFRDQTGNQGGSDSFLSTGFCSWNKADRLDQHVGLDVNSFH 138
Query: 159 VDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEAD 218
+ LM QSIK A QT + +Y + + S+ +++LL GL FRG DE++
Sbjct: 139 NNAKRKCEDLMRQGQSIKHALHKQTDVVKNDYRIWLNASIDVSRHLLHQGLPFRGHDESE 198
Query: 219 DSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIVCD 277
+S KG F+ELL T +N +L +AP N+ MT P IQKD+ + E+TR I+ +
Sbjct: 199 ESTNKGNFVELLKYTAGQNEVVKKVVLKNAPKNNQMTSPPIQKDIVHCFSKEVTRSIIEE 258
Query: 278 IADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETM 337
+ +DVF +L+DES D S +EQMAVV R+VD G+VKERF+G+ V+ETS+ SLK A++++
Sbjct: 259 MDNDVFGLLVDESADASDKEQMAVVFRFVDKYGVVKERFIGVIHVQETSSLSLKSAIDSL 318
Query: 338 LSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKT 397
+ GLS +RGQGYDGASNM+G F GL++LI E+ SA+YVHCFAHQLQL ++A AK
Sbjct: 319 FAKYGLSLKKLRGQGYDGASNMKGEFNGLRSLILKESSSAYYVHCFAHQLQLVVMAVAKK 378
Query: 398 HKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARA 457
H V FF +++L+N + AS KR++ +R+ + + I GEI+TG+GLNQ+ S+ R
Sbjct: 379 HVEVGEFFYMISVLLNVVGASCKRKDKIREIHRQKVEEKISNGEIKTGTGLNQELSLQRP 438
Query: 458 GDTRWGSHFRTLTSLMTLYGAIV---EVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFML 514
G+TRWGSH++TL L L+ +IV E I + G D + K + +L +FDF+F L
Sbjct: 439 GNTRWGSHYKTLLRLEELFSSIVIVLEYIQDEGTDTT--KRQQAYGILKYFHTFDFVFYL 496
Query: 515 YMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKH 574
+M+ ++G+T+ LS ALQR+DQD+LNAISLV TK QLQ++R++GW+ +++V + K+
Sbjct: 497 ELMLLVMGLTDSLSKALQRKDQDILNAISLVKTTKCQLQKVRDDGWDAFMAKVSSFSEKN 556
Query: 575 EIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELL 634
+ M+ +++ ++PR+ + ++NLHHYK DC ++VLD+QLQE N RFDE N+ELL
Sbjct: 557 NTGMLKMEEEFVDSRRPRK---KTGITNLHHYKVDCFYTVLDMQLQEFNDRFDEVNSELL 613
Query: 635 QCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLK 694
C+S LSP SF FD + L+R+ E YP+DF V L QL Y+ NVK+D +F LK
Sbjct: 614 ICMSSLSPIDSFRQFDKSMLVRLTEFYPDDFSFVERRSLDHQLEIYLDNVKNDERFTDLK 673
Query: 695 GLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQW 754
L DL ++VET K + LV++LLK++L+LPVATA+VER FSAM VK+ LRN++GD +
Sbjct: 674 CLGDLARVMVETRKHLSHPLVYRLLKVSLILPVATATVERCFSAMNFVKTTLRNRIGDMF 733
Query: 755 LNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
L+D LV FIE+ L T++ + ++ FQ M +RR L
Sbjct: 734 LSDCLVCFIEKQALNTVTNESVIKRFQDMSERRVHL 769
>pir||C86326 hypothetical protein T29M8.13 [imported] - Arabidopsis thaliana
gi|8954063|gb|AAF82236.1| Contains similarity to a
transposable element Tip100 protein for transposase from
Ipomoea purpurea gb|4063769 and is a member of the
transmembrane 4 family PF|00335. [Arabidopsis thaliana]
Length = 811
Score = 632 bits (1630), Expect = e-179
Identities = 345/756 (45%), Positives = 491/756 (64%), Gaps = 15/756 (1%)
Query: 44 LETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKNWFNLY 103
L +DP R I SYHP+ +DEVR+ YL IR P + RRFN WF+LY
Sbjct: 62 LPSDPAKRKSILSYHPNQRDEVRREYL-IRGPCQPRGHKFKQIAIGKVLRRFNPKWFDLY 120
Query: 104 -DWLEYSESKNLAFCLPCFLFKN-VSNYGG-DHFVGDGFGDWKNPRKLANHA--TSNNSH 158
DWLEYS K AFCL C+LF++ N GG D F+ GF W +L H N+ H
Sbjct: 121 GDWLEYSVEKEKAFCLYCYLFRDQTGNQGGSDSFLSTGFCSWNKADRLDQHVGLDVNSFH 180
Query: 159 VDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEAD 218
+ LM QSIK A QT + +Y + + S+ +++LL GL FRG DE++
Sbjct: 181 NNAKRKCEDLMRQGQSIKHALHKQTDVVKNDYRIWLNASIDVSRHLLHQGLPFRGHDESE 240
Query: 219 DSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIVCD 277
+S KG F+ELL T +N +L +AP N+ MT P IQKD+ + E+TR I+ +
Sbjct: 241 ESTNKGNFVELLKYTAGQNEVVKKVVLKNAPKNNQMTSPPIQKDIVHCFSKEVTRSIIEE 300
Query: 278 IADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETM 337
+ +DVF +L+DES D S +EQMAVV R+VD G+VKERF+G+ V+ETS+ SLK A++++
Sbjct: 301 MDNDVFGLLVDESADASDKEQMAVVFRFVDKYGVVKERFIGVIHVQETSSLSLKSAIDSL 360
Query: 338 LSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKT 397
+ GLS +RGQGYDGASNM+G F GL++LI E+ SA+YVHCFAHQLQL ++A AK
Sbjct: 361 FAKYGLSLKKLRGQGYDGASNMKGEFNGLRSLILKESSSAYYVHCFAHQLQLVVMAVAKK 420
Query: 398 HKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARA 457
H V FF +++L+N + AS KR++ +R+ + + I GEI+TG+GLNQ+ S+ R
Sbjct: 421 HVEVGEFFYMISVLLNVVGASCKRKDKIREIHRQKVEEKISNGEIKTGTGLNQELSLQRP 480
Query: 458 GDTRWGSHFRTLTSLMTLYGAIV---EVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFML 514
G+TRWGSH++TL L L+ +IV E I + G D + K + +L +FDF+F L
Sbjct: 481 GNTRWGSHYKTLLRLEELFSSIVIVLEYIQDEGTDTT--KRQQAYGILKYFHTFDFVFYL 538
Query: 515 YMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKH 574
+M+ ++G+T+ LS ALQR+DQD+LNAISLV TK QLQ++R++GW+ +++V + K+
Sbjct: 539 ELMLLVMGLTDSLSKALQRKDQDILNAISLVKTTKCQLQKVRDDGWDAFMAKVSSFSEKN 598
Query: 575 EIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELL 634
+ M+ +++ ++PR+ + ++NLHHYK DC ++VLD+QLQE N RFDE N+ELL
Sbjct: 599 NTGMLKMEEEFVDSRRPRK---KTGITNLHHYKVDCFYTVLDMQLQEFNDRFDEVNSELL 655
Query: 635 QCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLK 694
C+S LSP SF FD + L+R+ E YP+DF V L QL Y+ NVK+D +F LK
Sbjct: 656 ICMSSLSPIDSFRQFDKSMLVRLTEFYPDDFSFVERRSLDHQLEIYLDNVKNDERFTDLK 715
Query: 695 GLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQW 754
L DL ++VET K + LV++LLK++L+LPVATA+VER FSAM VK+ LRN++GD +
Sbjct: 716 CLGDLARVMVETRKHLSHPLVYRLLKVSLILPVATATVERCFSAMNFVKTTLRNRIGDMF 775
Query: 755 LNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
L+D LV FIE+ L T++ + ++ FQ M +RR L
Sbjct: 776 LSDCLVCFIEKQALNTVTNESVIKRFQDMSERRVHL 811
>gb|AAP55096.1| putative transposase [Oryza sativa (japonica cultivar-group)]
gi|37537014|ref|NP_922809.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
gi|19225003|gb|AAL86479.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
Length = 811
Score = 618 bits (1593), Expect = e-175
Identities = 334/765 (43%), Positives = 486/765 (62%), Gaps = 13/765 (1%)
Query: 28 APNSTNEGVKVVDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSD 87
A N + D L+ DPG+R PI+SY + QD VR+AY+ +Q ++ +P +
Sbjct: 51 ATNEEQDAPPEYDADHLQYDPGLRSPIASYDVNEQDAVRRAYILKGPNQCYAHD-FPVRE 109
Query: 88 FRGTRRRFNKNWFNLYDWLEYSESKNLAFCLPCFLFKNVSNYGGDHFVGDGFGDWKNPRK 147
G +R FN WF+ Y WLEYS +K+ FC+ C+LF + G +FV DG+ +W
Sbjct: 110 IYGKKRHFNFVWFHKYQWLEYSVAKDAGFCMVCYLFGS----GTSNFVTDGWRNWNKADA 165
Query: 148 LANHATSNNSHVDCVHMGYAL-MNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLR 206
L H S + Y L ++ SI V + + + Y R+ SL +YLL
Sbjct: 166 LDKHVGGITSAHNKAQEKYNLFVSGGPSIDNVIVKVSNESEIHYKARLTYSLRCLRYLLN 225
Query: 207 CGLAFRGSDEADDSLYKGPFLELLDTLKENNSDV-ATILDSAPGNSLMTCPKIQKDLASA 265
GLAFRG DE+++S +G F+ELL L ENN +V +L +APGN ++TC IQ+++
Sbjct: 226 QGLAFRGHDESEESSNRGNFIELLKWLAENNKEVDRLVLKNAPGNCILTCSSIQREIIHC 285
