

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.15 + phase: 0
(536 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
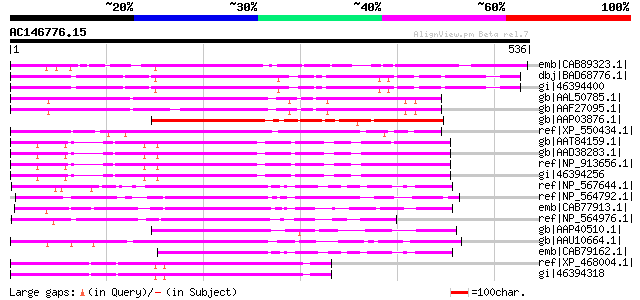
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB89323.1| putative protein [Arabidopsis thaliana] gi|15242... 336 1e-90
dbj|BAD68776.1| putative WRKY transcription factor [Oryza sativa... 335 2e-90
gi|46394400 TPA: WRKY transcription factor 73 [Oryza sativa (ind... 333 6e-90
gb|AAL50785.1| WRKY transcription factor 61 [Arabidopsis thalian... 308 2e-82
gb|AAF27095.1| Hypothetical protein [Arabidopsis thaliana] 285 3e-75
gb|AAP03876.1| Avr9/Cf-9 rapidly elicited protein 126 [Nicotiana... 275 3e-72
ref|XP_550434.1| WRKY transcription factor 6 -like [Oryza sativa... 262 2e-68
gb|AAT84159.1| transcription factor WRKY10 [Oryza sativa (indica... 222 2e-56
gb|AAD38283.1| putative WRKY DNA binding protein [Oryza sativa (... 222 3e-56
ref|NP_913656.1| putative WRKY DNA binding protein [Oryza sativa... 222 3e-56
gi|46394256 TPA: WRKY transcription factor 1 [Oryza sativa (japo... 222 3e-56
ref|NP_567644.1| WRKY family transcription factor [Arabidopsis t... 218 4e-55
ref|NP_564792.1| WRKY family transcription factor [Arabidopsis t... 215 3e-54
emb|CAB77913.1| putative DNA-binding protein [Arabidopsis thalia... 212 3e-53
ref|NP_564976.1| WRKY family transcription factor [Arabidopsis t... 208 3e-52
gb|AAP40510.1| putative WRKY family transcription factor [Arabid... 208 3e-52
gb|AAU10664.1| putative WRKY transcription factor [Oryza sativa ... 204 7e-51
emb|CAB79162.1| putative protein [Arabidopsis thaliana] gi|29613... 203 1e-50
ref|XP_468004.1| putative WRKY transcription factor [Oryza sativ... 188 3e-46
gi|46394318 TPA: WRKY transcription factor 32 [Oryza sativa (jap... 188 3e-46
>emb|CAB89323.1| putative protein [Arabidopsis thaliana]
gi|15242221|ref|NP_197017.1| WRKY family transcription
factor [Arabidopsis thaliana]
gi|29839650|sp|Q9LXG8|WRK72_ARATH Probable WRKY
transcription factor 72 (WRKY DNA-binding protein 72)
Length = 548
Score = 336 bits (862), Expect = 1e-90
Identities = 230/560 (41%), Positives = 307/560 (54%), Gaps = 85/560 (15%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQE-----TKKNNDNNHEE-------- 47
M EV+EEN++LK L +I ++Y++L+++F ++++QE TK N +H +
Sbjct: 43 MSEVKEENEKLKGMLERIESDYKSLKLRFFDIIQQEPSNTATKNQNMVDHPKPTTTDLSS 102
Query: 48 MNAESDLVSLSLGR---VPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFET 104
+ E +LVSLSLGR PS+++PK + EK + +S +N DEE K L+LG+
Sbjct: 103 FDQERELVSLSLGRRSSSPSDSVPKKE-EKTDAISA-EVNADEELTKAGLTLGIN----N 156
Query: 105 SKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMR-------DAEDEVAQ 157
G EGL + SE E W P K K DA+ E Q
Sbjct: 157 GNGGEPKEGLSMENRANSGSE----------EAWAPGKVTGKRSSPAPASGGDADGEAGQ 206
Query: 158 QTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQR 217
Q K+ARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAP CPVRKQVQR
Sbjct: 207 QNHVKRARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPGCPVRKQVQR 266
Query: 218 CVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLH 277
C +DMSILITTYEGTH+HSLPLSAT MASTTSAAASMLLSGSS+S P+A+ +
Sbjct: 267 CADDMSILITTYEGTHSHSLPLSATTMASTTSAAASMLLSGSSSS-----PAAE----MI 317
Query: 278 GLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQ 337
G N Y +N +P L S HPT+TLDLT+ +SS+SS + N + +N
Sbjct: 318 GNNLYDNSRFNNNNKSFYSPTLHSP-LHPTVTLDLTAPQHSSSSSSSLLSLNFN-KFSNS 375
Query: 338 LPRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQ 397
R+PS++L+FSS S S+N + NL + +G + + Y
Sbjct: 376 FQRFPSTSLNFSSTSST------------------SSNPSTLNLPAI-WGNGYSSYTPY- 415
Query: 398 TYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNTTNQG--NQN 455
Y +SN+ + V S TKA+T+DP+F S +AAA+S+++G+ Q
Sbjct: 416 PYNNVQFGTSNLGKTVQNSQSLTETLTKALTSDPSFHSVIAAAISTMVGSNGEQQIVGPR 475
Query: 456 QSAGENLSQKMKWAEMFQVSSTSLPSSSSKVNGCASSFLNKTAPVNNTQNGSLMLLSPSL 515
S N+ Q T+ +++ G SS L +N SL S L
Sbjct: 476 HSISNNIQQ------------TNTTNNNKGCGGYFSSLLMSNIMASNQTGASLDQPSSQL 523
Query: 516 -PFSATKSASTSPGGDNSDN 534
PFS K++S+S N N
Sbjct: 524 PPFSMFKNSSSSSSTTNFVN 543
>dbj|BAD68776.1| putative WRKY transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 624
Score = 335 bits (859), Expect = 2e-90
Identities = 235/560 (41%), Positives = 313/560 (54%), Gaps = 74/560 (13%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 60
MGEVREEN+RLK L++I +YR+L+ F ++++Q K ++ E +LVSL LG
Sbjct: 98 MGEVREENERLKTLLSRISHDYRSLQTHFYDVLQQGRAKKLPDSPATDIEEPELVSLRLG 157
Query: 61 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEE--LSLGL-ECKFETSKSGSTTEGLPNI 117
S K +E + S + E+F K + LSLGL +C+ + + S + +
Sbjct: 158 TSTS----KCKKEDKSTTSSEVKGSTEDFLKIKGGLSLGLSDCRVDANNSEKVQPDVMTL 213
Query: 118 PSPVNSSEVVPIKNDEVVETWPPSKTLNKTMR----DAEDEVAQQTPAKKARVCVRARCD 173
SP S E E E WPPSK L K +R +AED++A Q KKARV VRARCD
Sbjct: 214 -SPEGSFEDARDDTAETTEQWPPSKML-KNLRSVGAEAEDDIAPQPQVKKARVSVRARCD 271
