

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.4 - phase: 0
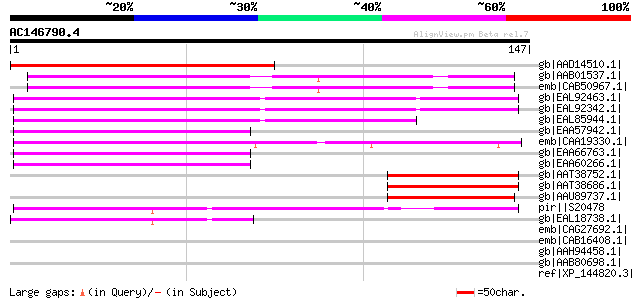
(147 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD14510.1| putative CENP-B/ARS binding protein-like protein ... 138 3e-32
gb|AAB01537.1| ARS binding protein 1 64 8e-10
emb|CAB50967.1| abp1 [Schizosaccharomyces pombe] gi|19113252|ref... 63 1e-09
gb|EAL92463.1| DNA binding protein [Aspergillus fumigatus Af293] 63 2e-09
gb|EAL92342.1| DNA binding protein [Aspergillus fumigatus Af293] 62 4e-09
gb|EAL85944.1| DNA binding protein [Aspergillus fumigatus Af293]... 57 9e-08
gb|EAA57942.1| hypothetical protein AN6156.2 [Aspergillus nidula... 54 6e-07
emb|CAA19330.1| SPBC14F5.12c [Schizosaccharomyces pombe] gi|1911... 54 8e-07
gb|EAA66763.1| hypothetical protein AN9495.2 [Aspergillus nidula... 52 4e-06
gb|EAA60266.1| hypothetical protein AN8717.2 [Aspergillus nidula... 52 4e-06
gb|AAT38752.1| hypothetical protein PGEC568H16.8 [Solanum demissum] 51 5e-06
gb|AAT38686.1| hypothetical protein PGEC568D21.5 [Solanum demissum] 49 2e-05
gb|AAU89737.1| hypothetical protein [Solanum tuberosum] 49 3e-05
pir||S20478 hypothetical protein - fruit fly (Drosophila melanog... 47 1e-04
gb|EAL18738.1| hypothetical protein CNBI3240 [Cryptococcus neofo... 44 8e-04
emb|CAG27692.1| tigger transposable element derived 14 [Equus ca... 39 0.020
emb|CAB16408.1| cbh [Schizosaccharomyces pombe] gi|19115495|ref|... 39 0.020
gb|AAH94458.1| Tigd2 protein [Mus musculus] 39 0.020
gb|AAB80698.1| Cbhp [Schizosaccharomyces pombe] 39 0.020
ref|XP_144820.3| PREDICTED: tigger transposable element derived ... 39 0.020
>gb|AAD14510.1| putative CENP-B/ARS binding protein-like protein [Arabidopsis
thaliana] gi|25411247|pir||B84479 hypothetical protein
At2g06660 [imported] - Arabidopsis thaliana
Length = 283
Score = 138 bits (347), Expect = 3e-32
Identities = 63/75 (84%), Positives = 69/75 (92%)
Query: 1 MTSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVR 60
MTSKIQPCDAGIIRAFKMHYRRRFYR+ILEGYE+GQSDPG INVLDAI+ A+ WTI+VR
Sbjct: 208 MTSKIQPCDAGIIRAFKMHYRRRFYREILEGYELGQSDPGTINVLDAISFAVSAWTINVR 267
Query: 61 KETIANCFRHCKIRS 75
+ETI NCFRHCKI S
Sbjct: 268 RETILNCFRHCKIHS 282
>gb|AAB01537.1| ARS binding protein 1
Length = 522
Score = 63.9 bits (154), Expect = 8e-10
Identities = 45/143 (31%), Positives = 69/143 (47%), Gaps = 15/143 (10%)
Query: 6 QPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRKETIA 65
QPC G+I A K YR+ + + ILE E+G++ NVL AI + WT D+ E I
Sbjct: 323 QPCSEGVIYALKASYRKHWVQYILEQNELGRNPYNTTNVLRAILWLVKAWTTDISPEIIE 382
Query: 66 NCFRHCKIRSASDVVGNLDES-----TFDEETQDLQTMINQCGYRNKMDIDNLMNYPGEN 120
N F + S V+G +ES DE L+ ++++ + M I++ ++ EN
Sbjct: 383 NAF------NLSGVLGLFNESAVTSRALDEMIHPLRELVSEFSVQAAMRIEDFISPSEEN 436
Query: 121 EACSEVQSLEDIVCTIIENNAED 143
V S EDI+ I +D
Sbjct: 437 ----IVDSSEDIINQIASQYMDD 455
>emb|CAB50967.1| abp1 [Schizosaccharomyces pombe] gi|19113252|ref|NP_596460.1|