Query: 266 CACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKET 325
CA E T+ I+ ++ DD + +L DE D+S +EQ+A+ +RY+D G V ERFLG+ V T
Sbjct: 286 CADETTKRIIEELGDDHYAILADECSDLSHKEQLALCVRYIDKLGRVCERFLGVVHVAST 345
Query: 326 SAKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAH 385
++ +LK A+ T+LS + LS S IRGQGYDGASNM+G+ GLKTLI E+PSA+Y+HCFAH
Sbjct: 346 TSAALKKAILTLLSDHHLSPSQIRGQGYDGASNMKGKINGLKTLIMKESPSAYYIHCFAH 405
Query: 386 QLQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETG 445
QLQL LV+ AK + FF +V+ L+N + S KR +MLRD + + + +E GEIE+G
Sbjct: 406 QLQLVLVSVAKGNDDCVWFFTQVSHLLNIVGTSCKRHDMLRDVRAQKIMEALELGEIESG 465
Query: 446 SGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPS-FDKFGETVLLLDV 504
GLNQ+ +AR GDTRWGSH++T+ ++ +Y I EV++ +G DP+ D + ++
Sbjct: 466 VGLNQEMGLARPGDTRWGSHYKTILHIIGMYPTIHEVLITLGKDPTQRDDWPRIHAVVGA 525
Query: 505 LQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELI 564
+SFDF+F ++M+ ILG TN+L L LQ+RDQD++NA+SLV K+++Q++R+EGWEE
Sbjct: 526 FESFDFVFSAHLMLVILGYTNELCLCLQKRDQDIVNAMSLVTLAKERMQKLRSEGWEEFF 585
Query: 565 -SRVVTICTKHEIDVPDMDAPYM-EGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQEL 622
VV+ C KH I VP +D Y+ G+ PR P +N H++ + V+D QEL
Sbjct: 586 QGTVVSFCNKHSIQVPTLDGKYVPHGRSPRFYP---DQTNDDHFRREVYIGVIDKISQEL 642
Query: 623 NARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVR 682
N+RFDE N ELL C+S L+P SF+++D ++L++A+ YP +F + + L QL ++
Sbjct: 643 NSRFDEVNMELLICMSALNPFNSFASYDAQQVLKLAKFYPKEFSTMDLIRLEMQLGTFID 702
Query: 683 NVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIV 742
+++ D +F L+ L++L LVETNK + V+ LLKL L+LPVATASVERVFSA+ +V
Sbjct: 703 DMRRDERFKGLETLAELSIKLVETNKHVLYDWVYLLLKLVLILPVATASVERVFSALSVV 762
Query: 743 KSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRR 787
KS LRN M D+ LND L+TFIERDV +S + I +F M RR
Sbjct: 763 KSKLRNSMCDKLLNDCLITFIERDVFSQVSEEDIKKNFMSMKTRR 807
>emb|CAE04315.1| OSJNBb0016D16.6 [Oryza sativa (japonica cultivar-group)]
gi|50928319|ref|XP_473687.1| OSJNBb0016D16.6 [Oryza
sativa (japonica cultivar-group)]
Length = 897
Score = 616 bits (1589), Expect = e-175
Identities = 331/755 (43%), Positives = 481/755 (62%), Gaps = 8/755 (1%)
Query: 38 VVDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNK 97
+ D LL DPG R I +Y + ++ V + Y+ + QP S+ +P G RRF
Sbjct: 131 IYDIDLLCHDPGRRIAIKNYDVNERNSVIRGYIALGPCQPRSHN-FPIRKIGGKPRRFLP 189
Query: 98 NWFNLYDWLEYSESKNLAFCLPCFLFKN-VSNYGGDHFVGDGFGDWKNPRKLANHATSNN 156
+W++ + WLEYS ++ AFC C+LFK+ ++N GGD FV GF +W +++A H
Sbjct: 190 SWYDEFKWLEYSVEQDAAFCFICYLFKHKINNSGGDAFVNRGFRNWHMRKRIAKHVGGMT 249
Query: 157 SHVDCVHMGYA-LMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
S + Y + P +I +F +Q Y R+ S+ K+LLR GLA RG
Sbjct: 250 SFHNVAQDKYNHFLAPKTNIVESFAATNEQDKARYMARLTYSMKCLKFLLRQGLAARGHY 309
Query: 216 EADDSLYKGPFLELLDTLKENNSDVA-TILDSAPGNSLMTCPKIQKDLASACACEITREI 274
E++ SL KG FLE+L L EN +V +L++AP N +T P+IQK +A+ CA E T+ I
Sbjct: 310 ESEKSLNKGNFLEMLSMLAENFEEVGKVVLNNAPKNCKLTAPEIQKQIANCCAKETTKLI 369
Query: 275 VCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDAL 334
+ D+ D+ F +L DES DV +EQ+A+ LRYVD G V ERFLG+ V+ T++ +LK A+
Sbjct: 370 MEDLGDEYFAILADESSDVYQKEQLALCLRYVDKKGRVVERFLGVVHVENTTSLTLKAAI 429
Query: 335 ETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVAC 394
E++L + LS S +RGQGYDGASNM+G GLK LI +E PSA+YVHCFAHQLQL LVA
Sbjct: 430 ESLLMEHSLSLSKVRGQGYDGASNMKGHANGLKKLIMDECPSAYYVHCFAHQLQLTLVAV 489
Query: 395 AKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSI 454
AK + FF ++ L+N + S K+ EMLR Q + + ++ G+IETG GLNQ+ +
Sbjct: 490 AKENPDCVWFFEQLRFLLNLLGNSCKKTEMLRVAQAQRIVEELDLGDIETGKGLNQEMGL 549
Query: 455 ARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD-KFGETVLLLDVLQSFDFIFM 513
R DTRWGSH++T+ +++LY +I +V++ VG D SF + +L + QSF+F+FM
Sbjct: 550 GRPADTRWGSHYKTVMHVLSLYPSIQKVLIMVGKDRSFGAECANAQTVLTIFQSFEFVFM 609
Query: 514 LYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNE-GWEELISRVVTICT 572
++M +LG T+DL+ ALQ+RDQD++NA+ L+ TK QLQ++R + GW++ + V + C
Sbjct: 610 AHLMQTVLGFTSDLNHALQKRDQDIVNAVGLILLTKFQLQQLREDPGWDDFLQEVQSFCV 669
Query: 573 KHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTE 632
KH+I +PDMD+ Y + RR + N+H + D SV+D QLQELN RFDE NT+
Sbjct: 670 KHKIKIPDMDSFYRPVGRDRRF--FIKIKNMHRFHVDMFLSVIDRQLQELNERFDEVNTD 727
Query: 633 LLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAK 692
LL C++ SP +F+++D +KL+++A YPNDF L L ++ ++ D +F K
Sbjct: 728 LLLCMAAFSPIDNFASWDKDKLIKLARFYPNDFSSTEMNHLPSALKLFLTEMRIDERFRK 787
Query: 693 LKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGD 752
+K L++L ++VET +V+KLLKL L+LPVATASVER+FSAM VK+ LRN+MGD
Sbjct: 788 VKNLANLSIMIVETKLHNRHEIVYKLLKLVLVLPVATASVERIFSAMNYVKNKLRNRMGD 847
Query: 753 QWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRR 787
Q+LND LVTFIER++ + I+ FQ M +RR
Sbjct: 848 QYLNDCLVTFIEREMFLKVKECDIINRFQAMKERR 882
>ref|NP_918185.1| putative transposase [Oryza sativa (japonica cultivar-group)]
Length = 897
Score = 614 bits (1583), Expect = e-174
Identities = 330/755 (43%), Positives = 480/755 (62%), Gaps = 8/755 (1%)
Query: 38 VVDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNK 97
+ D LL DPG R I +Y + ++ V + Y+ + QP S+ +P G RRF
Sbjct: 131 IYDIDLLCHDPGRRIAIKNYDVNERNSVIRGYIALGPCQPRSHN-FPIRKIGGKPRRFLP 189
Query: 98 NWFNLYDWLEYSESKNLAFCLPCFLFKN-VSNYGGDHFVGDGFGDWKNPRKLANHATSNN 156
+W++ + WLEYS ++ AFC C+LFK+ ++N GGD FV GF +W +++A H
Sbjct: 190 SWYDEFKWLEYSVEQDAAFCFICYLFKHKINNSGGDAFVNRGFRNWHMRKRIAKHVGGMT 249
Query: 157 SHVDCVHMGYA-LMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
S + Y + P +I +F +Q Y R+ S+ K+LLR GLA RG
Sbjct: 250 SFHNVAQDKYNHFLAPKTNIVESFAATNEQDKARYMARLTYSMKCLKFLLRQGLAARGHY 309
Query: 216 EADDSLYKGPFLELLDTLKENNSDVA-TILDSAPGNSLMTCPKIQKDLASACACEITREI 274
E++ SL KG FLE+L L EN +V +L++AP N +T P+IQK +A+ CA E T+ I
Sbjct: 310 ESEKSLNKGNFLEMLSMLAENFEEVGKVVLNNAPKNCKLTAPEIQKQIANCCAKETTKLI 369
Query: 275 VCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDAL 334
+ D+ D+ F +L DES DV +EQ+A+ LRYVD G V ERFLG+ V+ T++ +LK A+
Sbjct: 370 MEDLGDEYFAILADESSDVYQKEQLALCLRYVDKKGRVVERFLGVVHVENTTSLTLKAAI 429
Query: 335 ETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVAC 394
E++L + LS S +RGQGYDGASNM+G GLK LI +E PSA+YVHCFAHQLQL LVA
Sbjct: 430 ESLLMEHSLSLSKVRGQGYDGASNMKGHANGLKKLIMDECPSAYYVHCFAHQLQLTLVAV 489
Query: 395 AKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSI 454
AK + FF ++ L+N + S K+ EMLR Q + + ++ G+IETG GLNQ+ +
Sbjct: 490 AKENPDCVWFFEQLRFLLNLLGNSCKKTEMLRVAQAQRIVEELDLGDIETGKGLNQEMGL 549
Query: 455 ARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD-KFGETVLLLDVLQSFDFIFM 513
R DTRWGSH++T+ +++LY +I +V++ VG D SF + +L + QSF+F+FM