Query: 174 TPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTH 233
PTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVA CPVRKQVQRC +DMSILITTYEGTH
Sbjct: 272 APTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAAGCPVRKQVQRCADDMSILITTYEGTH 331
Query: 234 NHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNL-----------HGLNFY 282
NH L +SATAMASTTSAAASML+SGSS+++ + P+A + L G F+
Sbjct: 332 NHPLSVSATAMASTTSAAASMLISGSSSTSLAAYPAAAASPALAFDASSKPPLIGGRPFF 391
Query: 283 LPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRYP 342
LP + A++S S+PTITLDLTS P+ ++TSS S +N RYP
Sbjct: 392 LPTAAAA--------AITSTPSYPTITLDLTS-PAAAATSS---HAAFSLSNRFSHTRYP 439
Query: 343 SSTLSFSSPESNPMHWNSFLNY-ATTQNQPYSNNRNNNN------LSTLNFGRQ------ 389
S+ +FS + W +L+Y A+ PY+ ++ LS++N RQ
Sbjct: 440 STGFTFSGSGPSSAPWPGYLSYGASLSAHPYNAGGGKSSSSFEAALSSINGSRQQGGGGG 499
Query: 390 -NTMESIYQTYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNT 448
+ +YQ MQ+ ++ + D+ KAITADP+F +ALAAA++S +G
Sbjct: 500 GGSAPPLYQ--MQQKAAAAAPPPPSVITDT----IAKAITADPSFHTALAAAITSYVG-- 551
Query: 449 TNQGNQNQSAGENLSQKMKWAEMFQVSSTSLPSSSSKVNGCASSFLNKTAPVNNTQNGSL 508
+G+ S GE+ +KW E + T S++ +SS S
Sbjct: 552 -KKGSPPASGGEDSKVGLKWGEHLGLGLTHSSLSTAAAAAASSS--------------SQ 596
Query: 509 MLLSPSLPFS-ATKSASTSP 527
M L PSL S +T SASTSP
Sbjct: 597 MFLQPSLGLSGSTTSASTSP 616
>gi|46394400 TPA: WRKY transcription factor 73 [Oryza sativa (indica
cultivar-group)]
Length = 527
Score = 333 bits (855), Expect = 6e-90
Identities = 234/560 (41%), Positives = 312/560 (54%), Gaps = 74/560 (13%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 60
MGEVREEN+RLK L++I +YR+L+ F ++++Q K ++ E + VSL LG
Sbjct: 1 MGEVREENERLKTLLSRISHDYRSLQTHFYDVLQQGRAKKLPDSPATDIEEPEFVSLRLG 60
Query: 61 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEE--LSLGL-ECKFETSKSGSTTEGLPNI 117
S K +E + S + E+F K + LSLGL +C+ + + S + +
Sbjct: 61 TSTS----KCKKEDKSTTSSEVKGSTEDFLKIKGGLSLGLSDCRVDANNSEKVQPDVMTL 116
Query: 118 PSPVNSSEVVPIKNDEVVETWPPSKTLNKTMR----DAEDEVAQQTPAKKARVCVRARCD 173
SP S E E E WPPSK L K +R +AED++A Q KKARV VRARCD
Sbjct: 117 -SPEGSFEDARDDTAETTEQWPPSKML-KNLRSVGAEAEDDIAPQPQVKKARVSVRARCD 174
Query: 174 TPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTH 233
PTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVA CPVRKQVQRC +DMSILITTYEGTH
Sbjct: 175 APTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAAGCPVRKQVQRCADDMSILITTYEGTH 234
Query: 234 NHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNL-----------HGLNFY 282
NH L +SATAMASTTSAAASML+SGSS+++ + P+A + L G F+
Sbjct: 235 NHPLSVSATAMASTTSAAASMLISGSSSTSLAAYPAAAASPALAFDASSKPPLIGGRPFF 294
Query: 283 LPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRYP 342
LP + A++S S+PTITLDLTS P+ ++TSS S +N RYP
Sbjct: 295 LPTAAAA--------AITSTPSYPTITLDLTS-PAAAATSS---HAAFSLSNRFSHTRYP 342
Query: 343 SSTLSFSSPESNPMHWNSFLNY-ATTQNQPYSNNRNNNN------LSTLNFGRQ------ 389
S+ +FS + W +L+Y A+ PY+ ++ LS++N RQ
Sbjct: 343 STGFTFSGSGPSSAPWPGYLSYGASLSAHPYNAGGGKSSSSFEAALSSINGSRQQGGGGG 402
Query: 390 -NTMESIYQTYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNT 448
+ +YQ MQ+ ++ + D+ KAITADP+F +ALAAA++S +G
Sbjct: 403 GGSAPPLYQ--MQQKAAAAAPPPPSVITDT----IAKAITADPSFHTALAAAITSYVG-- 454
Query: 449 TNQGNQNQSAGENLSQKMKWAEMFQVSSTSLPSSSSKVNGCASSFLNKTAPVNNTQNGSL 508
+G+ S GE+ +KW E + T S++ +SS S
Sbjct: 455 -KKGSPPASGGEDSKVGLKWGEHLGLGLTHSSLSTAAAAAASSS--------------SQ 499
Query: 509 MLLSPSLPFS-ATKSASTSP 527
M L PSL S +T SASTSP
Sbjct: 500 MFLQPSLGLSGSTTSASTSP 519
>gb|AAL50785.1| WRKY transcription factor 61 [Arabidopsis thaliana]
gi|30686070|ref|NP_173320.2| WRKY family transcription
factor [Arabidopsis thaliana]
gi|20978774|sp|Q8VWV6|WRK61_ARATH Probable WRKY
transcription factor 61 (WRKY DNA-binding protein 61)
Length = 480
Score = 308 bits (790), Expect = 2e-82
Identities = 196/478 (41%), Positives = 273/478 (57%), Gaps = 48/478 (10%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETK------------KNNDNNHEEM 48
M E +EEN+RLK L+KI ++ L+ Q+N ++ + + K D + E++
Sbjct: 1 MDEAKEENRRLKSSLSKIKKDFDILQTQYNQLMAKHNEPTKFQSKGHHQDKGEDEDREKV 60
Query: 49 NAESDLVSLSLGRVPSNNIPK--NDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSK 106
N +LVSLSLGR ++ +P N +EK V + + + + N++ GL E
Sbjct: 61 NEREELVSLSLGRRLNSEVPSGSNKEEKNKDVEEAEGDRNYDDNEKSSIQGLSMGIEYKA 120
Query: 107 SGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNK-TMRDAEDEVAQQTPAKKAR 165
+ E L + S + I N+ + + N + EDE+ Q KK R
Sbjct: 121 LSNPNEKLEIDHNQETMS--LEISNNNKIRSQNSFGFKNDGDDHEDEDEILPQNLVKKTR 178
Query: 166 VCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSIL 225
V VR+RC+TPTMNDGCQWRKYGQKIAKGNPCPRAYYRCT+A SCPVRKQVQRC EDMSIL
Sbjct: 179 VSVRSRCETPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTIAASCPVRKQVQRCSEDMSIL 238
Query: 226 ITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPD 285
I+TYEGTHNH LP+SATAMAS TSAAASMLLSG+S+S+S + +LHGLNF L