hypothetical protein SPBC1105.04c [Schizosaccharomyces
pombe 972h-] gi|7490121|pir||T39281 ars binding protein
1 - fission yeast (Schizosaccharomyces pombe)
gi|19860259|sp|P49777|ABP1_SCHPO ARS-binding protein 1
Length = 522
Score = 63.2 bits (152), Expect = 1e-09
Identities = 45/143 (31%), Positives = 69/143 (47%), Gaps = 15/143 (10%)
Query: 6 QPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRKETIA 65
QPC G+I A K YR+ + + ILE E+G++ NVL AI + WT D+ E I
Sbjct: 323 QPCSEGVIYALKACYRKHWVQYILEQNELGRNPYNTTNVLRAILWLVKAWTTDISPEIIE 382
Query: 66 NCFRHCKIRSASDVVGNLDES-----TFDEETQDLQTMINQCGYRNKMDIDNLMNYPGEN 120
N F + S V+G +ES DE L+ ++++ + M I++ ++ EN
Sbjct: 383 NAF------NLSGVLGLFNESAVTSRALDEMIHPLRELVSEFSVQAAMRIEDFISPSEEN 436
Query: 121 EACSEVQSLEDIVCTIIENNAED 143
V S EDI+ I +D
Sbjct: 437 ----IVDSSEDIINQIASQYMDD 455
>gb|EAL92463.1| DNA binding protein [Aspergillus fumigatus Af293]
Length = 518
Score = 62.8 bits (151), Expect = 2e-09
Identities = 37/143 (25%), Positives = 67/143 (45%), Gaps = 2/143 (1%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
TS+ QP D GII +K +++R + R IL+ +E+ + +N+L A+ I +W D+
Sbjct: 311 TSRYQPLDQGIIHCWKSYWKRYWIRFILQEFELNRDPIASMNILRAVRWGIQSWEFDLSG 370
Query: 62 ETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGENE 121
+ I NCF+ + S +D + D+ + ++ MDID +N P E
Sbjct: 371 QVIQNCFQKA-LDSQPSYREPVDPAVLDDIQNAFSLLKLSTPIQDLMDIDTFLN-PAEEA 428
Query: 122 ACSEVQSLEDIVCTIIENNAEDD 144
++ +E + +DD
Sbjct: 429 VQDTLEDIESQILAQYGPELDDD 451
>gb|EAL92342.1| DNA binding protein [Aspergillus fumigatus Af293]
Length = 518
Score = 61.6 bits (148), Expect = 4e-09
Identities = 37/143 (25%), Positives = 66/143 (45%), Gaps = 2/143 (1%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
TS+ QP D GII +K +++R + R IL+ +E+ + +N+L A+ I +W D+
Sbjct: 311 TSRYQPLDQGIIHCWKSYWKRYWIRFILQEFELNRDPIASMNILRAVRWGIQSWEFDLSG 370
Query: 62 ETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGENE 121
+ I NCF+ + S +D + D+ + ++ MDID +N P E
Sbjct: 371 QVIQNCFQKA-LDSQPSYREPVDPAVLDDIQNAFSLLKLSTPIQDLMDIDTFLN-PAEEA 428
Query: 122 ACSEVQSLEDIVCTIIENNAEDD 144
+ +E + +DD
Sbjct: 429 VQDTPEDIESQILAQYGPELDDD 451
>gb|EAL85944.1| DNA binding protein [Aspergillus fumigatus Af293]
gi|66844340|gb|EAL84678.1| DNA binding protein
[Aspergillus fumigatus Af293]
Length = 515
Score = 57.0 bits (136), Expect = 9e-08
Identities = 33/114 (28%), Positives = 56/114 (48%), Gaps = 1/114 (0%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
TS+ QP D GII +K +++R + IL+ +E+ +NVL AI I +W D+
Sbjct: 311 TSRYQPLDQGIIFCWKSYWKRYWIHFILQEFELNHDPIASMNVLRAIRWGIQSWEFDLSG 370
Query: 62 ETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMN 115
+ I NCFR + S +D + D+ + + ++ MDI+ +N
Sbjct: 371 QVIQNCFRKA-LYSQPSYQEPVDPAVLDDIEKAFSLLKVSTPIQDLMDINTFLN 423
>gb|EAA57942.1| hypothetical protein AN6156.2 [Aspergillus nidulans FGSC A4]
