

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.10 - phase: 1 /pseudo
(406 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
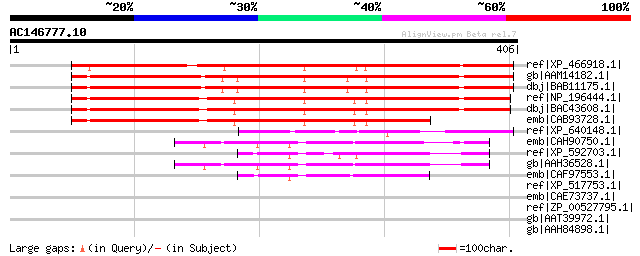
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_466918.1| unknown protein [Oryza sativa (japonica cultiva... 382 e-105
gb|AAM14182.1| unknown protein [Arabidopsis thaliana] gi|1849115... 372 e-101
dbj|BAB11175.1| unnamed protein product [Arabidopsis thaliana] 372 e-101
ref|NP_196444.1| expressed protein [Arabidopsis thaliana] 348 1e-94
dbj|BAC43608.1| unknown protein [Arabidopsis thaliana] 347 3e-94
emb|CAB93728.1| hypothetical protein (fragment at BAC end) [Arab... 295 1e-78
ref|XP_640148.1| hypothetical protein DDB0204860 [Dictyostelium ... 99 3e-19
emb|CAH90750.1| hypothetical protein [Pongo pygmaeus] 62 3e-08
ref|XP_592703.1| PREDICTED: similar to hypothetical protein MGC3... 60 2e-07
gb|AAH36528.1| Hypothetical protein MGC33648 [Homo sapiens] gi|2... 60 2e-07
emb|CAF97553.1| unnamed protein product [Tetraodon nigroviridis] 57 8e-07
ref|XP_517753.1| PREDICTED: similar to hypothetical protein MGC3... 40 0.17
emb|CAE73737.1| Hypothetical protein CBG21263 [Caenorhabditis br... 37 1.1
ref|ZP_00527795.1| Phage integrase:Phage integrase, N-terminal S... 35 3.2
gb|AAT39972.1| putative Sas10/Utp3 family protein [Solanum demis... 35 3.2
gb|AAH84898.1| Hypothetical LOC496567 [Xenopus tropicalis] gi|58... 35 5.4
>ref|XP_466918.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|49388185|dbj|BAD25311.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 405
Score = 382 bits (982), Expect = e-105
Identities = 212/398 (53%), Positives = 257/398 (64%), Gaps = 52/398 (13%)
Query: 50 QAVKTLAKSPTFA----FARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIAT 105
+AVKTLAK+P FA FARDPR LQ++ D+NRLFLYTSY RLG NA E DAEEII++A+
Sbjct: 10 EAVKTLAKNPMFANNRLFARDPRHLQFEADVNRLFLYTSYYRLGANAEEKDAEEIIDMAS 69
Query: 106 KASVADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSA 165
KASV++QQ VQENVH Q+ C MD++ LP+ K + + P+ S L A
Sbjct: 70 KASVSEQQKQVQENVHYQLTNMCQAMDSILLPDTKNG-------ASEANNYPRRSGLSFA 122
Query: 166 NGKFV-------------IHQLTIL-KLKDELGYTLNVKPSQISHKDAGQGLFLDGVVDV 211
G V +++ + K +D YTL+++PSQI HKDAGQGLFL G +
Sbjct: 123 VGTGVASANKPDVPSTRPLNRAELSNKFRDHFQYTLDIRPSQIPHKDAGQGLFLSGETNA 182
Query: 212 GAVVAFYPGVVYSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGR 269
