

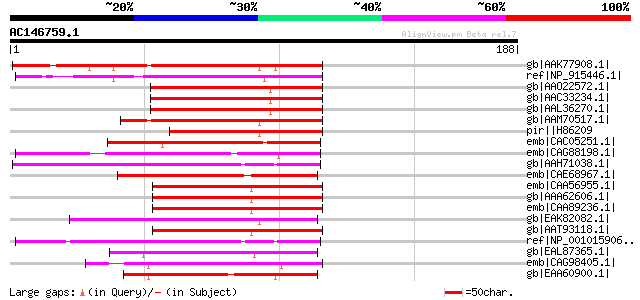
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146759.1 + phase: 0 /pseudo
(188 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK77908.1| AAA-metalloprotease FtsH [Pisum sativum] 99 8e-20
ref|NP_915446.1| putative AAA-metalloprotease [Oryza sativa (jap... 87 2e-16
gb|AAO22572.1| putative AAA-type ATPase [Arabidopsis thaliana] g... 80 4e-14
gb|AAC33234.1| putative AAA-type ATPase [Arabidopsis thaliana] g... 80 4e-14
gb|AAL36270.1| putative AAA-type ATPase [Arabidopsis thaliana] 79 6e-14
gb|AAM70517.1| At1g07510/F22G5_9 [Arabidopsis thaliana] gi|17381... 75 9e-13
pir||H86209 protein F22G5.10 [imported] - Arabidopsis thaliana g... 69 9e-11
emb|CAC05251.1| SPBC543.09 [Schizosaccharomyces pombe] gi|191135... 62 6e-09
emb|CAG88198.1| unnamed protein product [Debaryomyces hansenii C... 55 1e-06
gb|AAH71038.1| LOC432063 protein [Xenopus laevis] 55 1e-06
emb|CAE68967.1| Hypothetical protein CBG14947 [Caenorhabditis br... 55 1e-06
emb|CAA56955.1| YTA12 (=RCA1) [Saccharomyces cerevisiae] 54 2e-06
gb|AAA62606.1| Rca1p 54 2e-06
emb|CAA89236.1| Rca1p [Saccharomyces cerevisiae] gi|6323736|ref|... 54 2e-06
gb|EAK82082.1| hypothetical protein UM00898.1 [Ustilago maydis 5... 54 2e-06
gb|AAT93118.1| YMR089C [Saccharomyces cerevisiae] 54 2e-06
ref|NP_001015906.1| hypothetical protein LOC548660 [Xenopus trop... 53 4e-06
gb|EAL87365.1| matrix AAA protease MAP-1 [Aspergillus fumigatus ... 52 7e-06
emb|CAG98405.1| unnamed protein product [Kluyveromyces lactis NR... 52 7e-06
gb|EAA60900.1| hypothetical protein AN4557.2 [Aspergillus nidula... 52 7e-06
>gb|AAK77908.1| AAA-metalloprotease FtsH [Pisum sativum]
Length = 810
Score = 98.6 bits (244), Expect = 8e-20
Identities = 65/134 (48%), Positives = 83/134 (61%), Gaps = 22/134 (16%)
Query: 2 GNDKKKTNSQSLAQWMKVAVVGSFQEA-LKKLGTLMT--LIGGLILSFFYFRPFGKNQEI 58
G DKK + + G FQEA +K+ + +T L+ GL LS F F P + Q+I
Sbjct: 107 GGDKKNESKEDSKS--NTEDQGGFQEAFMKQFQSFLTPLLVMGLFLSSFSFGP-REQQQI 163
Query: 59 SFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNN-----ESEVAK-----------YKYYF 102
SFQEFK K+LEPGLVDHIVVSNK +AKIYV+N+ +SEV + YKYYF
Sbjct: 164 SFQEFKNKLLEPGLVDHIVVSNKSVAKIYVRNSPRDQADSEVLQGNLPAKGSSGHYKYYF 223
Query: 103 KIGSVDSFERKLKK 116
IGSV+SFE KL++
Sbjct: 224 NIGSVESFEEKLEE 237
>ref|NP_915446.1| putative AAA-metalloprotease [Oryza sativa (japonica
cultivar-group)] gi|19571026|dbj|BAB86453.1| putative
AAA-metalloprotease FtsH [Oryza sativa (japonica
cultivar-group)]
Length = 802
Score = 87.4 bits (215), Expect = 2e-16
Identities = 58/137 (42%), Positives = 77/137 (55%), Gaps = 34/137 (24%)
Query: 3 NDKKKTNSQSLAQWMKVAVVGSFQEALKKLGTLMT---LIGGLILSFFYFRPFGKNQEIS 59
+D KK +S QW +F+E++K+ ++ L G L+LS QEIS
Sbjct: 109 SDSKKQSSSG-DQW-------NFEESIKQFKDMIAPLFLFGLLLLSA---SASSSEQEIS 157