gi|67539972|ref|XP_663760.1| hypothetical protein
AN6156_2 [Aspergillus nidulans FGSC A4]
gi|49097666|ref|XP_410293.1| hypothetical protein
AN6156.2 [Aspergillus nidulans FGSC A4]
Length = 467
Score = 54.3 bits (129), Expect = 6e-07
Identities = 26/67 (38%), Positives = 37/67 (54%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
TS+ QP D GII+ K++YRR++ R +L YE + +LD I + W DV+
Sbjct: 259 TSRFQPLDQGIIQNLKIYYRRQWLRYMLSHYERNLDPLQSVTILDCIRWLVRAWHHDVQS 318
Query: 62 ETIANCF 68
TI CF
Sbjct: 319 STILACF 325
>emb|CAA19330.1| SPBC14F5.12c [Schizosaccharomyces pombe]
gi|19113530|ref|NP_596738.1| hypothetical protein
SPBC14F5.12c [Schizosaccharomyces pombe 972h-]
gi|7490450|pir||T39458 DNA binding protein - fission
yeast (Schizosaccharomyces pombe)
gi|26391975|sp|O60108|CBH2_SCHPO CENP-B homolog protein
2 (CBHP-2)
Length = 514
Score = 53.9 bits (128), Expect = 8e-07
Identities = 40/147 (27%), Positives = 67/147 (45%), Gaps = 5/147 (3%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
T PC I+ AFK +YR+ + +LE +G++ +NVL A+ I +W +D
Sbjct: 318 TEIYHPCGQEIVYAFKSYYRKYWLNYMLEEIRLGKNPSKTMNVLKAVRWMIRSWNVDFEP 377
Query: 62 ETIANCF-RHCKIRSASDVVGNLDESTFDEETQDLQTMINQ-CGYRNKMDIDNLMNYPGE 119
I NCF R ++ + G E+ +LQ +I + G ++ I+N +N E
Sbjct: 378 SIIYNCFLRSGLFQNQQPLTGPSPETI--RIAVNLQELIGKYLGDKDIFQIENFINPIEE 435
Query: 120 NEACSEVQSLEDIVCTII-ENNAEDDE 145
N A + + + + E E DE
Sbjct: 436 NSADTNDDIVNQVAAQFLDEREFETDE 462
>gb|EAA66763.1| hypothetical protein AN9495.2 [Aspergillus nidulans FGSC A4]
gi|49140522|ref|XP_413632.1| hypothetical protein
AN9495.2 [Aspergillus nidulans FGSC A4]
Length = 513
Score = 51.6 bits (122), Expect = 4e-06
Identities = 25/67 (37%), Positives = 37/67 (54%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
TS+ QP D GII+ K++YR+++ R +L YE + +LD I + +W DV
Sbjct: 308 TSRYQPLDQGIIQNLKIYYRKQWLRYMLSHYERDLDPLESVTILDCIRWLVRSWHHDVLS 367
Query: 62 ETIANCF 68
TI CF
Sbjct: 368 STILACF 374
>gb|EAA60266.1| hypothetical protein AN8717.2 [Aspergillus nidulans FGSC A4]
gi|67903460|ref|XP_681986.1| hypothetical protein
AN8717_2 [Aspergillus nidulans FGSC A4]
gi|49128522|ref|XP_412854.1| hypothetical protein
AN8717.2 [Aspergillus nidulans FGSC A4]
Length = 524
Score = 51.6 bits (122), Expect = 4e-06
Identities = 25/67 (37%), Positives = 37/67 (54%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
TS+ QP D GII+ K++YR+++ R +L YE + +LD I + +W DV
Sbjct: 308 TSRYQPLDQGIIQNLKIYYRKQWLRYMLSHYERDLDPLESVTILDCIRWLVRSWHHDVLS 367
Query: 62 ETIANCF 68
TI CF
Sbjct: 368 STILACF 374
>gb|AAT38752.1| hypothetical protein PGEC568H16.8 [Solanum demissum]
Length = 106
Score = 51.2 bits (121), Expect = 5e-06
Identities = 22/37 (59%), Positives = 32/37 (86%)
Query: 108 MDIDNLMNYPGENEACSEVQSLEDIVCTIIENNAEDD 144
MD++NL++YPGE++ACSEVQ LE+IV TI + N +D+
Sbjct: 1 MDVNNLLDYPGESDACSEVQRLEEIVDTIGKCNVDDE 37
>gb|AAT38686.1| hypothetical protein PGEC568D21.5 [Solanum demissum]
Length = 104