GAV+A YPGVVYSPAYY +IPGY +D N YLITR+DG +IDA+ W GGD +E+W+G
Sbjct: 183 GAVLAIYPGVVYSPAYYRYIPGYPKIDACNNYLITRYDGTIIDAKPWQLGGDSREIWDGS 242
Query: 270 KMVDEKG----SQVDNSD-------------------DVLERRNPLALAHFANHPSKGML 306
+VD SQ NSD +VLERRNPLA HFANHP KG
Sbjct: 243 DLVDYNAVPSKSQESNSDRAWRMLSKPLKKGHTENFGEVLERRNPLAFGHFANHPPKGST 302
Query: 307 PNVMICPYDFPLIENDMRAYIPNVLFGNAAEENT-ERFGSFWFKSRVPRNNESHVPTTLK 365
PNVMICPYDFPL E DMR YIPN+ FG E T +RFGSFWFKS N E P LK
Sbjct: 303 PNVMICPYDFPLTEKDMRVYIPNITFGGEEEPVTMKRFGSFWFKSGRSGNQEGESP-VLK 361
Query: 366 TVVLVATRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
T+VLV+TR++ DEEL LNYR N+K+ PEWY PVDEEE
Sbjct: 362 TLVLVSTRSICDEELFLNYRYSNSKKRPEWYIPVDEEE 399
>gb|AAM14182.1| unknown protein [Arabidopsis thaliana] gi|18491157|gb|AAL69481.1|
unknown protein [Arabidopsis thaliana]
gi|22327005|ref|NP_197711.2| expressed protein
[Arabidopsis thaliana]
Length = 399
Score = 372 bits (955), Expect = e-101
Identities = 202/392 (51%), Positives = 256/392 (64%), Gaps = 46/392 (11%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
QAV LAKS TFA ++PRQLQ++ D+N+LF+YTSYNRLG+ A E DAEEIIE+A KA++
Sbjct: 10 QAVGVLAKSTTFA--KNPRQLQFEADVNKLFMYTSYNRLGREAEETDAEEIIEMAGKATL 67
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ MD + LP+ ++ S S + P+ S L A G
Sbjct: 68 SEQQKQVQENIHYQVEKFCSLMDGILLPDVRRNESGSQSTSPRP---PRRSGLTFAIGSN 124
Query: 170 ---------VIHQLTILKLKD-------ELGYTLNVKPSQISHKDAGQGLFLDGVVDVGA 213
++ + LKL D ++GYTL KPS I HKDAGQG F+ G DVG
Sbjct: 125 NAFPHADQPLVPETKPLKLNDVSQRLMDQMGYTLETKPSVIPHKDAGQGCFIKGEADVGT 184
Query: 214 VVAFYPGVVYSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGR-- 269
V+AFYPGV+YSPA+Y +IPGY +D QN YLITR+DG VI+AQ WG GG+ +E WNG
Sbjct: 185 VLAFYPGVIYSPAFYKYIPGYPKVDSQNSYLITRYDGTVINAQPWGLGGESREAWNGSYT 244
Query: 270 KMVDEKGSQVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMI 311
V +N D VLERRNPLA H ANHP+K M PNVMI
Sbjct: 245 PAVKANTKTAENGSDRLWKALSKPLEGSGKGKEVLERRNPLAFGHLANHPAKEMTPNVMI 304
Query: 312 CPYDFPLIENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVA 371
CPYDFPL+ D+R YIPN+ FG++ E +RFGSFWFK+ E+ V LKT+VLVA
Sbjct: 305 CPYDFPLMAKDLRPYIPNISFGDSGEIKMKRFGSFWFKTGSKNGLEAPV---LKTLVLVA 361
Query: 372 TRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
TR+L +EELLLNYRL N+KR P+WY PV+EEE
Sbjct: 362 TRSLCNEELLLNYRLSNSKRRPDWYTPVNEEE 393