Query: 60 FQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNES--------------------EVAKYK 99
FQEFK K+LEPGLVDHIVVSNK +AK+YV+++ S + YK
Sbjct: 158 FQEFKNKLLEPGLVDHIVVSNKSIAKVYVRSSPSIDRIQDSDIHITTSHLPGIESPSSYK 217
Query: 100 YYFKIGSVDSFERKLKK 116
YYF IGSVDSFE KL++
Sbjct: 218 YYFNIGSVDSFEEKLQE 234
>gb|AAO22572.1| putative AAA-type ATPase [Arabidopsis thaliana]
gi|30684118|ref|NP_850129.1| FtsH protease, putative
[Arabidopsis thaliana]
Length = 809
Score = 79.7 bits (195), Expect = 4e-14
Identities = 42/83 (50%), Positives = 53/83 (63%), Gaps = 19/83 (22%)
Query: 53 GKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEV----------------- 95
G+ Q+ISFQEFK K+LEPGLVDHI VSNK +AK+YV++ +
Sbjct: 154 GEQQQISFQEFKNKLLEPGLVDHIDVSNKSVAKVYVRSTPKDQQTTDVVHGNGNGIPAKR 213
Query: 96 --AKYKYYFKIGSVDSFERKLKK 116
+YKYYF IGSVDSFE KL++
Sbjct: 214 TGGQYKYYFNIGSVDSFEEKLEE 236
>gb|AAC33234.1| putative AAA-type ATPase [Arabidopsis thaliana]
gi|7487874|pir||T02738 probable AAA-type ATPase
[imported] - Arabidopsis thaliana
Length = 807
Score = 79.7 bits (195), Expect = 4e-14
Identities = 42/83 (50%), Positives = 53/83 (63%), Gaps = 19/83 (22%)
Query: 53 GKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEV----------------- 95
G+ Q+ISFQEFK K+LEPGLVDHI VSNK +AK+YV++ +
Sbjct: 152 GEQQQISFQEFKNKLLEPGLVDHIDVSNKSVAKVYVRSTPKDQQTTDVVHGNGNGIPAKR 211
Query: 96 --AKYKYYFKIGSVDSFERKLKK 116
+YKYYF IGSVDSFE KL++
Sbjct: 212 TGGQYKYYFNIGSVDSFEEKLEE 234
>gb|AAL36270.1| putative AAA-type ATPase [Arabidopsis thaliana]
Length = 809
Score = 79.0 bits (193), Expect = 6e-14
Identities = 42/83 (50%), Positives = 52/83 (62%), Gaps = 19/83 (22%)
Query: 53 GKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEV----------------- 95
G Q+ISFQEFK K+LEPGLVDHI VSNK +AK+YV++ +
Sbjct: 154 GDQQQISFQEFKNKLLEPGLVDHIDVSNKSVAKVYVRSTPKDQQTTDVVHGNGNGIPAKR 213
Query: 96 --AKYKYYFKIGSVDSFERKLKK 116
+YKYYF IGSVDSFE KL++
Sbjct: 214 TGGQYKYYFNIGSVDSFEEKLEE 236
>gb|AAM70517.1| At1g07510/F22G5_9 [Arabidopsis thaliana] gi|17381253|gb|AAL36045.1|
At1g07510/F22G5_9 [Arabidopsis thaliana]
gi|22329400|ref|NP_172231.2| FtsH protease, putative
[Arabidopsis thaliana]
Length = 813
Score = 75.1 bits (183), Expect = 9e-13
Identities = 45/94 (47%), Positives = 58/94 (60%), Gaps = 20/94 (21%)
Query: 42 LILSFFYFRPFGKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNN---------- 91
LILS F + Q+ISFQEFK K+LE GLVDHI VSNKE+AK+YV+++
Sbjct: 148 LILSTFSLGS-REQQQISFQEFKNKLLEAGLVDHIDVSNKEVAKVYVRSSPKSQTTEEVV 206
Query: 92 ---------ESEVAKYKYYFKIGSVDSFERKLKK 116
+ +YKYYF IGSV+SFE KL++
Sbjct: 207 QGPGNGVPAKGRGGQYKYYFNIGSVESFEEKLEE 240
>pir||H86209 protein F22G5.10 [imported] - Arabidopsis thaliana
gi|8778569|gb|AAF79577.1| F22G5.10 [Arabidopsis
thaliana]
Length = 843
Score = 68.6 bits (166), Expect = 9e-11
Identities = 37/76 (48%), Positives = 48/76 (62%), Gaps = 19/76 (25%)