Sbjct: 239 ISTYEGTHNHPLPMSATAMASATSAAASMLLSGASSSSSAAA-------DLHGLNFSLSG 291
Query: 286 GT---KSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFV----RFNSSYNNNNQL 338
K +L +P S HPT+TLDLT ++SS+ PF+ RF+S +N ++
Sbjct: 292 NNITPKPKTHFLQSP---SSSGHPTVTLDLT---TSSSSQQPFLSMLNRFSSPPSNVSRS 345
Query: 339 PRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQT 398
YPS+ L+FS+ + M+W N + Y N + + T S +
Sbjct: 346 NSYPSTNLNFSNNTNTLMNWGGGGNPSDQYRAAYGNINTHQQSPYHKIIQTRTAGSSFDP 405
Query: 399 YMQKNNNSS---NISQHVGLQD-------STISAATKAITADPTFQSALAAALSSLIG 446
+ + +++ S N+ H+G+++ S + KAIT DP+FQSALA ALSS++G
Sbjct: 406 FGRSSSSHSPQINL-DHIGIKNIISHQVPSLPAETIKAITTDPSFQSALATALSSIMG 462
>gb|AAF27095.1| Hypothetical protein [Arabidopsis thaliana]
Length = 471
Score = 285 bits (729), Expect = 3e-75
Identities = 189/478 (39%), Positives = 265/478 (54%), Gaps = 57/478 (11%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETK------------KNNDNNHEEM 48
M E +EEN+RLK L+KI ++ L+ Q+N ++ + + K D + E++
Sbjct: 1 MDEAKEENRRLKSSLSKIKKDFDILQTQYNQLMAKHNEPTKFQSKGHHQDKGEDEDREKV 60
Query: 49 NAESDLVSLSLGRVPSNNIPK--NDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSK 106
N +LVSLSLGR ++ +P N +EK V + + + + N++ GL E
Sbjct: 61 NEREELVSLSLGRRLNSEVPSGSNKEEKNKDVEEAEGDRNYDDNEKSSIQGLSMGIEYKA 120
Query: 107 SGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNK-TMRDAEDEVAQQTPAKKAR 165
+ E L + S + I N+ + + N + EDE+ Q KK R
Sbjct: 121 LSNPNEKLEIDHNQETMS--LEISNNNKIRSQNSFGFKNDGDDHEDEDEILPQNLVKKTR 178
Query: 166 VCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSIL 225
V +MNDGCQWRKYGQKIAKGNPCPRAYYRCT+A SCPVRKQVQRC EDMSIL
Sbjct: 179 V---------SMNDGCQWRKYGQKIAKGNPCPRAYYRCTIAASCPVRKQVQRCSEDMSIL 229
Query: 226 ITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPD 285
I+TYEGTHNH LP+SATAMAS TSAAASMLLSG+S+S+S + +LHGLNF L
Sbjct: 230 ISTYEGTHNHPLPMSATAMASATSAAASMLLSGASSSSSAAA-------DLHGLNFSLSG 282
Query: 286 GT---KSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFV----RFNSSYNNNNQL 338
K +L +P S HPT+TLDLT ++SS+ PF+ RF+S +N ++
Sbjct: 283 NNITPKPKTHFLQSP---SSSGHPTVTLDLT---TSSSSQQPFLSMLNRFSSPPSNVSRS 336
Query: 339 PRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQT 398
YPS+ L+FS+ + M+W N + Y N + + T S +
Sbjct: 337 NSYPSTNLNFSNNTNTLMNWGGGGNPSDQYRAAYGNINTHQQSPYHKIIQTRTAGSSFDP 396
Query: 399 YMQKNNNSS---NISQHVGLQD-------STISAATKAITADPTFQSALAAALSSLIG 446
+ + +++ S N+ H+G+++ S + KAIT DP+FQSALA ALSS++G
Sbjct: 397 FGRSSSSHSPQINL-DHIGIKNIISHQVPSLPAETIKAITTDPSFQSALATALSSIMG 453
>gb|AAP03876.1| Avr9/Cf-9 rapidly elicited protein 126 [Nicotiana tabacum]
Length = 303
Score = 275 bits (702), Expect = 3e-72
Identities = 156/307 (50%), Positives = 208/307 (66%), Gaps = 24/307 (7%)
Query: 147 TMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVA 206
+++ A D Q K+ARV VR +CD PT+NDGCQWRKYGQKI++GNPCPR+YYRC+VA
Sbjct: 14 SVKRAGDVEVSQPNVKRARVSVRTKCDYPTINDGCQWRKYGQKISRGNPCPRSYYRCSVA 73
Query: 207 PSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGS 266
P CPVRKQVQRCVEDMS+LITTYEGTHNHSLP+ ATAMASTTSAAASMLLSGSS+S S
Sbjct: 74 PLCPVRKQVQRCVEDMSVLITTYEGTHNHSLPIEATAMASTTSAAASMLLSGSSSSQS-- 131
Query: 267 MPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFV 326
N +L LP+ +K+ LYLSNP S+ + PTITLD T+ P+ SS +S
Sbjct: 132 -----ANKDLRN----LPNNSKTTPLYLSNPP-SNSNPFPTITLDFTTFPTTSSFTS--F 179
Query: 327 RFNSSYNNNNQLPRYPSSTLSFSSPESNPMH---WNSFLNYATTQNQPYSNNRNNNNLST 383
F S++ +N + S++L+FSSPES+ + + +++Y T + PYS ++ N+ +
Sbjct: 180 NFPSNFQSNT---GFLSNSLNFSSPESDTLSKILGSGYVDYDPTTSLPYS--KSLTNIGS 234
Query: 384 LNFGRQNTMESIY-QTYMQKNNNS-SNISQHVGLQDSTISAATKAITADPTFQSALAAAL 441
N G+ + + Q + K+ NS SN + Q + TKAI +DP+FQS LAAA+
Sbjct: 235 SNLGKPSPAPKQFDQPVLGKSKNSISNNLKEESSQQAPTETLTKAIASDPSFQSVLAAAI 294
Query: 442 SSLIGNT 448
SS++G T
Sbjct: 295 SSMVGAT 301
>ref|XP_550434.1| WRKY transcription factor 6 -like [Oryza sativa (japonica
cultivar-group)] gi|55295932|dbj|BAD67800.1| WRKY
transcription factor 6 -like [Oryza sativa (japonica
cultivar-group)]
Length = 507
Score = 262 bits (669), Expect = 2e-68
Identities = 179/468 (38%), Positives = 251/468 (53%), Gaps = 48/468 (10%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAE-SDLVSLSL 59
MGEVREEN+RLK L +I+++Y++L F ++VK + + + + ++ + E DLVSLSL
Sbjct: 56 MGEVREENERLKTMLTRIVSDYKSLHTHFLDVVKVKEQTAAELSGDDDDDEPDDLVSLSL 115
Query: 60 GRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLEC------KFETSKSGSTTEG 113
P N ++ + D+ LSLGL C + ++
Sbjct: 116 CTRP--NAAATRRKGHERTPSSGGGGDDG----RLSLGLSCARGGVASDDDDDKQASRRA 169
Query: 114 LPNIP----SPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVR 169
LP +P S +S + E + S++ + A+DEV QQ AKKARV VR
Sbjct: 170 LPPMPVLNLSSDSSGDAAGAGAGEPTQPNKASRSSSGGGDGADDEVLQQQQAKKARVSVR 229
Query: 170 ARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTY 229
+CDTPTMNDGCQWRKYGQKI+KGNPCPRAYYRCTVAP+CPVRKQVQRC +DMSILITTY
Sbjct: 230 VKCDTPTMNDGCQWRKYGQKISKGNPCPRAYYRCTVAPNCPVRKQVQRCADDMSILITTY 289