>dbj|BAB11175.1| unnamed protein product [Arabidopsis thaliana]
Length = 437
Score = 372 bits (955), Expect = e-101
Identities = 202/392 (51%), Positives = 256/392 (64%), Gaps = 46/392 (11%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
QAV LAKS TFA ++PRQLQ++ D+N+LF+YTSYNRLG+ A E DAEEIIE+A KA++
Sbjct: 48 QAVGVLAKSTTFA--KNPRQLQFEADVNKLFMYTSYNRLGREAEETDAEEIIEMAGKATL 105
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ MD + LP+ ++ S S + P+ S L A G
Sbjct: 106 SEQQKQVQENIHYQVEKFCSLMDGILLPDVRRNESGSQSTSPRP---PRRSGLTFAIGSN 162
Query: 170 ---------VIHQLTILKLKD-------ELGYTLNVKPSQISHKDAGQGLFLDGVVDVGA 213
++ + LKL D ++GYTL KPS I HKDAGQG F+ G DVG
Sbjct: 163 NAFPHADQPLVPETKPLKLNDVSQRLMDQMGYTLETKPSVIPHKDAGQGCFIKGEADVGT 222
Query: 214 VVAFYPGVVYSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGR-- 269
V+AFYPGV+YSPA+Y +IPGY +D QN YLITR+DG VI+AQ WG GG+ +E WNG
Sbjct: 223 VLAFYPGVIYSPAFYKYIPGYPKVDSQNSYLITRYDGTVINAQPWGLGGESREAWNGSYT 282
Query: 270 KMVDEKGSQVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMI 311
V +N D VLERRNPLA H ANHP+K M PNVMI
Sbjct: 283 PAVKANTKTAENGSDRLWKALSKPLEGSGKGKEVLERRNPLAFGHLANHPAKEMTPNVMI 342
Query: 312 CPYDFPLIENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVA 371
CPYDFPL+ D+R YIPN+ FG++ E +RFGSFWFK+ E+ V LKT+VLVA
Sbjct: 343 CPYDFPLMAKDLRPYIPNISFGDSGEIKMKRFGSFWFKTGSKNGLEAPV---LKTLVLVA 399
Query: 372 TRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
TR+L +EELLLNYRL N+KR P+WY PV+EEE
Sbjct: 400 TRSLCNEELLLNYRLSNSKRRPDWYTPVNEEE 431
>ref|NP_196444.1| expressed protein [Arabidopsis thaliana]
Length = 386
Score = 348 bits (894), Expect = 1e-94
Identities = 190/382 (49%), Positives = 252/382 (65%), Gaps = 40/382 (10%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
Q V LAKS TFA ++PRQLQ++ D+N+LF++TSYNRLG++A EADAEEIIE+A KA+
Sbjct: 15 QGVGVLAKSTTFA--KNPRQLQFEADINKLFMFTSYNRLGRDAEEADAEEIIEMAGKATF 72
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ M+ + L N K D++ S+ + ++ A K
Sbjct: 73 SEQQKQVQENIHYQLQNFCSSMNEILLTNIDKTKDLNEPGSES-----RRDNIVPAADKP 127
Query: 170 VIHQLTILK-------LKDELGYTLNVKPSQISHKDAGQGLFLDGVVDVGAVVAFYPGVV 222
++ + LK LKD +GYTL +KPS I HKDAGQG F++G DVGAV+AFYPGV+
Sbjct: 128 IVPETKPLKLDEVSHRLKDRVGYTLEIKPSLIPHKDAGQGCFIEGEADVGAVLAFYPGVI 187
Query: 223 YSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDE---KGS 277
YSPA++ +IPGY +D QN YLIT +DG VI+AQ WGRGG +E+WNG E + +