Query: 60 FQEFKIKVLEPGLVDHIVVSNKELAKIYVKNN-------------------ESEVAKYKY 100
FQEFK K+LE GLVDHI VSNKE+AK+YV+++ + +YKY
Sbjct: 173 FQEFKNKLLEAGLVDHIDVSNKEVAKVYVRSSPKSQTTEEVVQGPGNGVPAKGRGGQYKY 232
Query: 101 YFKIGSVDSFERKLKK 116
YF IGSV+SFE KL++
Sbjct: 233 YFNIGSVESFEEKLEE 248
>emb|CAC05251.1| SPBC543.09 [Schizosaccharomyces pombe] gi|19113589|ref|NP_596797.1|
hypothetical protein SPBC543.09 [Schizosaccharomyces
pombe 972h-]
Length = 773
Score = 62.4 bits (150), Expect = 6e-09
Identities = 28/82 (34%), Positives = 55/82 (66%), Gaps = 4/82 (4%)
Query: 37 TLIGGLILSFFYFRPFGKN---QEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNES 93
T++GG+++++ + N QEI++Q+F+ + L+ GLV+ +VV N+ + ++ ++ +
Sbjct: 135 TILGGILVAYILYNVLSPNANMQEITWQDFRQQFLDKGLVERLVVVNRNMVRVILRGGVA 194
Query: 94 EVAKYKYYFKIGSVDSFERKLK 115
+YYF IGS+DSF+RKL+
Sbjct: 195 S-GSGQYYFSIGSIDSFDRKLE 215
>emb|CAG88198.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50422751|ref|XP_459952.1| unnamed protein product
[Debaryomyces hansenii]
Length = 864
Score = 54.7 bits (130), Expect = 1e-06
Identities = 36/118 (30%), Positives = 60/118 (50%), Gaps = 12/118 (10%)
Query: 3 NDKKKTNSQSLAQWMKVAVVGSFQEALKKLGTLMTLIGGLILSFFYFRPFGKNQEISFQE 62
N K K N + ++ + GS + + + L+GGL++ Y + EISFQ+
Sbjct: 203 NGKNKNNKDKKSMEFEINLNGSPMKYFQ-----LGLLGGLLMYVLYNLSQDSDNEISFQK 257
Query: 63 FKIKVLEPGLVDHIVVSNKELAKIYVKNNESEVAKY-----KYYFKIGSVDSFERKLK 115
F L LV +VV N + + V+ N++ A+Y + YF +GS++SFER L+
Sbjct: 258 FAADFLSKNLVSRLVVVNNK--TVIVELNDNGKAQYGNHQGRLYFNVGSIESFERSLR 313
>gb|AAH71038.1| LOC432063 protein [Xenopus laevis]
Length = 788
Score = 54.7 bits (130), Expect = 1e-06
Identities = 36/114 (31%), Positives = 56/114 (48%), Gaps = 2/114 (1%)
Query: 2 GNDKKKTNSQSLAQWMKVAVVGSFQEALKKLGTLMTLIGGLILSFFYFRPFGKNQEISFQ 61
G + +K +Q + W + G F K L L G+ F +F +EI+++
Sbjct: 109 GPNNRKDGNQEESTWWRRLQKGDFPWDDKDFRNLAILAAGIASGFLFFYLRDPGREINWK 168
Query: 62 EFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEVAKYKYYFKIGSVDSFERKLK 115
+F L G+VD + V NK+ ++ + S KY +F IGSVDSFER L+
Sbjct: 169 DFVHLYLARGVVDRLEVVNKQFVRV-IPTAGSTSEKY-VWFNIGSVDSFERNLE 220
>emb|CAE68967.1| Hypothetical protein CBG14947 [Caenorhabditis briggsae]
Length = 779
Score = 54.7 bits (130), Expect = 1e-06
Identities = 29/74 (39%), Positives = 45/74 (60%), Gaps = 2/74 (2%)
Query: 41 GLILSFFYFRPFGKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEVAKYKY 100
G++++ + F + +EIS++EF LEPGLV+ + V +K +I ++ + A
Sbjct: 130 GILIALYLFMDYQSYREISWKEFYSDFLEPGLVERLEVVDKRWVRIV--SSSGKYAGQTC 187
Query: 101 YFKIGSVDSFERKL 114
YF IGSVDSFER L
Sbjct: 188 YFNIGSVDSFERSL 201
>emb|CAA56955.1| YTA12 (=RCA1) [Saccharomyces cerevisiae]
Length = 825
Score = 54.3 bits (129), Expect = 2e-06
Identities = 27/67 (40%), Positives = 41/67 (60%), Gaps = 4/67 (5%)