Sbjct: 550 GRPADTRWGSHYKTVMHVLSLYPSIQKVLIMVGKDRSFGAECANAQTVLTIFQSFEFVFM 609
Query: 514 LYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNE-GWEELISRVVTICT 572
++M +LG T+DL+ ALQ+RDQD++NA+ L+ TK QLQ++R + GW++ + V + C
Sbjct: 610 AHLMQTVLGFTSDLNHALQKRDQDIVNAVGLILLTKFQLQQLREDPGWDDFLQEVQSFCV 669
Query: 573 KHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTE 632
KH+I +PDMD+ Y + RR + N+H + D SV+D QLQELN RFDE NT+
Sbjct: 670 KHKIKIPDMDSFYRPVGRDRRF--FIKIKNMHRFHVDMFLSVIDRQLQELNERFDEVNTD 727
Query: 633 LLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAK 692
LL C++ SP +F+++D +KL+++A YPNDF L L ++ ++ D +F K
Sbjct: 728 LLLCMAAFSPIDNFASWDKDKLIKLARFYPNDFSSTEMNHLPSALKLFLTEMRIDERFRK 787
Query: 693 LKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGD 752
+K L++L ++VET ++ KLLKL L+LPVATASVER+FSAM VK+ LRN+MGD
Sbjct: 788 VKNLANLSIMIVETKLHNRHEIISKLLKLVLVLPVATASVERIFSAMNYVKNKLRNRMGD 847
Query: 753 QWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRR 787
Q+LND LVTFIER++ + I+ FQ M +RR
Sbjct: 848 QYLNDCLVTFIEREMFLKVKECDIINRFQAMKERR 882
>ref|NP_908521.1| putative transposable element Tip100 protein [Oryza sativa
(japonica cultivar-group)]
Length = 802
Score = 613 bits (1580), Expect = e-174
Identities = 333/778 (42%), Positives = 491/778 (62%), Gaps = 14/778 (1%)
Query: 20 ASSSTTDIAPNSTNEGVKV-----VDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRR 74
+SSS TD +P++ N + DPG+R PI +Y P+I+D+V++AY
Sbjct: 11 SSSSNTDPSPSTENLSQPKRPRAEFSQSDIIGDPGLRKPIEAYPPEIRDQVKRAYALSGP 70
Query: 75 HQPPSNFVYPWSDFRGTRRRFNKNWFNLYDWLEYSESKNLAFCLPCFLF---KNVSNYGG 131
QP ++P G R F K WFN +DWLEYS SK+ A+CL C++F +G
Sbjct: 71 TQPNIT-IFPRKWQGGEWRSFQKTWFNEFDWLEYSVSKDAAYCLYCYIFFEPGKPEKFGS 129
Query: 132 DHFVGDGFGDWKNPRKLANHATSNNSHVDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYC 191
F +G+ +WK + ++ +H D + MN S+ +K+ + Y
Sbjct: 130 AVFAKEGYVNWKKGKDRLTVHSNCKTHNDARNKCEDFMNQRTSVSKKIEIVSKEEEIRYK 189
Query: 192 VRVKTSLLATKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENNSDVATILDSAPGNS 251
+R+ +SL ++L+ G AFRG DE+D SL KG F E++D K+ +V + N
Sbjct: 190 IRLTSSLDVVRFLIEQGDAFRGHDESDTSLSKGKFKEMVDWYKDKVPEVKDAYEKGLKNC 249
Query: 252 LMTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGL 311
M IQKDL ACA ++T I+ +I + F VLIDES DVS +EQM V+LR+V+ +G
Sbjct: 250 QMVSHHIQKDLTKACAEKVTAVIMDEIGNRNFSVLIDESRDVSIKEQMGVILRFVNDEGK 309
Query: 312 VKERFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQ 371
V ERFLG+ ++ +A +LK+AL M+S + L+ S IRGQGYDGASNMRG F G++ LI+
Sbjct: 310 VMERFLGLQHIERCTAIALKEALFGMISSHKLTISKIRGQGYDGASNMRGEFNGVQKLIR 369
Query: 372 NENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLA 431
+ENP A YVHCFAHQLQL +VA + + ++ FF V ++VN + AS R++ L K
Sbjct: 370 DENPYAFYVHCFAHQLQLVVVAVSTSTPAIADFFNYVPLIVNTVGASCMRKDALLAKHHD 429
Query: 432 QFAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPS 491
+ +E GEI TG GLNQ+SS+AR GDTRWGSH +TL ++ ++ AI++V+ V D +
Sbjct: 430 VLLEKVENGEITTGRGLNQESSLARPGDTRWGSHLKTLLRILVMWEAIIDVLEIVKKDST 489
Query: 492 FDKF-GETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKK 550
F G L+ +QSFDF+F++++M+++L IT+DLS ALQR+DQD++ A+SL+ D K+
Sbjct: 490 KPTFNGGAFGLMGKMQSFDFVFIMHLMIDMLSITDDLSRALQRKDQDIVEAMSLLIDVKE 549
Query: 551 QLQEMRNEGWEELISRVVTICTKHEIDVPDMDAPYME-GKKPRRVPPVSSVSNLHHYKND 609
LQ+MR GWE L++RV++ C KHEI VP MD E G R V+N H+Y +
Sbjct: 550 LLQDMRENGWEPLLNRVISFCNKHEIKVPKMDKEVNERGTSTHR---RHKVTNKHYYHVE 606
Query: 610 CLFSVLDLQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVS 669
+ +D L E+N RF E ++ELL C+S L+P SFS FDV+KL+R+AE+Y DF+
Sbjct: 607 IYLAAIDAILVEMNHRFSEVSSELLVCMSSLNPRNSFSNFDVDKLVRLAEIYAEDFLVGD 666
Query: 670 EVELRRQLHNYVRNVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVAT 729
+ LR QL N++ NV+ +F K L+ + ++V+T K +T+ LV++L++L+L+LPVAT
Sbjct: 667 LMLLRTQLGNFISNVRRSKEFLGCKDLAKVAELMVQTGKNRTYHLVYRLIELSLILPVAT 726
Query: 730 ASVERVFSAMKIVKSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRR 787
ASVERVFSAM ++K LRNKMGD+WLND ++ + E+ + +IS + I+ HF++M RR
Sbjct: 727 ASVERVFSAMSLIKIDLRNKMGDEWLNDLMICYTEKQIFRSISDEKIIQHFEEMKKRR 784
>gb|AAT38794.1| putative hAT family dimerisation domain containing protein [Solanum
demissum]
Length = 805
Score = 603 bits (1555), Expect = e-171
Identities = 339/811 (41%), Positives = 498/811 (60%), Gaps = 27/811 (3%)
Query: 1 MVFINYRMKKFFPVL------------SRDKASSSTTDIAPNSTNEGVKVVDYKLLETDP 48
M Y +KK+F + S K +++ ++++ +S+ E D L+ DP
Sbjct: 1 MATSQYSVKKYFTQVPKSTLASQSHSQSNQKENTNHSEVSLDSSQE----FDLSSLKFDP 56
Query: 49 GIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFV-YPWSDFRGTRRRFNKNWFN--LYDW 105
G R I +YHP+ +D +R+AYL QP YP ++ G+ RRFN WF+ +DW
Sbjct: 57 GERTSILNYHPNHRDVIRRAYLLNGPCQPRLAVQKYPQTNIFGSMRRFNHEWFDDIYHDW 116
Query: 106 LEYSESKNLAFCLPCFLFK--NVSNYGGDHFVGDGFGDWKNPRKLANHATSNNS-HVDCV 162
LEYS SK+ +CL C+LFK N + GG+ F GF W L H NS H
Sbjct: 117 LEYSVSKDAVYCLYCYLFKDHNTNQGGGETFSTIGFKSWNKKSGLDKHIGLPNSIHNQSK 176
Query: 163 HMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEADDSLY 222
L+ +SI+ AF Q+ Q+ Y +R+ S+ + L+ G AFRG DE+ SL
Sbjct: 177 KKCQDLLQQRRSIQFAFERQSNQLKHRYYMRLSASVDVVRLLISRGFAFRGHDESKSSLS 236
Query: 223 KGPFLELLDTLKENNSDVAT-ILDSAPGNSLMTCPKIQKDLASACACEITREIVCDIADD 281
+G FLE+L + + IL+ AP N MT P IQKD+ SAC E + I+ ++ D
Sbjct: 237 RGNFLEILSWYAKRCDKIRDYILEHAPVNDQMTSPMIQKDIVSACKIETVKAILEELNGD 296
Query: 282 VFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETMLSIN 341
F +L+DES DVS +EQMA++ RY+D G V ER + I VK+TSA SLK+ + +L+ +
Sbjct: 297 YFALLVDESFDVSHKEQMAIIFRYIDRMGFVMERLIDIVHVKDTSASSLKETIFNLLAQH 356
Query: 342 GLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTHKPV 401
LS SS+RGQ YDGASNM+G GLK LI+ E+ SAH +HCFAHQLQL LV +K V
Sbjct: 357 SLSPSSVRGQCYDGASNMQGEINGLKMLIRRESKSAHSIHCFAHQLQLTLVGVSKKCVEV 416
Query: 402 SGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTR 461
++ + N + +S KR + LRD Q + ++ GE+ TGSGLNQ ++RA DTR
Sbjct: 417 GKLVVLISNIFNVLGSSFKRMDNLRDSQKSTIQVALDMGELTTGSGLNQQLGLSRACDTR 476
Query: 462 WGSHFRTLTSLMTLYGAIVEVIVEVGNDP-SFDKFGETVLLLDVLQSFDFIFMLYMMVEI 520
WGSH+++ + + ++G+I+EV+ + D S D+ + + L+ Q+F+ FML++M ++
Sbjct: 477 WGSHYKSFNNFIIMFGSILEVLESLALDARSMDERAKAMGHLEACQTFEIAFMLHLMRDV 536
Query: 521 LGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEIDVPD 580
L ITN+L+ LQ+++QD+ NA+ LV K++LQ +R++ W+ LI++V T C KH I +P+
Sbjct: 537 LAITNELNKCLQKKEQDIANAMLLVEVAKRRLQVLRDDEWDSLIAKVSTFCIKHNILIPN 596
Query: 581 MDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELLQCVSCL 640
+ PY+ + RR +++ + LHHY+ + +++D QLQELN RFDE T+LL ++CL
Sbjct: 597 FEEPYVSSLRSRR--KLANYTILHHYRVEVFCNIIDWQLQELNDRFDEVTTDLLHGIACL 654
Query: 641 SPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKS-DPKFAKLKGLSDL 699