Sbjct: 188 YSPAFHRYIPGYPNVDAQNSYLITGYDGTVINAQPWGRGGISREVWNGSFTTPEIRTEAT 247
Query: 278 QVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMICPYDFPLI 319
+NS D V+E RNPLA HF NHP K M NVMICPYDF L
Sbjct: 248 TTENSSDKVWKMLSRPLEGSGGVEAVIEMRNPLAFGHFINHPGKEMESNVMICPYDFLLS 307
Query: 320 ENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVATRALQDEE 379
E +MRAYIPNV FGN + RF SF ++ + ++HV LKT+VLVAT+AL +EE
Sbjct: 308 ETEMRAYIPNVAFGNTGDVKMGRFESFLSRTGSNSSLDAHV---LKTIVLVATKALCNEE 364
Query: 380 LLLNYRLGNTKRCPEWYAPVDE 401
L+LNYRL N+++ EWY P+DE
Sbjct: 365 LMLNYRLSNSEKRLEWYIPLDE 386
>dbj|BAC43608.1| unknown protein [Arabidopsis thaliana]
Length = 386
Score = 347 bits (891), Expect = 3e-94
Identities = 189/382 (49%), Positives = 252/382 (65%), Gaps = 40/382 (10%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
Q V LAKS TFA ++PRQLQ++ D+N+LF++TSYNRLG++A EADAEEIIE+A KA+
Sbjct: 15 QGVGVLAKSTTFA--KNPRQLQFEADINKLFMFTSYNRLGRDAEEADAEEIIEMAGKATF 72
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ M+ + L N K D++ S+ + ++ A K
Sbjct: 73 SEQQKQVQENIHYQLQNFCSSMNEILLTNIDKTKDLNEPGSES-----RRDNIVPAADKP 127
Query: 170 VIHQLTILK-------LKDELGYTLNVKPSQISHKDAGQGLFLDGVVDVGAVVAFYPGVV 222
++ + LK LKD +GYTL +KPS I HKDAGQG F++G DVGAV+AFYPGV+
Sbjct: 128 IVPETKPLKLDEVSHRLKDRVGYTLEIKPSLIPHKDAGQGCFIEGEADVGAVLAFYPGVI 187
Query: 223 YSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDE---KGS 277
Y+PA++ +IPGY +D QN YLIT +DG VI+AQ WGRGG +E+WNG E + +
Sbjct: 188 YTPAFHRYIPGYPNVDAQNSYLITGYDGTVINAQPWGRGGISREVWNGSFTTPEIRTEAT 247
Query: 278 QVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMICPYDFPLI 319
+NS D V+E RNPLA HF NHP K M NVMICPYDF L
Sbjct: 248 TTENSSDKVWKMLSRPLEGSGGVEAVIEMRNPLAFGHFINHPGKEMESNVMICPYDFLLS 307
Query: 320 ENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVATRALQDEE 379
E +MRAYIPNV FGN + RF SF ++ + ++HV LKT+VLVAT+AL +EE
Sbjct: 308 ETEMRAYIPNVAFGNTGDVKMGRFESFLSRTGSNSSLDAHV---LKTIVLVATKALCNEE 364
Query: 380 LLLNYRLGNTKRCPEWYAPVDE 401
L+LNYRL N+++ EWY P+DE
Sbjct: 365 LMLNYRLSNSEKRLEWYIPLDE 386
>emb|CAB93728.1| hypothetical protein (fragment at BAC end) [Arabidopsis thaliana]
gi|11358190|pir||T50512 hypothetical protein T22D6.210 -
Arabidopsis thaliana (fragment)
Length = 325
Score = 295 bits (756), Expect = 1e-78
Identities = 159/318 (50%), Positives = 208/318 (65%), Gaps = 37/318 (11%)
Query: 50 QAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIEIATKASV 109
