

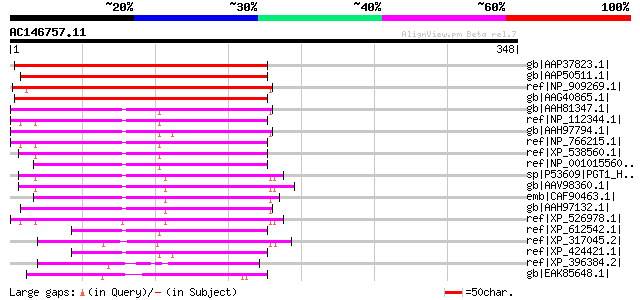
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.11 - phase: 0 /pseudo
(348 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP37823.1| At2g39550 [Arabidopsis thaliana] gi|27311719|gb|A... 206 6e-52
gb|AAP50511.1| geranylgeranyltransferase type I beta subunit [Ca... 198 2e-49
ref|NP_909269.1| putative geranylgeranyl-diphosphate geranylgera... 197 4e-49
gb|AAG40865.1| geranylgeranyltransferase beta subunit [Arabidops... 173 6e-42
gb|AAH81347.1| MGC89595 protein [Xenopus tropicalis] gi|56118392... 124 5e-27
ref|NP_112344.1| protein geranylgeranyltransferase type I, beta ... 116 9e-25
gb|AAH97794.1| Unknown (protein for MGC:115505) [Xenopus laevis] 115 2e-24
ref|NP_766215.1| geranylgeranyltransferase type I [Mus musculus... 115 3e-24
ref|XP_538560.1| PREDICTED: similar to protein geranylgeranyltra... 114 3e-24
ref|NP_001015560.1| protein geranylgeranyltransferase type I, be... 114 4e-24
sp|P53609|PGT1_HUMAN Geranylgeranyl transferase type I beta subu... 113 7e-24
gb|AAV98360.1| geranylgeranyltransferase type I beta subunit [Ho... 110 5e-23
emb|CAF90463.1| unnamed protein product [Tetraodon nigroviridis] 108 2e-22
gb|AAH97132.1| Unknown (protein for MGC:114070) [Danio rerio] 106 9e-22
ref|XP_526978.1| PREDICTED: protein geranylgeranyltransferase ty... 100 1e-19
ref|XP_612542.1| PREDICTED: similar to protein geranylgeranyltra... 96 1e-18
ref|XP_317045.2| ENSANGP00000019628 [Anopheles gambiae str. PEST... 96 2e-18
ref|XP_424421.1| PREDICTED: similar to tripartite motif protein ... 94 6e-18
ref|XP_396384.2| PREDICTED: similar to MGC89595 protein [Apis me... 88 4e-16
gb|EAK85648.1| hypothetical protein UM04373.1 [Ustilago maydis 5... 86 2e-15
>gb|AAP37823.1| At2g39550 [Arabidopsis thaliana] gi|27311719|gb|AAO00825.1|
putative geranylgeranyl transferase type I beta subunit
[Arabidopsis thaliana] gi|3355484|gb|AAC27846.1|
putative geranylgeranyl transferase type I beta subunit
[Arabidopsis thaliana] gi|15225494|ref|NP_181487.1|
geranylgeranyl transferase type I beta subunit (GGT-IB)
[Arabidopsis thaliana] gi|7484977|pir||T00565
geranylgeranyl-diphosphate geranylgeranyltransferase I
beta chain homolog At2g39550 - Arabidopsis thaliana
Length = 375
Score = 206 bits (525), Expect = 6e-52
Identities = 103/174 (59%), Positives = 125/174 (71%)
Query: 4 SEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE 63
S S + +KD H+ + +MMY LLP Y+SQEIN LTLA+F+IS L L + V+
Sbjct: 24 SPPVQSSPSANFEKDRHLMYLEMMYELLPYHYQSQEINRLTLAHFIISGLHFLGARDRVD 83
Query: 64 KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALA 123
K+ VA WVLSFQ +G+FYGF GS++SQFP DENG HN SHLASTYCALA
Sbjct: 84 KDVVAKWVLSFQAFPTNRVSLKDGEFYGFFGSRSSQFPIDENGDLKHNGSHLASTYCALA 143
Query: 124 ILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
ILK++G+DLS++DS+S+ SM NLQQ DGSFMPIHIGGETDLRFVYCA I M
Sbjct: 144 ILKVIGHDLSTIDSKSLLISMINLQQDDGSFMPIHIGGETDLRFVYCAAAICYM 197
Score = 34.3 bits (77), Expect = 5.7
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 27/100 (27%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPP 102
L Y + +L+S ++KE+ N++L+ Q G GF
Sbjct: 185 LRFVYCAAAICYMLDSWSGMDKESAKNYILNCQSYDG-----------GF---------- 223
Query: 103 DENGVFHHNNSHLASTYCALAILKIVGY---DLSSLDSES 139
G+ + SH +TYCA+A L+++GY DL S DS S
Sbjct: 224 ---GLIPGSESHGGATYCAIASLRLMGYIGVDLLSNDSSS 260