Score = 49.3 bits (116), Expect = 2e-05
Identities = 21/37 (56%), Positives = 31/37 (83%)
Query: 108 MDIDNLMNYPGENEACSEVQSLEDIVCTIIENNAEDD 144
MD++NL++YPGE++ C EVQSLE+IV TI + N +D+
Sbjct: 1 MDVNNLLDYPGESDTCLEVQSLEEIVDTIGKCNVDDE 37
>gb|AAU89737.1| hypothetical protein [Solanum tuberosum]
Length = 106
Score = 48.9 bits (115), Expect = 3e-05
Identities = 21/36 (58%), Positives = 30/36 (83%)
Query: 108 MDIDNLMNYPGENEACSEVQSLEDIVCTIIENNAED 143
MD++NL++YPGE++ CSEVQ LE+IV TI + N +D
Sbjct: 1 MDVNNLLDYPGESDTCSEVQRLEEIVDTIGKCNVDD 36
>pir||S20478 hypothetical protein - fruit fly (Drosophila melanogaster)
transposon pogoR11
Length = 499
Score = 46.6 bits (109), Expect = 1e-04
Identities = 39/146 (26%), Positives = 67/146 (45%), Gaps = 14/146 (9%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDP---GKINVLDAINLAIPTWTID 58
T+ +QP D GII +FK+ YRR ++ L G+S +++LDA+ W +
Sbjct: 296 TALLQPLDQGIIHSFKLEYRRILVKQQLIAVNCGKSTVEFLKSLSLLDALYFVNQGWK-N 354
Query: 59 VRKETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPG 118
V+ TI NCF+ + + + + E D+ + IN Y N +D D
Sbjct: 355 VKMLTIQNCFKKAGFKFSFENEDTIAEKDKQCVEVDIVSNINWNEYAN-VDAD------- 406
Query: 119 ENEACSEVQSLEDIVCTIIENNAEDD 144
EAC ++IV +++++ D
Sbjct: 407 --EACHGQLDDDEIVRSLVQDAKTSD 430
>gb|EAL18738.1| hypothetical protein CNBI3240 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57228776|gb|AAW45211.1|
hypothetical protein CNH03540 [Cryptococcus neoformans
var. neoformans JEC21] gi|58270724|ref|XP_572518.1|
hypothetical protein CNH03540 [Cryptococcus neoformans
var. neoformans JEC21]
Length = 329
Score = 43.9 bits (102), Expect = 8e-04
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query: 1 MTSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDP--GKINVLDAINLAIPTWTID 58
+ + +QP DAGII AFK YR + L YE G K+++ A+ +A +W +
Sbjct: 151 LPAHVQPMDAGIISAFKSRYRAKVIPFALANYEDGVPPHLCYKVDIKSAMEMADDSWN-E 209
Query: 59 VRKETIANCFR 69
+ ET+ NC++
Sbjct: 210 ITGETVENCWK 220
>emb|CAG27692.1| tigger transposable element derived 14 [Equus caballus]
Length = 524
Score = 39.3 bits (90), Expect = 0.020
Identities = 21/72 (29%), Positives = 37/72 (51%), Gaps = 2/72 (2%)
Query: 2 TSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRK 61
T+ +QP + GII K+ YR ++IL D GK+++ AI++ W V++
Sbjct: 308 TAVLQPLNLGIIHTMKVLYRSHLLKQILLKLN-SNEDQGKVDIKQAIDMIAAAWW-SVKQ 365
Query: 62 ETIANCFRHCKI 73
T+ C++ I
Sbjct: 366 STVVKCWQKAGI 377
>emb|CAB16408.1| cbh [Schizosaccharomyces pombe] gi|19115495|ref|NP_594583.1|
hypothetical protein SPAC9E9.10c [Schizosaccharomyces
pombe 972h-] gi|7490220|pir||T39217 centromere k-type
repeat binding protein Cbhp - fission yeast
(Schizosaccharomyces pombe)
gi|12643496|sp|O14423|CBH1_SCHPO CENP-B homolog protein
1 (CBHP-1)
Length = 514
Score = 39.3 bits (90), Expect = 0.020
Identities = 28/140 (20%), Positives = 56/140 (40%), Gaps = 1/140 (0%)
Query: 7 PCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRKETIAN 66