Q V LAKS TFA ++PRQLQ++ D+N+LF++TSYNRLG++A EADAEEIIE+A KA+
Sbjct: 15 QGVGVLAKSTTFA--KNPRQLQFEADINKLFMFTSYNRLGRDAEEADAEEIIEMAGKATF 72
Query: 110 ADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDLRSANGKF 169
++QQ VQEN+H Q++ FC+ M+ + L N K D++ S+ + ++ A K
Sbjct: 73 SEQQKQVQENIHYQLQNFCSSMNEILLTNIDKTKDLNEPGSES-----RRDNIVPAADKP 127
Query: 170 VIHQLTILK-------LKDELGYTLNVKPSQISHKDAGQGLFLDGVVDVGAVVAFYPGVV 222
++ + LK LKD +GYTL +KPS I HKDAGQG F++G DVGAV+AFYPGV+
Sbjct: 128 IVPETKPLKLDEVSHRLKDRVGYTLEIKPSLIPHKDAGQGCFIEGEADVGAVLAFYPGVI 187
Query: 223 YSPAYYHHIPGY--LDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDE---KGS 277
YSPA++ +IPGY +D QN YLIT +DG VI+AQ WGRGG +E+WNG E + +
Sbjct: 188 YSPAFHRYIPGYPNVDAQNSYLITGYDGTVINAQPWGRGGISREVWNGSFTTPEIRTEAT 247
Query: 278 QVDNSDD------------------VLERRNPLALAHFANHPSKGMLPNVMICPYDFPLI 319
+NS D V+E RNPLA HF NHP K M NVMICPYDF L
Sbjct: 248 TTENSSDKVWKMLSRPLEGSGGVEAVIEMRNPLAFGHFINHPGKEMESNVMICPYDFLLS 307
Query: 320 ENDMRAYIPNVLFGNAAE 337
E +MRAYIPNV FGN +
Sbjct: 308 ETEMRAYIPNVAFGNTGD 325
>ref|XP_640148.1| hypothetical protein DDB0204860 [Dictyostelium discoideum]
gi|60468149|gb|EAL66159.1| hypothetical protein
DDB0204860 [Dictyostelium discoideum]
Length = 378
Score = 98.6 bits (244), Expect = 3e-19
Identities = 67/232 (28%), Positives = 110/232 (46%), Gaps = 37/232 (15%)
Query: 184 GYTLNVKPSQISHKDAGQGLFLDGVVDVGAVVAFYPGVVYSPAYYHHIPGYLDEQNPYLI 243
G+ L+V+ S I+H++AG G+ ++G G V+A YPG Y +IP + N Y+I
Sbjct: 152 GFRLHVRKSDINHEEAGYGVHIEGECVPGTVIAIYPGDTYDG---DNIPSNVISDNDYMI 208
Query: 244 TRHDGNVIDAQLWGRGGDKKELWNGRKMVDEKGSQVDNSDDVLERRNPLALAHFANHP-- 301
+R+DG VID + W + D EL K + G++ + D+L +NP A+ +F NHP
Sbjct: 209 SRYDGTVIDGRSWNKRAD--ELILKEKFFKKLGTK-SSGTDLLCYKNPFAIGNFINHPPI 265
Query: 302 ---------SKGMLPNVMICPYDF-PLIENDMRAYIPNVLFGNAAEENTERFGSFWFKSR 351
+ PNV+ Y+F N + YIPN K+
Sbjct: 266 ITKDGDGDTDQTKEPNVIAYSYNFRSNFPNHLSQYIPN-------------------KTV 306
Query: 352 VPRNNESHVPTTLKTVVLVATRALQDEELLLNYRLGNTKRCPEWYAPVDEEE 403
+ + ++++L+A + ++++E+ LNYR P WY D EE
Sbjct: 307 TEHSIFNDTSVLRRSLLLIAFKPIKNQEVFLNYRFNPDLPYPNWYKQPDLEE 358
>emb|CAH90750.1| hypothetical protein [Pongo pygmaeus]
Length = 273
Score = 62.0 bits (149), Expect = 3e-08
Identities = 70/266 (26%), Positives = 114/266 (42%), Gaps = 47/266 (17%)
Query: 133 AVFLPNEKKVNDVSFELSQQTK------ILPQHSDLRSANGKFVIHQLTILKLKDELGYT 186