Query: 230 EGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKS 289
EGTH+H LP +A AMASTTSAAA+ML SGS+ S H L F G
Sbjct: 290 EGTHSHPLPPAAAAMASTTSAAAAMLTSGSTNSTMHGSGGVH-----HHLPFASAVGGGG 344
Query: 290 NQLYLSNPALSSQHSHPTITLDLTSNPS--NSSTSSPFVRFNSSYNNNNQLPR-YPSSTL 346
L +S+ S PT+TLDLT+ S + S++SP+ + Y ++ LP + S L
Sbjct: 345 GVGLLGPTTISTATSCPTVTLDLTAPHSLLHPSSASPYAAAAAGYESSRALPAAWSSGYL 404
Query: 347 SFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLN--------FGRQNTMESIYQT 398
++ + ++ + P+ ++ ++ FG Q T
Sbjct: 405 AYGGAAAAQPYYAKGV-----APSPFGHHFGMMGMAAAAARPAPEQLFGGQTT-----SP 454
Query: 399 YMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIG 446
Y+Q+ ++ + KAIT+DP+FQS LAAA++S +G
Sbjct: 455 YLQRAIGGGGVAPA-----AVTDTIAKAITSDPSFQSVLAAAITSYMG 497
>gb|AAT84159.1| transcription factor WRKY10 [Oryza sativa (indica cultivar-group)]
Length = 580
Score = 222 bits (566), Expect = 2e-56
Identities = 176/476 (36%), Positives = 218/476 (44%), Gaps = 83/476 (17%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEM-------QFNNMVKQETKKNNDNNHEEMNAESD 53
+G + EENQRL+ L ++ T Y+AL+M Q M++ T+ H++ AE
Sbjct: 137 LGRMNEENQRLRGMLTQVTTSYQALQMHLVALMQQRPQMMQPPTQPEPPPPHQDGKAEGA 196
Query: 54 LVS---LSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGST 110
+V L LG PS+ G E E S S ST
Sbjct: 197 VVPRQFLDLG--PSSGA-----------------------------GGEAAEEPSNS-ST 224
Query: 111 TEGLPNIPSPVNSSEVVPIKNDEVVET---WPPSKTLNKTMRDA------EDEVAQQTPA 161
G P S + + + + T W P + + M A D+ AQ
Sbjct: 225 EAGSPRRSSSTGNKDQERGDSPDAPSTAAAWLPGRAMAPQMGAAGAAGKSHDQQAQDANM 284
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
+KARV VRAR + P + DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED
Sbjct: 285 RKARVSVRARSEAPIIADGCQWRKYGQKMAKGNPCPRAYYRCTMATGCPVRKQVQRCAED 344
Query: 222 MSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF 281
SILITTYEGTHNH LP +A AMASTTSAAASMLL SGSMPSA L NF
Sbjct: 345 RSILITTYEGTHNHPLPPAAMAMASTTSAAASMLL-------SGSMPSADGAAGLMSSNF 397
Query: 282 YLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRY 341
+ S +S+ PT+TLDLT P + + P N P
Sbjct: 398 LARTVLPCSS---SMATISASAPFPTVTLDLTHAPPGAPNAVPL---------NAARPGA 445
Query: 342 PSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTYMQ 401
P+ P A P N + S L + + Q
Sbjct: 446 PAPQFQVPLPGG---------GMAPAFAVPPQVLYNQSKFSGLQMSSDSAEAA--AAAAQ 494
Query: 402 KNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIG--NTTNQGNQN 455
I Q G T+SAA AITADP F ALAAA++S+IG + GN N
Sbjct: 495 FAQPRPPIGQLPGPLSDTVSAAAAAITADPNFTVALAAAITSIIGGQHAAAAGNSN 550
>gb|AAD38283.1| putative WRKY DNA binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 470
Score = 222 bits (565), Expect = 3e-56
Identities = 176/477 (36%), Positives = 218/477 (44%), Gaps = 82/477 (17%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEM-------QFNNMVKQETKKNNDNNHEEMNAESD 53
+G + EENQRL+ L ++ T Y+AL+M Q M++ T+ H++ AE
Sbjct: 24 LGRMNEENQRLRGMLTQVTTSYQALQMHLVALMQQRPQMMQPPTQPEPPPPHQDGKAEGA 83
Query: 54 LVS---LSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGST 110
+V L LG PS+ G E E S S ST
Sbjct: 84 VVPRQFLDLG--PSSGA-----------------------------GGEAAEEPSNS-ST 111
Query: 111 TEGLPNIPSPVNSSEVVPIKNDEVVET---WPPSKTLNKTMRDA------EDEVAQQTPA 161
G P S + + + + T W P + + M A D+ AQ
Sbjct: 112 EAGSPRRSSSTGNKDQERGDSPDAPSTAAAWLPGRAMAPQMGAAGAAGKSHDQQAQDANM 171
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
+KARV VRAR + P + DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED
Sbjct: 172 RKARVSVRARSEAPIIADGCQWRKYGQKMAKGNPCPRAYYRCTMATGCPVRKQVQRCAED 231
Query: 222 MSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF 281
SILITTYEGTHNH LP +A AMASTTSAAASMLL SGSMPSA L NF
Sbjct: 232 RSILITTYEGTHNHPLPPAAMAMASTTSAAASMLL-------SGSMPSADGAAGLMSSNF 284
Query: 282 YLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRY 341
+ S +S+ PT+TLDLT P + + P N P
Sbjct: 285 LARTVLPCSS---SMATISASAPFPTVTLDLTHAPPGAPNAVPL---------NAARPGA 332
Query: 342 PSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNT-MESIYQTYM 400
P+ P A P N + S L + +
Sbjct: 333 PAPQFQVPLPGG---------GMAPAFAVPPQVLYNQSKFSGLQMSSDSAEAAAAAAAAA 383
Query: 401 QKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIG--NTTNQGNQN 455
Q I Q G T+SAA AITADP F ALAAA++S+IG + GN N
Sbjct: 384 QFAQPRPPIGQLPGPLSDTVSAAAAAITADPNFTVALAAAITSIIGGQHAAAAGNSN 440
>ref|NP_913656.1| putative WRKY DNA binding protein [Oryza sativa (japonica
cultivar-group)] gi|13486842|dbj|BAB40073.1| putative
WRKY DNA binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 635
Score = 222 bits (565), Expect = 3e-56
Identities = 176/477 (36%), Positives = 218/477 (44%), Gaps = 82/477 (17%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEM-------QFNNMVKQETKKNNDNNHEEMNAESD 53
+G + EENQRL+ L ++ T Y+AL+M Q M++ T+ H++ AE
Sbjct: 189 LGRMNEENQRLRGMLTQVTTSYQALQMHLVALMQQRPQMMQPPTQPEPPPPHQDGKAEGA 248
Query: 54 LVS---LSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGST 110