>gb|AAP50511.1| geranylgeranyltransferase type I beta subunit [Catharanthus roseus]
Length = 359
Score = 198 bits (503), Expect = 2e-49
Identities = 99/170 (58%), Positives = 121/170 (70%)
Query: 8 SSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAV 67
S E D+D HV F +MMY LLP+ Y+SQEINH+TLA+F I LDIL +L ++K+ V
Sbjct: 10 SDSELQFFDRDRHVQFLEMMYDLLPSRYQSQEINHITLAHFAIVGLDILGALDRIDKQEV 69
Query: 68 ANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKI 127
NWVLS Q ++ +NGQFYGFHGS++SQF ++ G N SHLAS+YCAL IL+
Sbjct: 70 INWVLSLQAHPKNADELDNGQFYGFHGSRSSQFQSNDEGDTVPNVSHLASSYCALTILRT 129
Query: 128 VGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
VGYD S L+S+ + SMKNLQQ DGSFMPIH G ETDLRFVYCA I M
Sbjct: 130 VGYDFSLLNSKLILESMKNLQQQDGSFMPIHSGAETDLRFVYCAAAICFM 179
>ref|NP_909269.1| putative geranylgeranyl-diphosphate geranylgeranyltransferase I
beta chain [Oryza sativa (japonica cultivar-group)]
Length = 358
Score = 197 bits (501), Expect = 4e-49
Identities = 102/183 (55%), Positives = 126/183 (68%), Gaps = 5/183 (2%)
Query: 3 SSEEASSM---ETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSL 59
+SE +M E ++ HV F + M LP Y SQE+NHLTLAYF ++ L +L L
Sbjct: 61 ASERGPAMAEPEQPEFARERHVLFLEAMASELPADYASQEVNHLTLAYFAVAGLSLLREL 120
Query: 60 HLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTY 119
V K+ +A W+LSFQV T N+ +NGQFYGF GS+T+QFP HN SHLASTY
Sbjct: 121 DSVNKDQIAKWILSFQVHPKTDNELDNGQFYGFCGSRTTQFPSTNMKDPCHNGSHLASTY 180
Query: 120 CALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM-- 177
ALAILKIVGYDL+++D++ + SSM+NLQQPDGSFMP HIG ETDLRFVYCA I M
Sbjct: 181 SALAILKIVGYDLANIDNKVLLSSMRNLQQPDGSFMPTHIGAETDLRFVYCAAAICSMLK 240
Query: 178 EWT 180
+WT
Sbjct: 241 DWT 243
>gb|AAG40865.1| geranylgeranyltransferase beta subunit [Arabidopsis thaliana]
Length = 376
Score = 173 bits (439), Expect = 6e-42
Identities = 94/175 (53%), Positives = 113/175 (63%), Gaps = 1/175 (0%)
Query: 4 SEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE 63
S S + +KD H+ + +MMY LLP Y+SQEIN LTLA+F+IS L L + V+
Sbjct: 24 SPPVQSSPSANFEKDRHLMYLEMMYELLPYHYQSQEINRLTLAHFIISGLHFLGARDRVD 83
Query: 64 KEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSK-TSQFPPDENGVFHHNNSHLASTYCAL 122
K VA WVLSFQ + F K FP +NG HN SHLASTYCAL
Sbjct: 84 KYVVAKWVLSFQAFPTNRVSLKRWRILWFLWFKGVLSFPLMKNGDLKHNGSHLASTYCAL 143
Query: 123 AILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
AILK++G+DLS++DS+S+ SM NLQQ DGSFMPIHIGGETDLRFVYCA I M
Sbjct: 144 AILKVIGHDLSTIDSKSLLISMINLQQDDGSFMPIHIGGETDLRFVYCAAAICYM 198
Score = 34.3 bits (77), Expect = 5.7
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 27/100 (27%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPP 102
L Y + +L+S ++KE+ N++L+ Q G GF
Sbjct: 186 LRFVYCAAAICYMLDSWSGMDKESAKNYILNCQSYDG-----------GF---------- 224
Query: 103 DENGVFHHNNSHLASTYCALAILKIVGY---DLSSLDSES 139
G+ + SH +TYCA+A L+++GY DL S DS S
Sbjct: 225 ---GLIPGSESHGGATYCAIASLRLMGYIGVDLLSNDSSS 261
>gb|AAH81347.1| MGC89595 protein [Xenopus tropicalis]
gi|56118392|ref|NP_001008137.1| MGC89595 protein
[Xenopus tropicalis]
Length = 372
Score = 124 bits (310), Expect = 5e-27
Identities = 73/186 (39%), Positives = 105/186 (56%), Gaps = 8/186 (4%)
Query: 1 MSSSEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLH 60
M+ E S E D HV + Q LLP S E N LT+A+F +S LD+L+SL+
Sbjct: 1 MAEDENGSPEERLLFLPDRHVRYFQRSLQLLPEQCASLETNRLTIAFFALSGLDMLDSLN 60