+P KSFS+FD+ K +RMAELYP+DF + + L QL +Y+ +V+ D +F L GL DL
Sbjct: 655 NPIKSFSSFDIRKKMRMAELYPDDFDESNMNILENQLASYIVDVRDVDERFFDLNGLCDL 714
Query: 700 CAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQWLNDRL 759
LV+T K + LVF+L+KLALLLPVATASVER FSAMK +K+ LR++M D + + L
Sbjct: 715 SKRLVQTKKHSNYPLVFRLVKLALLLPVATASVERAFSAMKFIKNDLRSQMSDDFFSGCL 774
Query: 760 VTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
V ++E+DV IS DVI+ FQ M RR L
Sbjct: 775 VPYLEKDVFDKISNDVIIKTFQDMKPRRIQL 805
>gb|AAX96758.1| hAT family dimerisation domain, putative [Oryza sativa (japonica
cultivar-group)]
Length = 1050
Score = 598 bits (1543), Expect = e-169
Identities = 319/756 (42%), Positives = 463/756 (61%), Gaps = 23/756 (3%)
Query: 47 DPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKNWFNLYDWL 106
DPG+R PI +H DI+D VR+ YL R P YP +R F+ +WF +++WL
Sbjct: 279 DPGLRKPIEQFHIDIRDAVRREYLS-RGPCQPIGHKYPQKLMGNRQRSFHDSWFKMHNWL 337
Query: 107 EYSESKNLAFCLPCFLFKNV--SNYGGDHFVGDGFGDWKNPRKLANHATSN-NSHVDCVH 163
EYS +K+ AFC C+LFK N+G D F GF +WK HAT + N HV V
Sbjct: 338 EYSIAKDAAFCFYCYLFKQPRPDNFGVDAFTSKGFTNWK-------HATGSFNEHVGKVD 390
Query: 164 MGYA--------LMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
+ N QS+ + +K++ +Y R+ L ++LL LAFRG D
Sbjct: 391 SFHNRARKHCEDFKNQRQSVSYKMDSGSKKLEQQYYGRITMILGVVRFLLLQALAFRGHD 450
Query: 216 EADDSLYKGPFLELLDTLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIV 275
E+ S KG FLE+++ KE + D +L ++PGN L+T PKIQK L ACA + T+ I+
Sbjct: 451 ESHGSSNKGNFLEMIEWYKEKDKDAQKLLGNSPGNHLLTSPKIQKHLCKACANKTTKAIL 510
Query: 276 CDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALE 335
DI + F +L+DES D S +EQMAV+LRY++ G V ERFLG+ V +T++ +LK AL+
Sbjct: 511 KDIGERNFAILVDESRDASIKEQMAVILRYINSKGQVIERFLGVEHVPDTTSVALKIALD 570
Query: 336 TMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACA 395
M+ +GLS +RGQGYDGASNMRG F GL+ + +ENP A Y+HCFAHQLQL +V+ A
Sbjct: 571 AMIVSHGLSMHKVRGQGYDGASNMRGEFHGLQRRVLDENPYAFYIHCFAHQLQLVVVSVA 630
Query: 396 KTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIA 455
K PVS FF M+VN + S KR + L + +E GEI +G G NQ +++A
Sbjct: 631 KCCSPVSDFFNYTPMIVNTVNGSCKRHDQLAQEHHDNLVSSLETGEIFSGRGKNQATNLA 690
Query: 456 RAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFMLY 515
R GDTRWGSH +TL L ++ A++EV+ + D K T LL ++SF+F+ +++
Sbjct: 691 RPGDTRWGSHHKTLCRLQHMWKAVLEVLENICEDNPTTKITATGLLKQ-MESFEFVLIMH 749
Query: 516 MMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHE 575
+M+ +LG TNDLS LQ++DQ+++ AI L+ T +++ +R GW+EL T C K+
Sbjct: 750 LMIRLLGKTNDLSQCLQKKDQNIIRAIGLIGTTLQKINHIRQHGWQELFEETKTFCLKYN 809
Query: 576 IDVPDM-DAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELL 634
I VPDM D G+ R + ++ HH+ N+ V D + ELN RF E +T+LL
Sbjct: 810 IIVPDMSDTTTTRGRSRGRGGQL--ITYQHHFNNEIFNVVHDQLIVELNNRFAERSTQLL 867
Query: 635 QCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLK 694
+C++CL P SF+ FD +KL+ +A +Y DF + L+ QL ++ +V++DP F
Sbjct: 868 RCIACLDPRNSFANFDEDKLIELARMYAADFSPYDCIVLKDQLETFIADVRADPHFLSCS 927
Query: 695 GLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQW 754
L +L +V++++ F LV++L++LAL+LPVATA+VER FSAM I+K+ LRNKM D+W
Sbjct: 928 DLGNLAMKMVQSDRHTVFPLVYRLIELALILPVATATVERAFSAMNIIKTELRNKMADEW 987
Query: 755 LNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
LN R+V +IERD+ +I D IL HFQ++ R+ L
Sbjct: 988 LNHRVVCYIERDIFASIDNDKILTHFQELRTRKMKL 1023
>gb|AAX96757.1| hAT family dimerisation domain, putative [Oryza sativa (japonica
cultivar-group)]
Length = 1071
Score = 597 bits (1540), Expect = e-169
Identities = 318/756 (42%), Positives = 463/756 (61%), Gaps = 23/756 (3%)
Query: 47 DPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKNWFNLYDWL 106
DPG+R PI +H DI+D VR+ YL R P YP +R F+ +WF +++WL
Sbjct: 103 DPGLRKPIEQFHIDIRDAVRREYLS-RGPCQPIGHKYPQKLMGNRQRSFHDSWFKMHNWL 161
Query: 107 EYSESKNLAFCLPCFLFKNV--SNYGGDHFVGDGFGDWKNPRKLANHATSN-NSHVDCVH 163
EYS +K+ AFC C+LFK N+G D F GF +WK HAT + N HV V
Sbjct: 162 EYSIAKDAAFCFYCYLFKQPRPDNFGVDAFTSKGFTNWK-------HATGSFNEHVGKVD 214
Query: 164 MGYA--------LMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
+ N QS+ + +K++ +Y R+ L ++LL LAFRG D
Sbjct: 215 SFHNRARKHCEDFKNQRQSVSYKMDSGSKKLEQQYYGRITMILGVVRFLLLQALAFRGHD 274
Query: 216 EADDSLYKGPFLELLDTLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIV 275
E+ S KG FLE+++ KE + D +L ++PGN L+T PKIQK L ACA + T+ I+
Sbjct: 275 ESHGSSNKGNFLEMIEWYKEKDKDAQKLLGNSPGNHLLTSPKIQKHLCKACANKTTKAIL 334
Query: 276 CDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALE 335
D+ + F +L+DES D S +EQMAV+LRY++ G V ERFLG+ V +T++ +LK AL+
Sbjct: 335 KDVGERNFAILVDESRDASIKEQMAVILRYINSKGQVIERFLGVEHVPDTTSVALKIALD 394
Query: 336 TMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACA 395
M+ +GLS +RGQGYDGASNMRG F GL+ + +ENP A Y+HCFAHQLQL +V+ A
Sbjct: 395 AMIVSHGLSMHKVRGQGYDGASNMRGEFHGLQRRVLDENPYAFYIHCFAHQLQLVVVSVA 454
Query: 396 KTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIA 455
K PVS FF M+VN + S KR + L + +E GEI +G G NQ +++A
Sbjct: 455 KCCSPVSDFFNYTPMIVNTVNGSCKRHDQLAQEHHDNLVSSLETGEIFSGRGKNQATNLA 514
Query: 456 RAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFMLY 515
R GDTRWGSH +TL L ++ A++EV+ + D K T LL ++SF+F+ +++
Sbjct: 515 RPGDTRWGSHHKTLCRLQHMWKAVLEVLENICEDNPTTKTTATGLLKQ-MESFEFVLIMH 573
Query: 516 MMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHE 575
+M+ +LG TNDLS LQ++DQ+++ AI L+ T +++ +R GW+EL T C K+
Sbjct: 574 LMIRLLGKTNDLSQCLQKKDQNIIRAIGLIGTTLQKINHIRQHGWQELFEETKTFCLKYN 633
Query: 576 IDVPDM-DAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELL 634
I VPDM D G+ R + ++ HH+ N+ V D + ELN RF E +T+LL
Sbjct: 634 IIVPDMSDTTTTRGRSRGRGGQL--ITYQHHFNNEIFNVVHDQLIVELNNRFAERSTQLL 691
Query: 635 QCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLK 694
+C++CL P SF+ FD +KL+ +A +Y DF + L+ QL ++ +V++DP F
Sbjct: 692 RCIACLDPRNSFANFDEDKLIELARMYAADFSPYDCIVLKDQLETFIADVRADPHFLSCS 751
Query: 695 GLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQW 754
L +L +V++++ F LV++L++LAL+LPVATA+VER FSAM I+K+ LRNKM D+W
Sbjct: 752 DLGNLAMKMVQSDRHTVFPLVYRLIELALILPVATATVERAFSAMNIIKTELRNKMADEW 811
Query: 755 LNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
LN R+V +IERD+ +I D IL HFQ++ R+ L
Sbjct: 812 LNHRVVCYIERDIFASIDNDKILTHFQELRTRKMKL 847
>ref|NP_914450.1| putative transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 796
Score = 585 bits (1507), Expect = e-165
Identities = 311/749 (41%), Positives = 466/749 (61%), Gaps = 10/749 (1%)
Query: 44 LETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKNWFNLY 103
++ DPG R I YHP+ ++ VR+ YL QP + + P ++ G R+FN W++ +