A+FL + K +++ L + K + +H ++ + Q + K ++ L T
Sbjct: 36 ALFLNDFNKQSEILTMLPESVKSKYQDLLAVEHQGVKLLENRH--QQQSTFKPEEILYKT 93
Query: 187 LNVKPSQISHK--DAGQGLFLD-GVVDVGAVVAFYPGVV---YSPAYYHHIPGYLDEQNP 240
L +Q + AG+G+F+ G+V GAVV+ YPG V Y P ++ I NP
Sbjct: 94 LGFSVAQATSSLISAGKGVFVTKGLVPKGAVVSMYPGTVYQKYEPIFFQSI------GNP 147
Query: 241 YLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDEKGSQVDNSDDVLERRNPLALAHFANH 300
++ DG +ID G NGR + D++ E NPLA+ + N+
Sbjct: 148 FIFRCLDGVLIDGNDKGISKVVYRSCNGRDRLGPL-KMSDSTWLTSEIHNPLAVGQYVNN 206
Query: 301 PSKGMLPNVMICPYDFPLI-ENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESH 359
S NV +D P + +++ Y+PN+ + + + H
Sbjct: 207 CSNDRAANVCYQEFDVPAVFPIELKQYLPNIAY----------------------SYDKH 244
Query: 360 VPTTLKTVVLVATRAL-QDEELLLNY 384
P L+ VVLVA R + Q EEL NY
Sbjct: 245 SP--LRCVVLVALRDINQGEELFSNY 268
>ref|XP_592703.1| PREDICTED: similar to hypothetical protein MGC33648 [Bos taurus]
Length = 299
Score = 59.7 bits (143), Expect = 2e-07
Identities = 60/213 (28%), Positives = 95/213 (44%), Gaps = 49/213 (23%)
Query: 183 LGYTLNVKPSQISHKDAGQGLFLD-GVVDVGAVVAFYPGVV---YSPAYYHHIPGYLDEQ 238
LG+++ PS + AG+G+F+ G+V GAVV+ YPG V Y P ++ I
Sbjct: 120 LGFSVARAPSSLI--SAGKGVFVTKGLVRKGAVVSMYPGTVYQKYEPIFFQSI------G 171
Query: 239 NPYLITRHDGNVIDAQLWGRGGDK---KELWNGRKMVDEKG--SQVDNSDDVLERRNPLA 293
NP++ DG +ID G DK K ++ D+ G D++ E NPLA
Sbjct: 172 NPFIFRCLDGVLID------GNDKGISKVVYRSCSGRDQLGPLKMSDSTWLTSEIHNPLA 225
Query: 294 LAHFANHPSKGMLPNVMICPYDFPLI-ENDMRAYIPNVLFGNAAEENTERFGSFWFKSRV 352
+ + N+ S NV +D P + +++ Y+PN+ + +
Sbjct: 226 IGQYVNNCSNDRAANVCYQEFDVPAVFPIELKQYLPNIAYSYDKQ--------------- 270
Query: 353 PRNNESHVPTTLKTVVLVATRAL-QDEELLLNY 384
+ L+ V+LVA R + Q EEL NY
Sbjct: 271 ---------SPLRCVILVALRDIEQGEELFSNY 294
>gb|AAH36528.1| Hypothetical protein MGC33648 [Homo sapiens]
gi|24308476|ref|NP_714917.1| hypothetical protein
LOC133383 [Homo sapiens]
Length = 299
Score = 59.7 bits (143), Expect = 2e-07
Identities = 68/266 (25%), Positives = 112/266 (41%), Gaps = 47/266 (17%)
Query: 133 AVFLPNEKKVNDVSFELSQQTK------ILPQHSDLRSANGKFVIHQLTILKLKDELGYT 186
A+FL + K +++ L + K + +H ++ + Q + K ++ L T
Sbjct: 62 ALFLNDFNKQSEILTMLPESVKSKYQDLLAVEHQGVKLRENRH--QQQSTFKPEEILYKT 119
Query: 187 LNVKPSQISHK--DAGQGLFLD-GVVDVGAVVAFYPGVV---YSPAYYHHIPGYLDEQNP 240
L +Q + AG+G+F+ G+V GAVV+ YPG V Y P ++ I NP