Query: 61 LVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDE-NGVFH-HNNSHLA 116
++ K + W+ S QV T D +N GF GS P P + +G++H H++ H+A
Sbjct: 61 VINKSEIIEWIYSLQVL--PTEDKSNLDRCGFRGSSCLGLPFNPSKGHGLYHPHDSGHIA 118
Query: 117 STYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITG 176
TY A+A L I+G DLS ++ E+ + ++ LQ PDGSF + G E D+RFVYCA I
Sbjct: 119 MTYTAIASLLILGDDLSRVNKEACLAGLRALQLPDGSFCAVPEGSENDMRFVYCAACICY 178
Query: 177 M--EWT 180
M +W+
Sbjct: 179 MLNDWS 184
>ref|NP_112344.1| protein geranylgeranyltransferase type I, beta subunit [Rattus
norvegicus] gi|1730527|sp|P53610|PGT1_RAT Geranylgeranyl
transferase type I beta subunit (Type I protein
geranyl-geranyltransferase beta subunit) (GGTase-I-beta)
gi|51247341|pdb|1S64|L Chain L, Rat Protein
Geranylgeranyltransferase Type-I Complexed With
L-778,123 And A Sulfate Anion gi|51247339|pdb|1S64|J
Chain J, Rat Protein Geranylgeranyltransferase Type-I
Complexed With L-778,123 And A Sulfate Anion
gi|51247337|pdb|1S64|H Chain H, Rat Protein
Geranylgeranyltransferase Type-I Complexed With
L-778,123 And A Sulfate Anion gi|51247335|pdb|1S64|F
Chain F, Rat Protein Geranylgeranyltransferase Type-I
Complexed With L-778,123 And A Sulfate Anion
gi|51247333|pdb|1S64|D Chain D, Rat Protein
Geranylgeranyltransferase Type-I Complexed With
L-778,123 And A Sulfate Anion gi|51247331|pdb|1S64|B
Chain B, Rat Protein Geranylgeranyltransferase Type-I
Complexed With L-778,123 And A Sulfate Anion
gi|39654228|pdb|1N4S|L Chain L, Protein
Geranylgeranyltransferase Type-I Complexed With Ggpp And
A Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654226|pdb|1N4S|J Chain J, Protein
Geranylgeranyltransferase Type-I Complexed With Ggpp And
A Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654224|pdb|1N4S|H Chain H, Protein
Geranylgeranyltransferase Type-I Complexed With Ggpp And
A Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654222|pdb|1N4S|F Chain F, Protein
Geranylgeranyltransferase Type-I Complexed With Ggpp And
A Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654220|pdb|1N4S|D Chain D, Protein
Geranylgeranyltransferase Type-I Complexed With Ggpp And
A Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654218|pdb|1N4S|B Chain B, Protein
Geranylgeranyltransferase Type-I Complexed With Ggpp And
A Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654210|pdb|1N4R|L Chain L, Protein
Geranylgeranyltransferase Type-I Complexed With A
Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654208|pdb|1N4R|J Chain J, Protein
Geranylgeranyltransferase Type-I Complexed With A
Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654206|pdb|1N4R|H Chain H, Protein
Geranylgeranyltransferase Type-I Complexed With A
Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654204|pdb|1N4R|F Chain F, Protein
Geranylgeranyltransferase Type-I Complexed With A
Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654202|pdb|1N4R|D Chain D, Protein
Geranylgeranyltransferase Type-I Complexed With A
Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654200|pdb|1N4R|B Chain B, Protein
Geranylgeranyltransferase Type-I Complexed With A
Geranylgeranylated Kkksktkcvil Peptide Product
gi|39654192|pdb|1N4Q|L Chain L, Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvil Peptide gi|39654190|pdb|1N4Q|J
Chain J, Protein Geranylgeranyltransferase Type-I