+V L LG PS+ G E E S S ST
Sbjct: 249 VVPRQFLDLG--PSSGA-----------------------------GGEAAEEPSNS-ST 276
Query: 111 TEGLPNIPSPVNSSEVVPIKNDEVVET---WPPSKTLNKTMRDA------EDEVAQQTPA 161
G P S + + + + T W P + + M A D+ AQ
Sbjct: 277 EAGSPRRSSSTGNKDQERGDSPDAPSTAAAWLPGRAMAPQMGAAGAAGKSHDQQAQDANM 336
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
+KARV VRAR + P + DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED
Sbjct: 337 RKARVSVRARSEAPIIADGCQWRKYGQKMAKGNPCPRAYYRCTMATGCPVRKQVQRCAED 396
Query: 222 MSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF 281
SILITTYEGTHNH LP +A AMASTTSAAASMLL SGSMPSA L NF
Sbjct: 397 RSILITTYEGTHNHPLPPAAMAMASTTSAAASMLL-------SGSMPSADGAAGLMSSNF 449
Query: 282 YLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRY 341
+ S +S+ PT+TLDLT P + + P N P
Sbjct: 450 LARTVLPCSS---SMATISASAPFPTVTLDLTHAPPGAPNAVPL---------NAARPGA 497
Query: 342 PSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNT-MESIYQTYM 400
P+ P A P N + S L + +
Sbjct: 498 PAPQFQVPLPGG---------GMAPAFAVPPQVLYNQSKFSGLQMSSDSAEAAAAAAAAA 548
Query: 401 QKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIG--NTTNQGNQN 455
Q I Q G T+SAA AITADP F ALAAA++S+IG + GN N
Sbjct: 549 QFAQPRPPIGQLPGPLSDTVSAAAAAITADPNFTVALAAAITSIIGGQHAAAAGNSN 605
>gi|46394256 TPA: WRKY transcription factor 1 [Oryza sativa (japonica
cultivar-group)]
Length = 593
Score = 222 bits (565), Expect = 3e-56
Identities = 176/477 (36%), Positives = 218/477 (44%), Gaps = 82/477 (17%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEM-------QFNNMVKQETKKNNDNNHEEMNAESD 53
+G + EENQRL+ L ++ T Y+AL+M Q M++ T+ H++ AE
Sbjct: 147 LGRMNEENQRLRGMLTQVTTSYQALQMHLVALMQQRPQMMQPPTQPEPPPPHQDGKAEGA 206
Query: 54 LVS---LSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGST 110
+V L LG PS+ G E E S S ST
Sbjct: 207 VVPRQFLDLG--PSSGA-----------------------------GGEAAEEPSNS-ST 234
Query: 111 TEGLPNIPSPVNSSEVVPIKNDEVVET---WPPSKTLNKTMRDA------EDEVAQQTPA 161
G P S + + + + T W P + + M A D+ AQ
Sbjct: 235 EAGSPRRSSSTGNKDQERGDSPDAPSTAAAWLPGRAMAPQMGAAGAAGKSHDQQAQDANM 294
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
+KARV VRAR + P + DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED
Sbjct: 295 RKARVSVRARSEAPIIADGCQWRKYGQKMAKGNPCPRAYYRCTMATGCPVRKQVQRCAED 354
Query: 222 MSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNF 281
SILITTYEGTHNH LP +A AMASTTSAAASMLL SGSMPSA L NF
Sbjct: 355 RSILITTYEGTHNHPLPPAAMAMASTTSAAASMLL-------SGSMPSADGAAGLMSSNF 407
Query: 282 YLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRY 341
+ S +S+ PT+TLDLT P + + P N P
Sbjct: 408 LARTVLPCSS---SMATISASAPFPTVTLDLTHAPPGAPNAVPL---------NAARPGA 455
Query: 342 PSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNT-MESIYQTYM 400
P+ P A P N + S L + +
Sbjct: 456 PAPQFQVPLPGG---------GMAPAFAVPPQVLYNQSKFSGLQMSSDSAEAAAAAAAAA 506
Query: 401 QKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIG--NTTNQGNQN 455
Q I Q G T+SAA AITADP F ALAAA++S+IG + GN N
Sbjct: 507 QFAQPRPPIGQLPGPLSDTVSAAAAAITADPNFTVALAAAITSIIGGQHAAAAGNSN 563
>ref|NP_567644.1| WRKY family transcription factor [Arabidopsis thaliana]
gi|15990590|gb|AAL11009.1| WRKY transcription factor 31
[Arabidopsis thaliana] gi|20978775|sp|Q93WT0|WRK31_ARATH
Probable WRKY transcription factor 31 (WRKY DNA-binding
protein 31)
Length = 538
Score = 218 bits (555), Expect = 4e-55
Identities = 169/478 (35%), Positives = 240/478 (49%), Gaps = 94/478 (19%)
Query: 3 EVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNH---EEMNAES------- 52
+++ ENQRL+ L++ T + AL+MQ +++Q+ ++N+ +H +E AE
Sbjct: 120 KMKIENQRLRDMLSQATTNFNALQMQLVAVMRQQEQRNSSQDHLLAQESKAEGRKRQELQ 179
Query: 53 --------DLVSLSLGRVPSNNIPKNDQEKVNKVSKLAL---NNDEEFNKEELSLGLECK 101
DL S + ++ V S +L +N E K LG E
Sbjct: 180 IMVPRQFMDLGPSSGAAEHGAEVSSEERTTVRSGSPPSLLESSNPRENGKR--LLGREES 237
Query: 102 FETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAE--DEVAQQT 159
E S+S + G PN VP N PS + + R+ D+ A +
Sbjct: 238 SEESESNAW--GNPN---------KVPKHN--------PSSSNSNGNRNGNVIDQSAAEA 278
Query: 160 PAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCV 219
+KARV VRAR + ++DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC
Sbjct: 279 TMRKARVSVRARSEAAMISDGCQWRKYGQKMAKGNPCPRAYYRCTMAGGCPVRKQVQRCA 338
Query: 220 EDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGL 279
ED SILITTYEG HNH LP +ATAMASTT+AAASMLLSGS +S G M TN +
Sbjct: 339 EDRSILITTYEGNHNHPLPPAATAMASTTTAAASMLLSGSMSSQDGLM--NPTNLLARAI 396
Query: 280 NFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLP 339
LP + S +S+ PTITLDLT++P+ + N + NN L
Sbjct: 397 ---LPCSS-------SMATISASAPFPTITLDLTNSPNGN---------NPNMTTNNPLM 437
Query: 340 RYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTY 399
++ P NP + Q NN+ + S L Q +
Sbjct: 438 QFA------QRPGFNPAVLPQVV------GQAMYNNQQQSKFSGLQLPAQPLQIAA---- 481
Query: 400 MQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQS 457