Sbjct: 120 LGFSVAQATSSLISAGKGVFVTKGLVPKGAVVSMYPGTVYQKYEPIFFQSI------GNP 173
Query: 241 YLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDEKGSQVDNSDDVLERRNPLALAHFANH 300
++ DG +ID G NGR + D++ E NPLA+ + N+
Sbjct: 174 FIFRCLDGVLIDGNDKGISKVVYRSCNGRDRLGPL-KMSDSTWLTSEIHNPLAVGQYVNN 232
Query: 301 PSKGMLPNVMICPYDFPLI-ENDMRAYIPNVLFGNAAEENTERFGSFWFKSRVPRNNESH 359
S NV +D P + +++ Y+PN+ + +
Sbjct: 233 CSNDRAANVCYQEFDVPAVFPIELKQYLPNIAYSYDKQ---------------------- 270
Query: 360 VPTTLKTVVLVATRAL-QDEELLLNY 384
+ L+ VVLVA R + Q EEL NY
Sbjct: 271 --SPLRCVVLVALRDINQGEELFSNY 294
>emb|CAF97553.1| unnamed protein product [Tetraodon nigroviridis]
Length = 279
Score = 57.4 bits (137), Expect = 8e-07
Identities = 51/159 (32%), Positives = 73/159 (45%), Gaps = 14/159 (8%)
Query: 183 LGYTLNVKPSQISHKDAGQGLFLD-GVVDVGAVVAFYPGVV---YSPAYYHHIPGYLDEQ 238
LG+ ++ KPS + AG G+F+ G V GA+VA YPG V Y P ++ I +
Sbjct: 101 LGFCIDRKPSSLPF--AGTGVFVTKGFVPKGAIVALYPGTVYQAYEPIFFQSI------R 152
Query: 239 NPYLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDEKGSQVDNSDDVLERRNPLALAHFA 298
NP++ DG +ID G +GR V D S + NPLA+ +
Sbjct: 153 NPFVFRCIDGILIDGNDKGISRMVFRSCSGRDRVG-FFMMSDISWLTSDPLNPLAVGQYV 211
Query: 299 NHPSKGMLPNVMICPYDFP-LIENDMRAYIPNVLFGNAA 336
N+ S NV YD P ++R ++PNV F A
Sbjct: 212 NNCSNQRPANVCYQEYDVPDAFPTELRQFLPNVNFSQDA 250
>ref|XP_517753.1| PREDICTED: similar to hypothetical protein MGC33648 [Pan
troglodytes]
Length = 161
Score = 39.7 bits (91), Expect = 0.17
Identities = 32/119 (26%), Positives = 51/119 (41%), Gaps = 11/119 (9%)
Query: 218 YPGVVYS---PAYYHHIPGYLDEQNPYLITRHDGNVIDAQLWGRGGDKKELWNGRKMVDE 274
YPG VY P ++ I NP++ DG +ID G NGR +
Sbjct: 2 YPGTVYQKYEPIFFQSIG------NPFIFRCLDGVLIDGNDKGISKVVYRSCNGRDRLGP 55
Query: 275 KGSQVDNSDDVLERRNPLALAHFANHPSKGMLPNVMICPYDFPLI-ENDMRAYIPNVLF 332
D++ E NPLA+ + N+ S NV +D P + +++ Y+PN+ +
Sbjct: 56 L-KMSDSTWLTSEIHNPLAVGQYVNNCSNDRAANVCYQEFDVPAVFPIELKQYLPNIAY 113
>emb|CAE73737.1| Hypothetical protein CBG21263 [Caenorhabditis briggsae]
Length = 514
Score = 37.0 bits (84), Expect = 1.1
Identities = 16/47 (34%), Positives = 26/47 (55%)
Query: 355 NNESHVPTTLKTVVLVATRALQDEELLLNYRLGNTKRCPEWYAPVDE 401
N +SH P + +VL +D+ + LGNTK CP+ AP+++
Sbjct: 272 NQDSHEPVPCRLLVLWTKNDQKDDAESFKWILGNTKECPKCQAPIEK 318
>ref|ZP_00527795.1| Phage integrase:Phage integrase, N-terminal SAM-like [Chlorobium
phaeobacteroides DSM 266] gi|67776262|gb|EAM35922.1|