Sbjct: 45 IQFDPGKRKRIEDYHPNQKEMVRRKYLDNGPCQPRPDDL-PVTNIGGRDRKFNPEWYDEF 103
Query: 104 -DWLEYSESKNLAFCLPCFLFK--NVSNYGGDHFVGDGFGDWKNPRKLANHATSNNS-HV 159
WLEYSESK+ +C CFLF+ N + G D FV G+ W +L H +S H
Sbjct: 104 GSWLEYSESKDKVYCFYCFLFRDPNKKSAGYDAFVTSGWSSWNKKHRLKEHVGDIDSVHN 163
Query: 160 DCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEADD 219
+ AL+ Q I + NQ+ + Y R+ S+ T+ LL+ G+ FRG DE+++
Sbjct: 164 QAMRDCDALLKKEQHIDVSLNNQSNAEKIAYYTRLNGSIDVTRILLKLGMPFRGHDESEE 223
Query: 220 SLYKGPFLELLDTLKENNSDVATILD-SAPGNSLMTCPKIQKDLASACACEITREIVCDI 278
SL KG F E E N + + +APGNS T KIQ D+ A E+ R I+ ++
Sbjct: 224 SLNKGNFKEFHAYTAEQNPALHKVAGKNAPGNSQYTSSKIQNDIIRCFAKEVLRYILEEV 283
Query: 279 ADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETML 338
+DVFC+L+DES DVS +EQMAVVLRYV G VKER +G+ VKET++ LK A++ +
Sbjct: 284 GNDVFCLLVDESRDVSYKEQMAVVLRYVGKCGSVKERLVGLVHVKETTSMYLKSAIDDLF 343
Query: 339 SINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTH 398
+ + LS +RGQGYDGASNMRG F GL++LI EN SA+Y+HCFAHQLQL +VA K H
Sbjct: 344 AEHKLSLKQVRGQGYDGASNMRGEFNGLQSLIMRENSSAYYIHCFAHQLQLVVVAIVKKH 403
Query: 399 KPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAG 458
K + FF +++L+N + S+KR++M+RD +E G + TGSGLNQ+ S+ R G
Sbjct: 404 KGIGNFFDMISLLLNVVGGSSKRRDMIRDINAEHVHDALECGLLITGSGLNQEISLQRPG 463
Query: 459 DTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFDKFG-ETVLLLDVLQSFDFIFMLYMM 517
DTRW SH++TL SL+ L+ V+V+ V + DK + LL Q F F+F L +M
Sbjct: 464 DTRWSSHYKTLKSLVQLFPTTVKVLEYVEENDRDDKNSRQAAGLLVYFQLFQFVFYLQLM 523
Query: 518 VEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEID 577
+ IL ITN S+ LQR+DQD++NA++ V T+ L E R GWE +++ V C K++I
Sbjct: 524 LNILAITNTFSVTLQRKDQDIVNAVNCVKSTRNHLDEFRRSGWESILAEVHNFCEKYDIS 583
Query: 578 VPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELLQCV 637
+M+ Y+ K+PR+ + ++N HH++ DC ++D LQEL+ RF+E+ ++LL C
Sbjct: 584 ELEMEDAYINPKRPRQ---KTGITNKHHFEVDCFNEIVDWLLQELDNRFNEKTSQLLICA 640
Query: 638 SCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLKGLS 697
+ LSP +SF F++ L+ +A+LYP DF D ++LR L Y+ +V+ D +FA ++ +
Sbjct: 641 AALSPRQSFHDFNLEHLMSLAKLYPKDFDDGELMDLRHHLSLYIADVRGDDRFANIETIC 700
Query: 698 DLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQWLND 757
L +V+T K + LV++LLKLAL+LPVATA+VER FSAMKIVK++LRN++ D+ L+D
Sbjct: 701 QLSQKMVDTRKHICYPLVYRLLKLALVLPVATATVERCFSAMKIVKTYLRNRISDEHLSD 760
Query: 758 RLVTFIERDVLFTISTDVILAHFQQMDDR 786
++ ++E+ L ++ ++ F+ + R
Sbjct: 761 CVICYVEKQELMKVTNSAVIHRFETLKAR 789
>gb|AAT39314.1| putative transposase [Solanum demissum]
Length = 705
Score = 571 bits (1472), Expect = e-161
Identities = 310/706 (43%), Positives = 448/706 (62%), Gaps = 10/706 (1%)
Query: 93 RRFNKNWFN--LYDWLEYSESKNLAFCLPCFLFK--NVSNYGGDHFVGDGFGDWKNPRKL 148
RRFN WF+ +DWLEYS SK+ +CL C+LFK N + GG+ F GF W L
Sbjct: 2 RRFNHEWFDDIYHDWLEYSVSKDAVYCLYCYLFKDHNTNQGGGETFSTIGFKSWNKKSGL 61
Query: 149 ANHATSNNS-HVDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRC 207
H NS H L+ +SI+ AF Q+ Q+ Y +R+ S+ + L+
Sbjct: 62 DKHIGLPNSIHNQSKKKCQDLLQQRRSIQFAFERQSNQLKHRYYMRLSASVDVVRLLISR 121
Query: 208 GLAFRGSDEADDSLYKGPFLELLDTLKENNSDVAT-ILDSAPGNSLMTCPKIQKDLASAC 266
G AFRG DE+ SL +G FLE+L + + IL+ AP N MT P IQKD+ SAC
Sbjct: 122 GFAFRGHDESKSSLSRGNFLEILSWYAKRCDKIRDYILEHAPVNDQMTSPMIQKDIVSAC 181
Query: 267 ACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETS 326
E + I+ ++ D F +L+DES DVS +EQMA++ RY+D G V ER + I VK TS
Sbjct: 182 KIETVKAILEELNGDYFALLVDESFDVSHKEQMAIIFRYIDRMGFVMERLIDIVHVKATS 241
Query: 327 AKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQ 386
A SLK+ + +L+ + LS SS+RGQ YDGASNM+G GLK LI+ E+ SAH +HCFAHQ
Sbjct: 242 ASSLKETIFNLLAQHSLSPSSVRGQCYDGASNMQGEINGLKMLIRRESKSAHSIHCFAHQ 301
Query: 387 LQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGS 446
LQL LV +K V ++ + N + +S KR + LRD Q + ++ GE+ TGS
Sbjct: 302 LQLTLVGVSKKCVEVGKLVVLISNIFNVLGSSFKRMDNLRDSQKSTIQVALDMGELTTGS 361
Query: 447 GLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDP-SFDKFGETVLLLDVL 505
GLNQ ++RA DTRWGSH+++ + + ++G+I+EV+ + D S D+ + + L+
Sbjct: 362 GLNQQLGLSRACDTRWGSHYKSFNNFIIMFGSILEVLESLALDARSMDERAKAMGHLEAC 421
Query: 506 QSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELIS 565
Q+F+ FML++M ++L ITN+L+ LQ+++QD+ NA+ LV K++LQ +R++ W+ LI+
Sbjct: 422 QTFEIAFMLHLMRDVLAITNELNKCLQKKEQDIANAMLLVEVAKRRLQVLRDDEWDSLIA 481
Query: 566 RVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNAR 625
+V T C KH I +P+ + PY+ + RR +++ + LHHY+ + +++D QLQELN R
Sbjct: 482 KVSTFCIKHNILIPNFEEPYVSSLRSRR--KLANYTILHHYRVEVFCNIIDWQLQELNDR 539
Query: 626 FDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVK 685
FDE T+LL ++CL+P KSFS+FD+ K++RMAELYP+DF + + L QL +Y+ +V+
Sbjct: 540 FDEVTTDLLHGIACLNPIKSFSSFDIRKIMRMAELYPDDFDESNMNILENQLASYIVDVR 599
Query: 686 S-DPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKS 744
D +F L GL DL LV+T K + LVF+L+KLALLLPVATASVER FSAMK +K+
Sbjct: 600 DVDERFFDLNGLCDLSKRLVQTKKHSNYPLVFRLVKLALLLPVATASVERAFSAMKFIKN 659
Query: 745 HLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFSL 790
LR++M D + + LV ++E+DV IS DVI+ FQ M RR L
Sbjct: 660 DLRSQMSDDFFSGCLVPYLEKDVFDKISNDVIIKTFQDMKPRRIQL 705
>dbj|BAA36225.1| transposase [Ipomoea purpurea]
Length = 808
Score = 553 bits (1426), Expect = e-156
Identities = 317/772 (41%), Positives = 467/772 (60%), Gaps = 41/772 (5%)
Query: 40 DYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQ------PPSNFVYPWSDFRGTRR 93
D + LE DPG+R PI + +DEVR+AY+K +Q P S +P R
Sbjct: 51 DIQALERDPGLRLPIWKCPIEKRDEVRRAYIKAGPYQCLLSKYPKSGEKHP--------R 102
Query: 94 RFNKNWFNLY-DWLEYSESKNLAFCLPCFLFKNVSNYGG-DHFVGDGFGDWKNPRKLANH 151
F +WF L+ WLEYS SK+ AFCLPC+LF G D F +GF WK R N
Sbjct: 103 SFQASWFKLFPSWLEYSPSKDAAFCLPCYLFHTQDGPSGLDAFTINGFRSWKKVRDGKNC 162
Query: 152 ATSNNSHVDCVHMGYALMNPNQSIKAA---FVNQ-----------TKQMNVEYCVRVKTS 197
+ H+G L +P+++ + A +NQ T Q E +R++TS
Sbjct: 163 SF-------LAHIGKDLTSPHRNAEKACADLMNQQSHIAQHIEKFTSQEIAENRLRLRTS 215
Query: 198 LLATKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENNSDVATILDSAPGNSLMTCPK 257
+ AT++L G +FRG DE+ S +G FLELL + N +V +LD AP N+ T P
Sbjct: 216 IEATRWLAFQGCSFRGHDESITSTNRGNFLELLSFIASFNDEVTKVLDKAPRNASYTSPT 275
Query: 258 IQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFL 317
IQK + A ++ I +I D FC+++DE+ D S +EQM++VLR+VD DG ++ERF
Sbjct: 276 IQKQILQVLATKVKSAIREEIGDAKFCIIVDEARDESKKEQMSIVLRFVDRDGFIQERFF 335
Query: 318 GITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSA 377
G+ VK+TSA +LK+ + ++LS + L +IRGQGYDGASNMRG + GLK LI +E P A
Sbjct: 336 GVVHVKDTSASTLKEGIFSILSRHNLDVQNIRGQGYDGASNMRGEWNGLKALILDECPYA 395
Query: 378 HYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLI 437