Query: 54 KNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYV----KNNESEVAKYKYYFKIGSVDS 109
+ EI++Q+F+ K+L G V ++V NK + K+ + KN + YYF IGS+DS
Sbjct: 202 EQSEITWQDFREKLLAKGYVAKLIVVNKSMVKVMLNDNGKNQADNYGRNFYYFTIGSIDS 261
Query: 110 FERKLKK 116
FE KL+K
Sbjct: 262 FEHKLQK 268
>gb|AAA62606.1| Rca1p
Length = 825
Score = 54.3 bits (129), Expect = 2e-06
Identities = 27/67 (40%), Positives = 41/67 (60%), Gaps = 4/67 (5%)
Query: 54 KNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYV----KNNESEVAKYKYYFKIGSVDS 109
+ EI++Q+F+ K+L G V ++V NK + K+ + KN + YYF IGS+DS
Sbjct: 202 EQSEITWQDFREKLLAKGYVAKLIVVNKSMVKVMLNDNGKNQADNYGRNFYYFTIGSIDS 261
Query: 110 FERKLKK 116
FE KL+K
Sbjct: 262 FEHKLQK 268
>emb|CAA89236.1| Rca1p [Saccharomyces cerevisiae] gi|6323736|ref|NP_013807.1|
Component, with Afg3p, of the mitochondrial inner
membrane m-AAA protease that mediates degradation of
misfolded or unassembled proteins and is also required
for correct assembly of mitochondrial enzyme complexes;
Yta12p [Saccharomyces cerevisiae]
gi|1710045|sp|P40341|RCA1_YEAST Mitochondrial
respiratory chain complexes assembly protein RCA1
(TAT-binding homolog 12)
Length = 825
Score = 54.3 bits (129), Expect = 2e-06
Identities = 27/67 (40%), Positives = 41/67 (60%), Gaps = 4/67 (5%)
Query: 54 KNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYV----KNNESEVAKYKYYFKIGSVDS 109
+ EI++Q+F+ K+L G V ++V NK + K+ + KN + YYF IGS+DS
Sbjct: 202 EQSEITWQDFREKLLAKGYVAKLIVVNKSMVKVMLNDNGKNQADNYGRNFYYFTIGSIDS 261
Query: 110 FERKLKK 116
FE KL+K
Sbjct: 262 FEHKLQK 268
>gb|EAK82082.1| hypothetical protein UM00898.1 [Ustilago maydis 521]
gi|49068448|ref|XP_398513.1| hypothetical protein
UM00898.1 [Ustilago maydis 521]
Length = 860
Score = 54.3 bits (129), Expect = 2e-06
Identities = 33/108 (30%), Positives = 52/108 (47%), Gaps = 16/108 (14%)
Query: 23 GSFQEALKKLGTLMTLIGGLILSFFYFRPFGKNQEISFQEFKIKVLEPGLVDHIVVSNKE 82
G F E T++ I L + P ++EI++QEF+ L+ GLVD +VV N+
Sbjct: 176 GQFTEIRINANTILATIVSTYLFYRLTSPDQPSREITWQEFRTAFLDKGLVDRLVVVNRS 235
Query: 83 LAKIYVKNN----------------ESEVAKYKYYFKIGSVDSFERKL 114
K+Y+ +N S Y+F +GSV++FER+L
Sbjct: 236 KVKVYLHSNATGSLYPSPNGGSSTPASGSGHAAYWFSVGSVEAFERRL 283
>gb|AAT93118.1| YMR089C [Saccharomyces cerevisiae]
Length = 825
Score = 54.3 bits (129), Expect = 2e-06
Identities = 27/67 (40%), Positives = 41/67 (60%), Gaps = 4/67 (5%)
Query: 54 KNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYV----KNNESEVAKYKYYFKIGSVDS 109
+ EI++Q+F+ K+L G V ++V NK + K+ + KN + YYF IGS+DS
Sbjct: 202 EQSEITWQDFREKLLAKGYVAKLIVVNKSMVKVMLNDNGKNQADNYGRNFYYFTIGSIDS 261
Query: 110 FERKLKK 116
FE KL+K
Sbjct: 262 FEHKLQK 268
>ref|NP_001015906.1| hypothetical protein LOC548660 [Xenopus tropicalis]
Length = 778
Score = 53.1 bits (126), Expect = 4e-06
Identities = 36/113 (31%), Positives = 57/113 (49%), Gaps = 3/113 (2%)