Phage integrase:Phage integrase, N-terminal SAM-like
[Chlorobium phaeobacteroides DSM 266]
Length = 328
Score = 35.4 bits (80), Expect = 3.2
Identities = 24/93 (25%), Positives = 46/93 (48%), Gaps = 3/93 (3%)
Query: 148 ELSQQTKILPQHSDLRSANGKFVIHQLTILKLKDELGYTLN--VKPSQISHKDAGQGLFL 205
+L Q + +H +L+ + F Q+T+ ++ +G L ++P I+ K A F
Sbjct: 45 DLDQFFSFMQRHLELQHMS-LFDPEQVTVASVRLFMGDLLQRGLQPKSIARKLASVKSFY 103
Query: 206 DGVVDVGAVVAFYPGVVYSPAYYHHIPGYLDEQ 238
+ + G + + +V +P Y H+PG+L EQ
Sbjct: 104 RYLQETGVIKSSVLSLVSTPRYPRHVPGFLTEQ 136
>gb|AAT39972.1| putative Sas10/Utp3 family protein [Solanum demissum]
Length = 746
Score = 35.4 bits (80), Expect = 3.2
Identities = 27/97 (27%), Positives = 47/97 (47%), Gaps = 7/97 (7%)
Query: 176 ILKLKDELGYTLNVKPSQISHKDAGQGLFLDGVVDVGAVV---AFYPGVVYS---PAYY- 228
++++K+ L T N+K ++ K+ F DGV ++ AFY + S P +Y
Sbjct: 351 LVEVKNLLNKTFNIKVLPLAVKNGKANNFFDGVWEIPTRTIHQAFYTVIAQSSIHPGFYM 410
Query: 229 HHIPGYLDEQNPYLITRHDGNVIDAQLWGRGGDKKEL 265
+ GYL + L+ ++ NV +L GR D + L
Sbjct: 411 KELDGYLPSKLEDLLHKNFDNVTGVKLAGRNLDSESL 447
>gb|AAH84898.1| Hypothetical LOC496567 [Xenopus tropicalis]
gi|58332000|ref|NP_001011149.1| hypothetical LOC496567
[Xenopus tropicalis]
Length = 312
Score = 34.7 bits (78), Expect = 5.4
Identities = 29/141 (20%), Positives = 69/141 (48%), Gaps = 10/141 (7%)
Query: 90 KNASEADAEEIIEIATKASVADQQMVVQENVHAQIKAFCTFMD-----AVFLPNEKKVND 144
+N +E+ ++ + A K ++ ++ + + + +++ ++D + LP E + D
Sbjct: 152 RNGTESVSDADLIQALKTTITNRYGLPDKYISVELETPLIYIDLKQNTSQKLPGEVDITD 211
Query: 145 VSFELSQQTKILPQHSDLRSANGKF-VIHQLTILKLKDELGYTLNVKPSQISHKDAGQGL 203
V++ + + K L AN +F ++ + +K+ + Y ++ KP +IS + G+
Sbjct: 212 VAYYMEKDVK----GDSLFPANNQFQILANGNKISVKEPMIYYIDEKPHEISMRHLTPGV 267
Query: 204 FLDGVVDVGAVVAFYPGVVYS 224
VV V AVVA ++++
Sbjct: 268 IAVIVVVVLAVVALIAVLIFT 288
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 696,416,195
Number of Sequences: 2540612
Number of extensions: 29949001
Number of successful extensions: 54106
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 54063
Number of HSP's gapped (non-prelim): 17
length of query: 406
length of database: 863,360,394
effective HSP length: 130
effective length of query: 276
effective length of database: 533,080,834
effective search space: 147130310184
effective search space used: 147130310184
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146777.10