P + GI+ FK +YRR + + L + + +NVL AI + +W V ++TI
Sbjct: 323 PFEQGIVDTFKANYRRYWLQYSLNQINILRDPLKAVNVLKAIRWMLWSWNFGVSEKTIQA 382
Query: 67 CFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGENEACSEV 126
F + ++ + ++ +I+ G I+ + E+E
Sbjct: 383 SFLRSGLLGPHSEAEGQGMESYQKAIMEIGNLISSKGSEAVKQINEYIRPSEEDETKLHQ 442
Query: 127 QSLEDIVCTIIENNA-EDDE 145
+++ + +E E DE
Sbjct: 443 DAVDTVAAEFLEERRFESDE 462
>gb|AAH94458.1| Tigd2 protein [Mus musculus]
Length = 504
Score = 39.3 bits (90), Expect = 0.020
Identities = 32/133 (24%), Positives = 58/133 (43%), Gaps = 3/133 (2%)
Query: 1 MTSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVR 60
+TS IQP G++ K +YR +K ++ ++ + VLDAI A W +R
Sbjct: 298 VTSLIQPMSQGVLTTVKRYYRAGLIQKYMDEGNDPKTFWKNLTVLDAIYEASRAWN-QIR 356
Query: 61 KETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGEN 120
TI ++ + + ++DE +L T++ ++I+N+ +
Sbjct: 357 SNTITRAWKKLFPGNEENPSVSIDEGAI--LAANLATVLQNTEDCEHVNIENIEQWFDSR 414
Query: 121 EACSEVQSLEDIV 133
+ S Q L DIV
Sbjct: 415 SSGSNCQVLADIV 427
>gb|AAB80698.1| Cbhp [Schizosaccharomyces pombe]
Length = 514
Score = 39.3 bits (90), Expect = 0.020
Identities = 28/140 (20%), Positives = 56/140 (40%), Gaps = 1/140 (0%)
Query: 7 PCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVRKETIAN 66
P + GI+ FK +YRR + + L + + +NVL AI + +W V ++TI
Sbjct: 323 PFEQGIVDTFKANYRRYWLQYSLNQINILRDPLKAVNVLKAIRWMLWSWNFGVSEKTIQA 382
Query: 67 CFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGENEACSEV 126
F + ++ + ++ +I+ G I+ + E+E
Sbjct: 383 SFLRSGLLGPHSEAEGQGMESYQKAIMEIGNLISSKGSEAVKQINEYIRPSEEDETKLHQ 442
Query: 127 QSLEDIVCTIIENNA-EDDE 145
+++ + +E E DE
Sbjct: 443 DAVDTVAAEFLEERRFESDE 462
>ref|XP_144820.3| PREDICTED: tigger transposable element derived 2 homolog [Mus
musculus]
Length = 525
Score = 39.3 bits (90), Expect = 0.020
Identities = 32/133 (24%), Positives = 58/133 (43%), Gaps = 3/133 (2%)
Query: 1 MTSKIQPCDAGIIRAFKMHYRRRFYRKILEGYEVGQSDPGKINVLDAINLAIPTWTIDVR 60
+TS IQP G++ K +YR +K ++ ++ + VLDAI A W +R
Sbjct: 319 VTSLIQPMSQGVLTTVKRYYRAGLIQKYMDEGNDPKTFWKNLTVLDAIYEASRAWN-QIR 377
Query: 61 KETIANCFRHCKIRSASDVVGNLDESTFDEETQDLQTMINQCGYRNKMDIDNLMNYPGEN 120
TI ++ + + ++DE +L T++ ++I+N+ +
Sbjct: 378 SNTITRAWKKLFPGNEENPSVSIDEGAI--LAANLATVLQNTEDCEHVNIENIEQWFDSR 435
Query: 121 EACSEVQSLEDIV 133
+ S Q L DIV
Sbjct: 436 SSGSNCQVLADIV 448
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 241,768,972
Number of Sequences: 2540612
Number of extensions: 8903799
Number of successful extensions: 19269
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 19234
Number of HSP's gapped (non-prelim): 97
length of query: 147
length of database: 863,360,394
effective HSP length: 123
effective length of query: 24
effective length of database: 550,865,118
effective search space: 13220762832
effective search space used: 13220762832
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146790.4