+YVHCFAH+LQLALVA +K PV FF K+N ++N + AS KR + L+ + + L+
Sbjct: 396 YYVHCFAHRLQLALVASSKEVIPVHQFFTKLNSIINVVGASCKRNDQLKAAHASNISHLL 455
Query: 438 EEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFDKF-G 496
E+E+G GLNQ S+ R GDTRW SH ++++SLM ++ A EV++ + D + G
Sbjct: 456 SIDELESGRGLNQIGSLQRPGDTRWSSHLKSISSLMRMFSATCEVLLNIIEDGTTHAHRG 515
Query: 497 ETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMR 556
+ +VL SF+F+F++++M ++L I+N L ALQ + QD+LNA+ LV+ TK +Q +R
Sbjct: 516 DADAAYEVLTSFEFVFIMHLMKKVLEISNMLCQALQLQSQDILNAMHLVSSTKLLIQTLR 575
Query: 557 NEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLD 616
+ GW+EL++ V + C I VPD DA Y+ ++ R ++ HHYK D ++V+D
Sbjct: 576 DSGWDELVASVKSFCETVNITVPDFDAQYI-ARRGRARHQQDELTIGHHYKVDIFYAVID 634
Query: 617 LQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAE-LYPNDFVDVSEVELRR 675
QLQEL+ RFD + EL+ S L P + +F ++ + ++ E YP DF D ++LR
Sbjct: 635 SQLQELSNRFDHKAMELIVLSSSLDPKEMRISFRIDDVCKLVEKFYPQDFEDYETLQLRV 694
Query: 676 QLHNYVRNVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERV 735
QL ++ +V+ P F L+ +SDLC LV+T K + LVF+++ L L LPV+TA+ ER
Sbjct: 695 QLEHF-EHVQQLPDFRTLESISDLCRWLVKTRKSNIYPLVFRVVTLVLTLPVSTATTERS 753
Query: 736 FSAMKIVKSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRR 787
FSAM IVK+ LRNKM D++L+D L+ +IE+ + S D I+ F+ M +RR
Sbjct: 754 FSAMNIVKTTLRNKMEDEFLSDCLLVYIEKQIAKQFSIDSIIDAFRDMQERR 805
>gb|AAP55079.1| putative transposase [Oryza sativa (japonica cultivar-group)]
gi|37536980|ref|NP_922792.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
gi|18854992|gb|AAL79684.1| putative transposase [Oryza
sativa]
Length = 1283
Score = 517 bits (1331), Expect = e-145
Identities = 277/657 (42%), Positives = 410/657 (62%), Gaps = 8/657 (1%)
Query: 38 VVDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNK 97
+ D LL DPG R I +Y + ++ V + Y+ + QP S+ +P G RRF
Sbjct: 271 IYDIDLLCHDPGRRIAIKNYDVNERNSVIRGYIALGPCQPRSHS-FPIRKIGGKPRRFLP 329
Query: 98 NWFNLYDWLEYSESKNLAFCLPCFLFKN-VSNYGGDHFVGDGFGDWKNPRKLANHATSNN 156
+W++ + WLEYS ++ AFC C+LFK+ ++N GGD FV GF +W +++A H
Sbjct: 330 SWYDEFKWLEYSVEQDAAFCFICYLFKHKINNSGGDAFVNRGFRNWHMRKRIAKHVGGMT 389
Query: 157 SHVDCVHMGYA-LMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
S + Y + P +I +F +Q Y R+ S+ K+LLR GLA RG
Sbjct: 390 SFHNVAQDKYNHFLAPKTNIVESFAATNEQDKARYMARLNYSMKCLKFLLRQGLAARGHY 449
Query: 216 EADDSLYKGPFLELLDTLKENNSDVA-TILDSAPGNSLMTCPKIQKDLASACACEITREI 274
E++ SL KG FLE+L L EN +V +L++AP N +T P+IQK +A+ CA E T+ I
Sbjct: 450 ESEKSLNKGNFLEMLSMLAENFEEVGKVVLNNAPKNCKLTAPEIQKQIANCCAKETTKLI 509
Query: 275 VCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDAL 334
+ D+ D+ F +L DES DV +EQ+A+ LRYVD G V ERFLG+ V+ T++ +LK A+
Sbjct: 510 MEDLGDEYFAILADESSDVYQKEQLALCLRYVDKKGRVVERFLGVVHVENTTSLTLKAAI 569
Query: 335 ETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVAC 394
E++L + LS S +RGQGYDGASNM+G GLK LI +E PSA+YVHCFAHQLQL LVA
Sbjct: 570 ESLLMEHSLSLSKVRGQGYDGASNMKGHANGLKKLIMDECPSAYYVHCFAHQLQLTLVAV 629
Query: 395 AKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSI 454
AK + FF ++ L+N + S K+ EMLR Q + + ++ G+IETG GLNQ+ +
Sbjct: 630 AKENPDCVWFFEQLRFLLNLLGNSCKKTEMLRVAQAQRIVEELDLGDIETGKGLNQEMGL 689
Query: 455 ARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD-KFGETVLLLDVLQSFDFIFM 513
R DTRWGSH++T+ +++LY +I +V++ VG D SF + +L + QSF+F+FM
Sbjct: 690 GRPADTRWGSHYKTVMHVLSLYPSIQKVLIMVGKDRSFGAECANAQTVLTIFQSFEFVFM 749
Query: 514 LYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNE-GWEELISRVVTICT 572
++M +LG T+DL+ ALQ+RDQD++NA+ L+ TK QLQ++R + GW++ + V + C
Sbjct: 750 AHLMQTVLGFTSDLNHALQKRDQDIVNAVGLILLTKFQLQQLREDPGWDDFLQEVQSFCV 809
Query: 573 KHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTE 632
KH+I +PDMD+ Y + RR + N+H + D SV+D QLQELN RFDE NT+
Sbjct: 810 KHKIKIPDMDSFYRPVGRDRRF--FIKIKNMHRFHVDMFLSVIDRQLQELNERFDEVNTD 867
Query: 633 LLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPK 689
LL C++ SP +F+++D +KL+++A YPNDF L L ++ ++ D +
Sbjct: 868 LLLCMAAFSPIDNFASWDKDKLIKLARFYPNDFSSTEMNHLPSALKLFLTEMRIDER 924
>emb|CAB78089.1| putative protein [Arabidopsis thaliana] gi|4538896|emb|CAB39633.1|
putative protein [Arabidopsis thaliana]
gi|15233987|ref|NP_192704.1| hypothetical protein
[Arabidopsis thaliana] gi|7485592|pir||T04013
hypothetical protein F17A8.10 - Arabidopsis thaliana
Length = 664
Score = 508 bits (1307), Expect = e-142
Identities = 295/707 (41%), Positives = 422/707 (58%), Gaps = 65/707 (9%)
Query: 39 VDYKLLETDPGIRPPISSYHPDIQDEVRKAYLKIRRHQPPSNFVYPWSDFRGTRRRFNKN 98
+++ L +DP R I SYH + +DEVR+ YL IR P + RRFN
Sbjct: 15 INFNKLPSDPAKRKSILSYHLNQRDEVRREYL-IRGPCQPRGHKFKQIVIGKVLRRFNPK 73
Query: 99 WFNLY-DWLEYSESKNLAFCLPCFLFKN-VSNYGG-DHFVGDGFGDWKNPRKLANHATSN 155
WF+LY DWLEYS K AFCL C+LF++ N GG D F+ GF W +L H +
Sbjct: 74 WFDLYGDWLEYSVEKEKAFCLYCYLFRDQAGNQGGSDSFLSTGFCSWNKADRLDQHVGLD 133
Query: 156 NSHVDCVHMGYALMNPNQSIKAAFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSD 215
+ +F N K+ + L+R GL+FRG D
Sbjct: 134 VN--------------------SFHNNAKRKCED--------------LMRQGLSFRGHD 159
Query: 216 EADDSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREI 274
E+++S KG FLELL T +N +L +AP N+ MT P IQKD+ + E+TR I
Sbjct: 160 ESEESTNKGNFLELLKYTAGQNEVVKKVVLKNAPKNNQMTSPPIQKDIVHCFSEEVTRSI 219
Query: 275 VCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDAL 334
+ ++ +DVF +L+DES D S +EQM VV R+VD G+VKERF+G+ VKETS+ SLK A+
Sbjct: 220 IEEMDNDVFGLLVDESADASNKEQMTVVFRFVDKYGVVKERFIGVIHVKETSSLSLKSAI 279
Query: 335 ETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVAC 394
+++ + GLS +RGQGYDGASNM+G F GL++LI E
Sbjct: 280 DSLFAKYGLSLKKLRGQGYDGASNMKGEFNGLRSLILKE--------------------I 319
Query: 395 AKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSI 454
AK H V FF +++L+N + AS R++ +R+ + + I GEI+TG+ LNQ+ S+
Sbjct: 320 AKKHVEVGEFFDMISVLLNVVGASCTRKDKIREIHRQKVEEKISNGEIKTGTRLNQELSL 379
Query: 455 ARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD--KFGETVLLLDVLQSFDFIF 512
R G+TRWGSH++TL L L+ +IV +++E D D K + +L +FDF+F
Sbjct: 380 QRPGNTRWGSHYKTLLRLEELFSSIV-IVLEYIQDEGTDTTKRQQAYGILKYFHTFDFVF 438
Query: 513 MLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICT 572
L +M+ ++G+T+ LS ALQR+DQD+LN ISLV TK QLQ++R++GW+ ++ V +
Sbjct: 439 YLELMLLVMGLTDSLSKALQRKDQDILNVISLVKTTKCQLQKVRDDGWDAFMAEVSSFSE 498
Query: 573 KHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTE 632
K+ + M+ +++ ++PR+ S ++NLHHYK DC ++VLD+QLQE N RFDE N+E
Sbjct: 499 KNNTAMLKMEEEFVDSRRPRKK---SGITNLHHYKVDCFYTVLDMQLQEFNDRFDEVNSE 555
Query: 633 LLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAK 692
LL C+S LSP SF FD + L+R+ E YP++F V L QL Y+ NVK+D +F