+S++++ ++SAA+ AI +DP F +ALAAA++S++ +++Q N +
Sbjct: 482 ------TSSVAE-------SVSAASAAIASDPNFAAALAAAITSIMNGSSHQNNNTNN 526
>ref|NP_564792.1| WRKY family transcription factor [Arabidopsis thaliana]
gi|20978780|sp|Q9C519|WRKY6_ARATH WRKY transcription
factor 6 (WRKY DNA-binding protein 6) (AtWRKY6)
gi|12658412|gb|AAK01128.1| transcription factor WRKY6
[Arabidopsis thaliana] gi|12658410|gb|AAK01127.1|
transcription factor WRKY6 [Arabidopsis thaliana]
Length = 553
Score = 215 bits (547), Expect = 3e-54
Identities = 155/460 (33%), Positives = 218/460 (46%), Gaps = 78/460 (16%)
Query: 7 ENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLGRVPSNN 66
+NQ+L+ L ++ Y +L+M ++++Q+ ++NN E +V P
Sbjct: 170 DNQKLRELLTQVSNSYTSLQMHLVSLMQQQQQQNNKVIEAAEKPEETIV-------PRQF 222
Query: 67 IPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSPVNSSEV 126
I V + ++ ++ E+ + S E + + G SP S
Sbjct: 223 IDLGPTRAVGEAEDVSNSSSEDRTRSGGSSAAERRSNGKRLGREE-------SPETESNK 275
Query: 127 VPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKY 186
+ + +N T D+ A+ T +KARV VRAR + P ++DGCQWRKY
Sbjct: 276 I--------------QKVNSTTPTTFDQTAEAT-MRKARVSVRARSEAPMISDGCQWRKY 320
Query: 187 GQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMAS 246
GQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED SILITTYEG HNH LP +A AMAS
Sbjct: 321 GQKMAKGNPCPRAYYRCTMATGCPVRKQVQRCAEDRSILITTYEGNHNHPLPPAAVAMAS 380
Query: 247 TTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHP 306
TT+AAA+MLLSGS +S+ G M N L LP T S +S+ P
Sbjct: 381 TTTAAANMLLSGSMSSHDGMM---NPTNLL--ARAVLPCST-------SMATISASAPFP 428
Query: 307 TITLDLTSN--PSNSSTSSPFVRFNSSYNNNNQLPRYPSSTLSFSSPESNPMHWNSFLNY 364
T+TLDLT + P N S S N+++N+ Q P+ ++ P P L
Sbjct: 429 TVTLDLTHSPPPPNGSNPSSSAATNNNHNSLMQRPQQQQQQMTNLPPGMLPHVIGQAL-- 486
Query: 365 ATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTYMQKNNNSSNISQHVGLQDSTISAAT 424
N + S L F + + + Q ++
Sbjct: 487 -----------YNQSKFSGLQFSGGSPSTAAFS------------------QSHAVADTI 517
Query: 425 KAITADPTFQSALAAALSSLIGNTTNQGNQNQSAGENLSQ 464
A+TADP F +ALAA +SS+I T N + G N +Q
Sbjct: 518 TALTADPNFTAALAAVISSMINGT----NHHDGEGNNKNQ 553
>emb|CAB77913.1| putative DNA-binding protein [Arabidopsis thaliana]
gi|15990594|gb|AAL11011.1| WRKY transcription factor 42
[Arabidopsis thaliana] gi|4773884|gb|AAD29757.1|
putative DNA-binding protein [Arabidopsis thaliana]
gi|20978795|sp|Q9XEC3|WRK42_ARATH Probable WRKY
transcription factor 42 (WRKY DNA-binding protein 42)
gi|15233516|ref|NP_192354.1| WRKY family transcription
factor [Arabidopsis thaliana]
Length = 528
Score = 212 bits (539), Expect = 3e-53
Identities = 162/462 (35%), Positives = 227/462 (49%), Gaps = 81/462 (17%)
Query: 6 EENQRLKMCLNKIMTEYRALEMQFNNMVKQE-------TKKNNDN--NHEEMNAESDLVS 56
E+NQRLK L++ + +L+MQ +++Q+ T +NNDN N E+
Sbjct: 125 EDNQRLKQMLSQTTNNFNSLQMQLVAVMRQQEDHHHLATTENNDNVKNRHEVPEMVPRQF 184
Query: 57 LSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPN 116
+ LG S+ + ++ V S +L E+ + + + + E+ ++ S PN
Sbjct: 185 IDLGP-HSDEVSSEERTTVRSGSPPSLL--EKSSSRQNGKRVLVREESPETESNGWRNPN 241
Query: 117 -IPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTP 175
+P SS + E N + + E A+ T +KARV VRAR + P
Sbjct: 242 KVPKHHASSSICGGNGSE-----------NASSKVIEQAAAEAT-MRKARVSVRARSEAP 289
Query: 176 TMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNH 235
++DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED +ILITTYEG HNH
Sbjct: 290 MLSDGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCAEDRTILITTYEGNHNH 349
Query: 236 SLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLS 295
LP +A MASTT+AAASMLLSGS+ SN + + NL LP + S
Sbjct: 350 PLPPAAMNMASTTTAAASMLLSGSTMSNQDGLMNP---TNLLARTI-LPCSS-------S 398
Query: 296 NPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRYPSSTLSFSSPESNP 355
+S+ PTITLDLT +P N NN P +NP
Sbjct: 399 MATISASAPFPTITLDLTESP----------------NGNN--------------PTNNP 428
Query: 356 MHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTYMQKNNNSSNISQHVGL 415
+ S + NQ + L + +Q+ ++ N S
Sbjct: 429 LMQFSQRSGLVELNQSVLPHMMGQ---ALYYNQQSKFSGLHMPSQPLNAGES-------- 477
Query: 416 QDSTISAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQS 457
+SAAT AI ++P F +ALAAA++S+I + NQ N N +
Sbjct: 478 ----VSAATAAIASNPNFAAALAAAITSIINGSNNQQNGNNN 515
>ref|NP_564976.1| WRKY family transcription factor [Arabidopsis thaliana]
gi|15384221|gb|AAK96197.1| WRKY transcription factor 36
[Arabidopsis thaliana] gi|20978784|sp|Q9CAR4|WRK36_ARATH
Probable WRKY transcription factor 36 (WRKY DNA-binding
protein 36) gi|12325232|gb|AAG52562.1| hypothetical
protein; 74231-76109 [Arabidopsis thaliana]
Length = 387
Score = 208 bits (530), Expect = 3e-52
Identities = 140/404 (34%), Positives = 218/404 (53%), Gaps = 66/404 (16%)
Query: 3 EVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDN----NHEEMNAESDL-VSL 57
+VREEN++LK+ L+ I+ Y +L+MQ + ++ Q+ ++ + ++ N + D+ +SL
Sbjct: 35 KVREENEKLKLLLSTILNNYNSLQMQVSKVLGQQQGASSMELDHIDRQDENNDYDVDISL 94