Query: 3 NDKKKTNSQSLAQWMKVAVVGSFQEALKKLGTLMTLIGGLILSFFYFRPFGKNQEISFQE 62
N+++ N + A W ++ G F K L L G+ F +F +EI+++E
Sbjct: 101 NNRRDGNKEESAWWRRLQK-GDFPWDDKDFRNLAILAAGIASGFLFFYLRDPGREINWKE 159
Query: 63 FKIKVLEPGLVDHIVVSNKELAKIYVKNNESEVAKYKYYFKIGSVDSFERKLK 115
F L G+VD + V NK+ ++ + + KY +F IGSVDSFER L+
Sbjct: 160 FVHLYLARGVVDRLEVVNKQFVRV-IPTAGTTSEKY-VWFNIGSVDSFERNLE 210
>gb|EAL87365.1| matrix AAA protease MAP-1 [Aspergillus fumigatus Af293]
Length = 885
Score = 52.4 bits (124), Expect = 7e-06
Identities = 31/88 (35%), Positives = 49/88 (55%), Gaps = 11/88 (12%)
Query: 38 LIGGLILSFFY--FRPFGKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVK------ 89
LI LI + Y F P +EI++QEF+ + GLV+ + V N+ + K+ +
Sbjct: 236 LITSLITYYVYRSFFPGENGKEITWQEFRANFFDKGLVEKLTVVNRAVVKVDLHRDALAR 295
Query: 90 ---NNESEVAKYKYYFKIGSVDSFERKL 114
++ + ++YYF IGSVDSFER+L
Sbjct: 296 VYPDSPALQPNFRYYFSIGSVDSFERRL 323
>emb|CAG98405.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50311345|ref|XP_455697.1| unnamed protein product
[Kluyveromyces lactis]
Length = 842
Score = 52.4 bits (124), Expect = 7e-06
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 11/94 (11%)
Query: 29 LKKLGTLMTLIGGLILSFFYFR--PFGKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKI 86
L K+G +T IL+F R + +E+++QEF+ ++L G V ++V N L K+
Sbjct: 169 LFKVGLTLT-----ILAFILHRLNSMEEQRELTWQEFRNQLLVKGYVSKLIVINNNLVKV 223
Query: 87 YVKNNESEVAKYK----YYFKIGSVDSFERKLKK 116
+ +N ++ Y+F IGSV+SFE KLKK
Sbjct: 224 ILNDNGKNQPEHMGHDFYFFTIGSVESFEHKLKK 257
>gb|EAA60900.1| hypothetical protein AN4557.2 [Aspergillus nidulans FGSC A4]
gi|67536774|ref|XP_662161.1| hypothetical protein
AN4557_2 [Aspergillus nidulans FGSC A4]
gi|49094466|ref|XP_408694.1| hypothetical protein
AN4557.2 [Aspergillus nidulans FGSC A4]
Length = 883
Score = 52.4 bits (124), Expect = 7e-06
Identities = 28/86 (32%), Positives = 53/86 (61%), Gaps = 16/86 (18%)
Query: 43 ILSFFYFR---PFGKNQEISFQEFKIKVLEPGLVDHIVVSNKELAKIYVKNNESEVAK-- 97
+++++ +R P +++I+++EF+ K L+ GLV+ + V+N+ ++ V+ N VA+
Sbjct: 238 VIAYYIYRSIFPGDNSKDITWEEFRSKFLDKGLVERLTVTNR--TRVRVELNRDAVARTY 295
Query: 98 ---------YKYYFKIGSVDSFERKL 114
+ YYF +GSV+SFERKL
Sbjct: 296 PDSPAASPNFYYYFTVGSVESFERKL 321
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.337 0.148 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 283,666,730
Number of Sequences: 2540612
Number of extensions: 10487493
Number of successful extensions: 42479
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 42374
Number of HSP's gapped (non-prelim): 89
length of query: 188
length of database: 863,360,394
effective HSP length: 120
effective length of query: 68
effective length of database: 558,486,954
effective search space: 37977112872
effective search space used: 37977112872
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146759.1