Sbjct: 556 LLICMSSLSPIDSFCQFDKSMLVRLTEFYPDEFSFVERRSLDHQLEIYLDNVKNDERFTD 615
Query: 693 LKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAM 739
LK DL ++VET K + LV++LLKL+L+LPVATA+VER FSAM
Sbjct: 616 LKCFGDLARVMVETRKHLSHPLVYRLLKLSLILPVATATVERCFSAM 662
>ref|NP_189621.1| hAT dimerisation domain-containing protein [Arabidopsis thaliana]
Length = 536
Score = 499 bits (1285), Expect = e-139
Identities = 259/541 (47%), Positives = 374/541 (68%), Gaps = 8/541 (1%)
Query: 253 MTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLV 312
MT P IQKD+ + E+TR I+ ++ +DVF +L+DES D S +EQMAVV R+VD G+V
Sbjct: 1 MTSPPIQKDIVHCFSEEVTRSIIEEMDNDVFGLLVDESADASDKEQMAVVFRFVDKYGVV 60
Query: 313 KERFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQN 372
KERF+G+ V+ETS+ SLK A++++ + GLS +RG+GYDGASNM+G+F GL++LI
Sbjct: 61 KERFIGVIHVQETSSLSLKSAIDSLFAKYGLSLKKLRGKGYDGASNMKGKFNGLRSLILK 120
Query: 373 ENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQ 432
EN SA+YVHCFAHQLQL ++A AK H V FF +++L+N + AS KR++ +R+ +
Sbjct: 121 ENSSAYYVHCFAHQLQLVVMAVAKKHVEVGEFFYMISVLLNVVGASCKRKDKIREIDRQK 180
Query: 433 FAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIV---EVIVEVGND 489
+ I GEI+TG+GLNQ+ S+ R G+TRWGSH++TL L L+ +IV E I + G D
Sbjct: 181 VEEKISNGEIKTGTGLNQELSLQRPGNTRWGSHYKTLLRLEELFSSIVIVLEYIQDEGTD 240
Query: 490 PSFDKFGETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTK 549
+ K + +L +FDF+F L +M+ ++G+T+ LS ALQR+DQD+LNAISLV TK
Sbjct: 241 TT--KRQQAYGILKYFHTFDFVFYLELMLLVMGLTDSLSKALQRKDQDILNAISLVKTTK 298
Query: 550 KQLQEMRNEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKND 609
QLQ++R++GW+ ++ V + K + M+ +++ ++PR+ + ++NLHHYK D
Sbjct: 299 CQLQKVRDDGWDAFMAEVSSFSEKTNTGMLKMEEEFVDSRRPRK---KTGITNLHHYKVD 355
Query: 610 CLFSVLDLQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVS 669
C ++VLD+QLQE N FDE N+ELL C+S LSP SF FD + L+R+ E YP+DF V
Sbjct: 356 CFYTVLDMQLQEFNDCFDELNSELLICMSSLSPIDSFRQFDKSMLVRLTEFYPDDFSFVE 415
Query: 670 EVELRRQLHNYVRNVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVAT 729
L QL Y+ NVK+D +F L+ L DL ++VET K + LV++LLKL+L+LPVAT
Sbjct: 416 RRSLDHQLEIYLDNVKNDERFTDLEHLGDLARVMVETRKHLSHPLVYRLLKLSLILPVAT 475
Query: 730 ASVERVFSAMKIVKSHLRNKMGDQWLNDRLVTFIERDVLFTISTDVILAHFQQMDDRRFS 789
A+V R FSAM VK+ LRN+ GD +L+D LV FIE+ VL T++ + ++ FQ M +RR
Sbjct: 476 ATVVRCFSAMNFVKTTLRNRFGDVFLSDCLVCFIEKQVLNTVTNESVIKRFQDMSERRVH 535
Query: 790 L 790
L
Sbjct: 536 L 536
>gb|AAL86462.1| putative transposase protein, 5'-partial [Oryza sativa (japonica
cultivar-group)]
Length = 881
Score = 465 bits (1197), Expect = e-129
Identities = 238/512 (46%), Positives = 346/512 (67%), Gaps = 4/512 (0%)
Query: 274 IVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDA 333
I+ D+ D+ F +L DES DV +EQ+A+ LRYVD G V ERFLG+ V+ T++ +LK A
Sbjct: 3 IMEDLGDEYFAILADESSDVYQKEQLALCLRYVDKKGRVVERFLGVVHVENTTSLTLKAA 62
Query: 334 LETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVA 393
+E++L + LS S +RGQGYDGASNM+G GLK LI +E PSA+YVHCFAHQLQL LVA
Sbjct: 63 IESLLMEHSLSLSKVRGQGYDGASNMKGHANGLKKLIMDECPSAYYVHCFAHQLQLTLVA 122
Query: 394 CAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSS 453
AK + FF ++ L+N + S K+ EMLR Q + + ++ G+IETG GLNQ+
Sbjct: 123 VAKENPDCVWFFEQLRFLLNLLGNSCKKTEMLRVAQAQRIVEELDLGDIETGKGLNQEMG 182
Query: 454 IARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFD-KFGETVLLLDVLQSFDFIF 512
+ R DTRWGSH++T+ +++LY +I +V++ VG D SF + +L + QSF+F+F
Sbjct: 183 LGRPADTRWGSHYKTVMHVLSLYPSIQKVLIMVGKDRSFGAECANAQTVLTIFQSFEFVF 242
Query: 513 MLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNE-GWEELISRVVTIC 571
M ++M +LG T+DL+ ALQ+RDQD++NA+ L+ TK QLQ++R + GW++ + V + C
Sbjct: 243 MAHLMQTVLGFTSDLNHALQKRDQDIVNAVGLILLTKFQLQQLREDPGWDDFLQEVQSFC 302
Query: 572 TKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENT 631
KH+I +PDMD+ Y + RR + N+H + D SV+D QLQELN RFDE NT
Sbjct: 303 VKHKIKIPDMDSFYRPVGRDRRF--FIKIKNMHRFHVDMFLSVIDRQLQELNERFDEVNT 360
Query: 632 ELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFA 691
+LL C++ SP +F+++D +KL+++A YPNDF L L ++ ++ D +F
Sbjct: 361 DLLLCMAAFSPIDNFASWDKDKLIKLARFYPNDFSSTEMNHLPSALKLFLTEMRIDERFR 420
Query: 692 KLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMG 751
K+K L++L ++VET +V+KLLKL L+LPVATASVER+FSAM VK+ LRN+MG
Sbjct: 421 KVKNLANLSIMIVETKLHNRHEIVYKLLKLVLVLPVATASVERIFSAMNYVKNKLRNRMG 480
Query: 752 DQWLNDRLVTFIERDVLFTISTDVILAHFQQM 783
DQ+LND LVTFIER++ + +L + + +
Sbjct: 481 DQYLNDCLVTFIEREMFLKVKELSVLTYCEPL 512
>emb|CAE03530.2| OSJNBa0061C06.15 [Oryza sativa (japonica cultivar-group)]
Length = 587
Score = 427 bits (1097), Expect = e-118
Identities = 238/565 (42%), Positives = 353/565 (62%), Gaps = 11/565 (1%)
Query: 125 NVSNYGGDHFVGDGFGDWKNPRKLANHATS-NNSHVDCVHMGYALMNPNQSIKAAFVNQT 183
+ +N+GGD FV GF +W + + HA + N++H + M P SI+ +F + +
Sbjct: 14 STNNHGGDAFVNGGFRNWNIKSRFSKHAGAVNSAHCEAEEKYNLFMQPKTSIRESFASNS 73
Query: 184 KQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENNSDV-AT 242
+ +Y R+ SL +YLLR LAFRG E+ DS KG F EL+ L N +V
Sbjct: 74 AEFKDQYLARLTWSLKCIRYLLRQELAFRGHVESKDSNNKGKFRELVQWLAGNFEEVNKV 133
Query: 243 ILDSAPGNSLMTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVV 302
+L +AP M KIQK L +CA + T+ ++ ++ D+ F +L D S D +EQ+A+
Sbjct: 134 VLGNAPTGCQMIDHKIQKQLIGSCAHKTTKLVIEELHDECFAILADGSSDAYQQEQLALC 193
Query: 303 LRYVDGDGLVKE----RFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGASN 358
LR+V+ G E RFLG+ V+ T++ +LK+A++++L L S + GQGYDGASN
Sbjct: 194 LRFVNTTGQPVEQPVERFLGLVHVEYTTSLTLKEAIKSLLIKYQLPLSKVHGQGYDGASN 253
Query: 359 MRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIRAS 418
M+G GLK LI +E+PSA+YVHCFAHQLQL LVA AK + + FFG++ L+N + S
Sbjct: 254 MKGHINGLKKLIVDESPSAYYVHCFAHQLQLTLVAVAKENIDCAWFFGQLAYLLNVLGMS 313
Query: 419 NKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGA 478
K+ MLR Q + ++ GEIE+G GLNQ+ +AR GDTRWGSH++T+ +M LY +
Sbjct: 314 CKKICMLRIAQAEYMIEALKLGEIESGQGLNQEMGLARPGDTRWGSHYKTIMHVMLLYPS 373
Query: 479 IVEVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDL 538
I +V+ +VG + + + +L V QSF+F+F+L+MM EI G T+D AL RR+QD+
Sbjct: 374 IKKVLFKVGKECNGAEAIGAQTMLQVFQSFEFVFLLHMMNEIFGYTSDFCNALLRREQDI 433
Query: 539 LNAISLVNDTKKQLQEMRNE-GWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPV 597
+NA+ L+ TK +L +R + G +E + +V + C KH++ V DMD Y ++ R+
Sbjct: 434 VNAMDLLEFTKAELDVLREDCGCKEFLGKVTSFCVKHKVKVVDMDGKYKPIQRSRKF--F 491
Query: 598 SSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRM 657
N H + D V+D QLQELN RFDE NTELL+C++ SPAKSFSAF+V+ L+++
Sbjct: 492 KDAVNYHRFHADMFLGVIDRQLQELNNRFDEVNTELLRCMASFSPAKSFSAFNVDNLVKL 551