Query: 58 SLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNI 117
LGR K +++ NKV K++ N EE + +LG + ++ ++ + +
Sbjct: 95 RLGRSEQ----KISKKEENKVDKISTKNVEESKDKRSALGFGFQIQSYEASKLDDLCRQV 150
Query: 118 PSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTM 177
++ V + D N+ +D +E +QT KK RVCV+A C+ P++
Sbjct: 151 KLANAENKCVSSRKDV-------KSVRNENHQDVLEE-HEQTGLKKTRVCVKASCEDPSI 202
Query: 178 NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRC-VEDMSILITTYEGTHNHS 236
NDGCQWRKYGQK AK NP PRAYYRC+++ +CPVRKQVQRC E+ S +TTYEG H+H
Sbjct: 203 NDGCQWRKYGQKTAKTNPLPRAYYRCSMSSNCPVRKQVQRCGEEETSAFMTTYEGNHDHP 262
Query: 237 LPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSN 296
LP+ A+ MA+ TSAAAS+L SGSS+S+S + S L+++ P +
Sbjct: 263 LPMEASHMAAGTSAAASLLQSGSSSSSSSTSAS---------LSYFFP---------FHH 304
Query: 297 PALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLP-RYPSSTLSFSSPESNP 355
++S+ +SHPT+TLDLT N NQLP YP S+ SFS
Sbjct: 305 FSISTTNSHPTVTLDLTRP-----------------NYPNQLPDDYPLSSSSFS------ 341
Query: 356 MHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQTY 399
LN+++ P S++ + N S L + +S+ Y
Sbjct: 342 ------LNFSSPDPPPPSSHDHTLNFSGLRTQAPLSTDSLLARY 379
>gb|AAP40510.1| putative WRKY family transcription factor [Arabidopsis thaliana]
gi|7268215|emb|CAB77742.1| putative DNA-binding protein
[Arabidopsis thaliana] gi|19172392|gb|AAL85881.1| WRKY
transcription factor 47 [Arabidopsis thaliana]
gi|15234284|ref|NP_192081.1| WRKY family transcription
factor [Arabidopsis thaliana]
gi|20978798|sp|Q9ZSI7|WRK47_ARATH Probable WRKY
transcription factor 47 (WRKY DNA-binding protein 47)
Length = 489
Score = 208 bits (530), Expect = 3e-52
Identities = 136/322 (42%), Positives = 160/322 (49%), Gaps = 74/322 (22%)
Query: 147 TMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVA 206
T + + Q P +KARV VRAR D T+NDGCQWRKYGQK+AKGNPCPRAYYRCT+A
Sbjct: 208 TNHEEQQNPHDQLPYRKARVSVRARSDATTVNDGCQWRKYGQKMAKGNPCPRAYYRCTMA 267
Query: 207 PSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGS 266
CPVRKQVQRC ED +IL TTYEG HNH LP SATAMA+TTSAAA+MLLSGSS+SN
Sbjct: 268 VGCPVRKQVQRCAEDTTILTTTYEGNHNHPLPPSATAMAATTSAAAAMLLSGSSSSNLHQ 327
Query: 267 MPSAQTNNNLHGLNFYLPDGTKSNQLYLSNP------ALSSQHSHPTITLDLTSNPSNSS 320
S+ P T S+ Y + P LS+ PTITLDLT+ P
Sbjct: 328 TLSS-------------PSATSSSSFYHNFPYTSTIATLSASAPFPTITLDLTNPP---R 371
Query: 321 TSSPFVRFNSSYNNNNQLPRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNN 380
P +F S Y LP NQ S N NN
Sbjct: 372 PLQPPPQFLSQYGPAAFLP---------------------------NANQIRSMNNNNQQ 404
Query: 381 LSTLN-FGRQNTMESIYQTYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAA 439
L N FG Q + + + AI DP F +ALAA
Sbjct: 405 LLIPNLFGPQAPPREM------------------------VDSVRAAIAMDPNFTAALAA 440
Query: 440 ALSSLIGNTTNQGNQNQSAGEN 461
A+S++IG N N N +N
Sbjct: 441 AISNIIGGGNNDNNNNTDINDN 462
>gb|AAU10664.1| putative WRKY transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 625
Score = 204 bits (518), Expect = 7e-51
Identities = 156/500 (31%), Positives = 225/500 (44%), Gaps = 86/500 (17%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQET----------------KKNNDNN 44
+G + EEN+RLK L+ + T+Y +L+MQF +++Q KK + +
Sbjct: 159 IGRLSEENKRLKNMLSNVTTKYNSLQMQFVTLMQQRRSVLAAPIHQQELLDPEKKEQEGS 218
Query: 45 HEEMNAESDLVSLSLGRV---PSNNIPKNDQEKVNKVSKLALNN--------------DE 87
++ +SLG P P + V + +N D
Sbjct: 219 QQQQQQLIPRQFISLGSASLQPDVEAPHSVVVVGGDVCAPSSSNPDAAVPAMMPLPHFDH 278
Query: 88 EFNKEELSLGLECKFETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKT 147
+ + G E +++ P P P + ++ +W P+ + +
Sbjct: 279 HNHHHPIHGGRERGSSPAEADHHRHHQQEQPPPP------PQQQQQLPPSWLPADKVPRF 332
Query: 148 MRDAEDE-VAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVA 206
+ E V + +KARV VRAR D P ++DGCQWRKYGQK+AKGNPCPRAYYRCT+A
Sbjct: 333 LPGKGPEPVPEAATMRKARVSVRARSDAPMISDGCQWRKYGQKMAKGNPCPRAYYRCTMA 392
Query: 207 PSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGS 266
CPVRKQVQRC ED ++LITTYEG HNH LP +A AMASTT+AAASMLLSGS S GS
Sbjct: 393 AGCPVRKQVQRCAEDRTVLITTYEGNHNHPLPPAAMAMASTTAAAASMLLSGSMPSADGS 452
Query: 267 MPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFV 326
+ + G NF + + +S+ PT+TLDLT
Sbjct: 453 L--------MAGSNFLARAVLPCSSTVAT---ISASAPFPTVTLDLTQTAP--------- 492
Query: 327 RFNSSYNNNNQLPRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNF 386
P P+S+ P P + L A +P + + L
Sbjct: 493 ------------PPPPASSTQPQPPRPEPAQLQAALAEAA---RPVALPQ----LFGQKL 533
Query: 387 GRQNTMESIYQTYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIG 446
Q+ + ++ K ++ G T++AAT AI +DP F + LAAAL+S IG
Sbjct: 534 YDQSKLSAVQAVAGTKGSDG-------GALADTVNAATAAIASDPNFTAVLAAALTSYIG 586
Query: 447 NTTNQGNQNQSAGENLSQKM 466
+ + G Q +
Sbjct: 587 SRSGSGGAGAGGSSGTVQPL 606
>emb|CAB79162.1| putative protein [Arabidopsis thaliana] gi|2961352|emb|CAA18110.1|
putative protein [Arabidopsis thaliana]
gi|11357339|pir||T49114 hypothetical protein AT4g22070 -