Query: 658 AELYPNDFVDVSEV-ELRRQLHNYV 681
A+ YP+DF DV E+ +L QL+ Y+
Sbjct: 552 AKFYPSDF-DVEEMNQLPFQLNRYI 575
>ref|NP_189620.1| hypothetical protein [Arabidopsis thaliana]
Length = 505
Score = 421 bits (1082), Expect = e-116
Identities = 221/484 (45%), Positives = 328/484 (67%), Gaps = 9/484 (1%)
Query: 178 AFVNQTKQMNVEYCVRVKTSLLATKYLLRCGLAFRGSDEADDSLYKGPFLELLDTLKENN 237
A QT + +Y +R+ S+ +++LL GL FRG DE+++S KG FLELL + N
Sbjct: 26 ALHKQTNVVKNDYRIRLNASIDVSRHLLHQGLPFRGHDESEESTNKGNFLELLKYIAGQN 85
Query: 238 SDVA-TILDSAPGNSLMTCPKIQKDLASACACEITREIVCDIADDVFCVLIDESGDVSGR 296
V +L +AP N+ MT P IQKD+ + E+TR I+ ++ +DVF +L+DES D S +
Sbjct: 86 EVVKKVVLKNAPKNNQMTSPPIQKDIVHCFSEEVTRSIIEEMDNDVFGLLVDESADASDK 145
Query: 297 EQMAVVLRYVDGDGLVKERFLGITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGA 356
EQMAVV R+VD G+VKERF+G+ V+ETS+ SLK A++++ + GLS +RG+GYDGA
Sbjct: 146 EQMAVVFRFVDKYGVVKERFIGVIHVQETSSLSLKSAIDSLFAKYGLSLKKLRGKGYDGA 205
Query: 357 SNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIR 416
SNM+G+F GL++LI EN SA+YVHCFAHQLQL ++A AK H V FF +++L+N +
Sbjct: 206 SNMKGKFNGLRSLILKENSSAYYVHCFAHQLQLVVMAVAKKHVEVGEFFYMISVLLNVVG 265
Query: 417 ASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLY 476
A KR++ +R+ + + I GEI+TG+ LNQ+ S+ R G+TRWGSH++TL L L+
Sbjct: 266 AFCKRKDKIREIDRQKVEEKISNGEIKTGTRLNQELSLQRPGNTRWGSHYKTLLRLEELF 325
Query: 477 GAIV---EVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQR 533
+IV E I + G D + K + +L +FDF+F L +M+ ++G+++ LS A QR
Sbjct: 326 SSIVIVLEYIQDEGTDTT--KRQQAYGILKYFHTFDFVFYLELMLLVMGLSDSLSKAWQR 383
Query: 534 RDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRR 593
+DQD+LNAISLV TK QLQ++R++GW+ ++ V + K I + M+ +++ ++PR+
Sbjct: 384 KDQDILNAISLVKTTKCQLQKVRDDGWDAFMAEVSSFSEKTNIGMLKMEEEFVDSRRPRK 443
Query: 594 VPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNK 653
+ ++NLHHYK DC ++VLD+QLQE N RFDE N+ELL C+S LSP SF FD +
Sbjct: 444 ---KTGITNLHHYKVDCFYTVLDMQLQEFNDRFDELNSELLICMSSLSPIDSFRQFDKSM 500
Query: 654 LLRM 657
L+R+
Sbjct: 501 LVRL 504
>ref|NP_174954.1| hypothetical protein [Arabidopsis thaliana]
Length = 496
Score = 409 bits (1050), Expect = e-112
Identities = 217/496 (43%), Positives = 322/496 (64%), Gaps = 11/496 (2%)
Query: 202 KYLLRCGLAFRGSDEADDSLYKGPFLELLD-TLKENNSDVATILDSAPGNSLMTCPKIQK 260
+YL+R L FRG DE+D+S +G FLEL+ T+ +N +L++AP N+ M ++ +
Sbjct: 5 RYLVRQRLPFRGHDESDESANRGNFLELVKYTVGQNEVISKVVLENAPKNNQMVFDEMSE 64
Query: 261 DLASACACEITRE-IVCDIADDVFCVLIDE-SGDVSGREQMAVVLRYVDGDGLVKERFLG 318
L + ++ D+ D V +VS +EQMAVV R+VD G VKERF+G
Sbjct: 65 PLIPETRDPVPSSFLLLDLILDYPVVWQQTVQSNVSDKEQMAVVFRFVDKHGTVKERFIG 124
Query: 319 ITSVKETSAKSLKDALETMLSINGLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAH 378
+ VKETS+ SLK A++++ + GLS +RGQGYDGA+NM+G F L++LI EN SA+
Sbjct: 125 LIHVKETSSASLKCAIDSLFAKRGLSMKQLRGQGYDGANNMKGEFNWLRSLILRENSSAY 184
Query: 379 YVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIE 438
Y+HCFAHQLQL +VA AK V FF ++ L+N + AS K ++ +R++ L + + I
Sbjct: 185 YIHCFAHQLQLVVVAVAKKQFEVGDFFDMISALLNVVGASCKGKDRIREEYLKEIEEGIN 244
Query: 439 EGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTLYGAIV---EVIVEVGNDPSFDKF 495
+GEI+TG GLNQ+ S+ R G+TRWG+H+ TL L L+ I+ E + E G D + K
Sbjct: 245 QGEIKTGKGLNQELSLQRPGNTRWGTHYTTLHRLAHLFSVIIKLLEFVEEEGTDST--KR 302
Query: 496 GETVLLLDVLQSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEM 555
+ LL +FDF F L +M+ +LG+TN LS+ALQ++DQD+LN +SLV TK+QL ++
Sbjct: 303 RQANGLLKYFNTFDFAFYLQLMLLLLGLTNSLSVALQKKDQDILNPMSLVKSTKQQLCKL 362
Query: 556 RNEGWEELISRVVTICTKHEIDVPDMDAPYMEGKKPRRVPPVSSVSNLHHYKNDCLFSVL 615
R++GW+ L++ V + C KH+I++ MD ++ + PR+ S+++N HHY+ +C +++L
Sbjct: 363 RDDGWDSLVNEVFSFCKKHDIELVIMDGEFVNPRNPRK---RSNMTNFHHYQVECFYTIL 419
Query: 616 DLQLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRR 675
D+Q+QE N RFDE NTELL CV+ LSP SF FD K+LR++E YP DF V L
Sbjct: 420 DMQIQEFNDRFDEVNTELLSCVASLSPIDSFHEFDQLKVLRLSEFYPQDFTHVDRRSLEH 479
Query: 676 QLHNYVRNVKSDPKFA 691
QL Y+ N++ D +FA
Sbjct: 480 QLGLYIDNIREDERFA 495
>ref|XP_474987.1| OSJNBa0065B15.7 [Oryza sativa (japonica cultivar-group)]
gi|38344561|emb|CAD39903.2| OSJNBa0065B15.7 [Oryza
sativa (japonica cultivar-group)]
Length = 639
Score = 394 bits (1012), Expect = e-108
Identities = 210/496 (42%), Positives = 322/496 (64%), Gaps = 23/496 (4%)
Query: 291 GDVSGREQMAVVLRYVDGDGLVKERFLGIT--SVKETSAKSLKDALETMLSINGLSFSSI 348
GDV G A ++ D D L K + + + SV+ET K LS +
Sbjct: 164 GDVGGSHYQA--MKKCD-DLLQKRQHIDVAFHSVRETELK--------------LSLKQV 206
Query: 349 RGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKV 408
RGQGYDGASNMRG F GL++LI E SA+YVHCFAHQLQL LV + HK VS FF K+
Sbjct: 207 RGQGYDGASNMRGEFNGLQSLIMREYSSAYYVHCFAHQLQLVLVDIVRKHKDVSDFFTKI 266
Query: 409 NMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRT 468
++L+N + S+KR++++RD + + +K + G+++TG+ LNQ+ + R GDTRW SH++T
Sbjct: 267 SILLNVVGGSSKRRDLIRDINVKEMSKALGSGQLQTGTRLNQEQCLQRPGDTRWSSHYKT 326
Query: 469 LTSLMTLYGAIVEVIVEVGNDPSFDKFGETVL-LLDVLQSFDFIFMLYMMVEILGITNDL 527
L SL+ ++ IV+V+ V D + K + LL+ QSFDF+F L++M+ IL ITN L
Sbjct: 327 LKSLVGMFATIVKVLEIVEKDKNDWKIRDQGSNLLEYFQSFDFVFYLHLMLTILTITNSL 386
Query: 528 SLALQRRDQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEIDVPDMDAPYME 587
SLALQR+DQD++NA+ V T+ L E+R E WE+++ V C ++I +++ Y++
Sbjct: 387 SLALQRKDQDIVNAMKCVKSTRLNLDELRREKWEKVLDEVSDFCDNYDIVKLEIEDTYID 446
Query: 588 GKKPRRVPPVSSVSNLHHYKNDCLFSVLDLQLQELNARFDEENTELLQCVSCLSPAKSFS 647
KK R S ++N H+Y+ DC V+D LQEL+ RF E +++LL C S SP SF
Sbjct: 447 PKKRRHK---SGITNKHYYQVDCFNDVIDWILQELDNRFSETSSQLLICSSAFSPRDSFH 503
Query: 648 AFDVNKLLRMAELYPNDFVDVSEVELRRQLHNYVRNVKSDPKFAKLKGLSDLCAILVETN 707
F++ L+ +A+LYP+DF + +L QL Y+ +V+ D +F+ ++ +++L I+VET
Sbjct: 504 DFNLENLMSLAKLYPSDFNSGNLRDLSHQLGLYIADVRDDGRFSNIQTIAELSQIMVETR 563
Query: 708 KCKTFALVFKLLKLALLLPVATASVERVFSAMKIVKSHLRNKMGDQWLNDRLVTFIERDV 767
K + LV++LLKL L+LPVATA+VER FSAMK VK++LRNK+GD++L+D L+ ++E++
Sbjct: 564 KHLCYPLVYQLLKLVLVLPVATATVERCFSAMKNVKTYLRNKIGDEYLSDSLICYVEKEE 623
Query: 768 LFTISTDVILAHFQQM 783
+ ++ + ++ F +M
Sbjct: 624 MKKVTNEAVVRRFMKM 639
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,288,440,530
Number of Sequences: 2540612
Number of extensions: 53548944
Number of successful extensions: 134162
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 133778
Number of HSP's gapped (non-prelim): 208
length of query: 790
length of database: 863,360,394
effective HSP length: 136
effective length of query: 654
effective length of database: 517,837,162
effective search space: 338665503948
effective search space used: 338665503948
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146806.1