Arabidopsis thaliana
Length = 458
Score = 203 bits (517), Expect = 1e-50
Identities = 128/305 (41%), Positives = 172/305 (55%), Gaps = 50/305 (16%)
Query: 153 DEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVR 212
D+ A + +KARV VRAR + ++DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVR
Sbjct: 192 DQSAAEATMRKARVSVRARSEAAMISDGCQWRKYGQKMAKGNPCPRAYYRCTMAGGCPVR 251
Query: 213 KQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQT 272
KQVQRC ED SILITTYEG HNH LP +ATAMASTT+AAASMLLSGS +S G M T
Sbjct: 252 KQVQRCAEDRSILITTYEGNHNHPLPPAATAMASTTTAAASMLLSGSMSSQDGLM--NPT 309
Query: 273 NNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSY 332
N + LP + S +S+ PTITLDLT++P+ + N +
Sbjct: 310 NLLARAI---LPCSS-------SMATISASAPFPTITLDLTNSPNGN---------NPNM 350
Query: 333 NNNNQLPRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTM 392
NN L ++ P NP + Q NN+ + S L Q
Sbjct: 351 TTNNPLMQFA------QRPGFNPAVLPQVV------GQAMYNNQQQSKFSGLQLPAQPLQ 398
Query: 393 ESIYQTYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNTTNQG 452
+ +S++++ ++SAA+ AI +DP F +ALAAA++S++ +++Q
Sbjct: 399 IAA----------TSSVAE-------SVSAASAAIASDPNFAAALAAAITSIMNGSSHQN 441
Query: 453 NQNQS 457
N +
Sbjct: 442 NNTNN 446
>ref|XP_468004.1| putative WRKY transcription factor [Oryza sativa (japonica
cultivar-group)] gi|46805418|dbj|BAD16920.1| putative
WRKY transcription factor [Oryza sativa (japonica
cultivar-group)] gi|46805321|dbj|BAD16840.1| putative
WRKY transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 637
Score = 188 bits (478), Expect = 3e-46
Identities = 126/348 (36%), Positives = 181/348 (51%), Gaps = 24/348 (6%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 60
M ++EEN+ L+ ++K + +Y L+M+ +Q + E + + S G
Sbjct: 213 MERMKEENRMLRRVVDKTVRDYYELQMKLAAYQQQPAAADEPKETEVFLSLGATAAASAG 272
Query: 61 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSP 120
K+ ++ + + ++D + KE+L L L +S + + P
Sbjct: 273 CGGGFPEAKSKEQAAWRRRSVG-SDDSDCGKEDLGLSLSLGASSSYDDDQ-KAVEARPHD 330
Query: 121 VNSSEVVPIKNDEVVETWPPSKTLNKTMR------DAEDEVAQQ--------TPA-KKAR 165
V+ + + + P L ++ + A E+A PA +K R
Sbjct: 331 VDGAAAAAMIGGDGSRPAPRGYALLESSKVQGGAAPAAGELAAAGGITSQSVNPANRKTR 390
Query: 166 VCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSIL 225
V VR RC PTMNDGCQWRKYGQK+AKGNPCPRAYYRCTVAP CPVRKQVQRC+EDMSIL
Sbjct: 391 VSVRVRCQGPTMNDGCQWRKYGQKVAKGNPCPRAYYRCTVAPGCPVRKQVQRCLEDMSIL 450
Query: 226 ITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPD 285
+TTYEGTHNH LP+ ATAMASTTSAAA+ +L S+TS+S ++ ++ + + L
Sbjct: 451 VTTYEGTHNHPLPVGATAMASTTSAAATFMLLSSTTSSSSVSDASAAPSSSYLSPYLLNS 510
Query: 286 GTKSNQLYLSNPALSSQHSHPTITLDLTSN-PSNSSTSSPFVRFNSSY 332
+ + QH L+L N PS+SS +P +S Y
Sbjct: 511 ASPLLMPGATGGGGGMQH------LNLFGNSPSSSSLLAPQAPGSSKY 552
>gi|46394318 TPA: WRKY transcription factor 32 [Oryza sativa (japonica
cultivar-group)]
Length = 604
Score = 188 bits (478), Expect = 3e-46
Identities = 126/348 (36%), Positives = 181/348 (51%), Gaps = 24/348 (6%)
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 60
M ++EEN+ L+ ++K + +Y L+M+ +Q + E + + S G
Sbjct: 151 MERMKEENRMLRRVVDKTVRDYYELQMKLAAYQQQPAAADEPKETEVFLSLGATAAASAG 210
Query: 61 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSP 120
K+ ++ + + ++D + KE+L L L +S + + P
Sbjct: 211 CGGGFPEAKSKEQAAWRRRSVG-SDDSDCGKEDLGLSLSLGASSSYDDDQ-KAVEARPHD 268
Query: 121 VNSSEVVPIKNDEVVETWPPSKTLNKTMR------DAEDEVAQQ--------TPA-KKAR 165
V+ + + + P L ++ + A E+A PA +K R
Sbjct: 269 VDGAAAAAMIGGDGSRPAPRGYALLESSKVQGGAAPAAGELAAAGGITSQSVNPANRKTR 328
Query: 166 VCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSIL 225
V VR RC PTMNDGCQWRKYGQK+AKGNPCPRAYYRCTVAP CPVRKQVQRC+EDMSIL
Sbjct: 329 VSVRVRCQGPTMNDGCQWRKYGQKVAKGNPCPRAYYRCTVAPGCPVRKQVQRCLEDMSIL 388
Query: 226 ITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPD 285
+TTYEGTHNH LP+ ATAMASTTSAAA+ +L S+TS+S ++ ++ + + L
Sbjct: 389 VTTYEGTHNHPLPVGATAMASTTSAAATFMLLSSTTSSSSVSDASAAPSSSYLSPYLLNS 448
Query: 286 GTKSNQLYLSNPALSSQHSHPTITLDLTSN-PSNSSTSSPFVRFNSSY 332
+ + QH L+L N PS+SS +P +S Y
Sbjct: 449 ASPLLMPGATGGGGGMQH------LNLFGNSPSSSSLLAPQAPGSSKY 490
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.306 0.121 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 923,621,833
Number of Sequences: 2540612
Number of extensions: 41103777
Number of successful extensions: 383155
Number of sequences better than 10.0: 4590
Number of HSP's better than 10.0 without gapping: 1269
Number of HSP's successfully gapped in prelim test: 3539
Number of HSP's that attempted gapping in prelim test: 214475
Number of HSP's gapped (non-prelim): 46297
length of query: 536
length of database: 863,360,394
effective HSP length: 133
effective length of query: 403
effective length of database: 525,458,998
effective search space: 211759976194
effective search space used: 211759976194
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146776.15