Complexed With A Ggpp Analog And A Kkksktkcvil Peptide
gi|39654188|pdb|1N4Q|H Chain H, Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvil Peptide gi|39654186|pdb|1N4Q|F
Chain F, Protein Geranylgeranyltransferase Type-I
Complexed With A Ggpp Analog And A Kkksktkcvil Peptide
gi|39654184|pdb|1N4Q|D Chain D, Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvil Peptide gi|39654182|pdb|1N4Q|B
Chain B, Protein Geranylgeranyltransferase Type-I
Complexed With A Ggpp Analog And A Kkksktkcvil Peptide
gi|39654178|pdb|1N4P|L Chain L, Protein
Geranylgeranyltransferase Type-I Complexed With
Geranylgeranyl Diphosphate gi|39654176|pdb|1N4P|J Chain
J, Protein Geranylgeranyltransferase Type-I Complexed
With Geranylgeranyl Diphosphate gi|39654174|pdb|1N4P|H
Chain H, Protein Geranylgeranyltransferase Type-I
Complexed With Geranylgeranyl Diphosphate
gi|39654172|pdb|1N4P|F Chain F, Protein
Geranylgeranyltransferase Type-I Complexed With
Geranylgeranyl Diphosphate gi|39654170|pdb|1N4P|D Chain
D, Protein Geranylgeranyltransferase Type-I Complexed
With Geranylgeranyl Diphosphate gi|39654168|pdb|1N4P|B
Chain B, Protein Geranylgeranyltransferase Type-I
Complexed With Geranylgeranyl Diphosphate
gi|56553995|pdb|1TNZ|L Chain L, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Rrcvll Peptide Derived From Cdc42 Splice
Isoform-2 gi|56553993|pdb|1TNZ|J Chain J, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Rrcvll Peptide Derived From Cdc42 Splice
Isoform-2 gi|56553991|pdb|1TNZ|H Chain H, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Rrcvll Peptide Derived From Cdc42 Splice
Isoform-2 gi|56553989|pdb|1TNZ|F Chain F, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Rrcvll Peptide Derived From Cdc42 Splice
Isoform-2 gi|56553987|pdb|1TNZ|D Chain D, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Rrcvll Peptide Derived From Cdc42 Splice
Isoform-2 gi|56553985|pdb|1TNZ|B Chain B, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Rrcvll Peptide Derived From Cdc42 Splice
Isoform-2 gi|56553977|pdb|1TNY|L Chain L, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Frekkffcail Peptide Derived From The
Heterotrimeric G Protein Gamma-2 Subunit
gi|56553975|pdb|1TNY|J Chain J, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Frekkffcail Peptide Derived From The
Heterotrimeric G Protein Gamma-2 Subunit
gi|56553973|pdb|1TNY|H Chain H, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Frekkffcail Peptide Derived From The
Heterotrimeric G Protein Gamma-2 Subunit
gi|56553971|pdb|1TNY|F Chain F, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Frekkffcail Peptide Derived From The
Heterotrimeric G Protein Gamma-2 Subunit
gi|56553969|pdb|1TNY|D Chain D, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Frekkffcail Peptide Derived From The
Heterotrimeric G Protein Gamma-2 Subunit
gi|56553967|pdb|1TNY|B Chain B, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Frekkffcail Peptide Derived From The
Heterotrimeric G Protein Gamma-2 Subunit
gi|56553957|pdb|1TNU|L Chain L, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Gcincckvl Peptide Derived From Rhob
gi|56553955|pdb|1TNU|J Chain J, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Gcincckvl Peptide Derived From Rhob
gi|56553953|pdb|1TNU|H Chain H, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Gcincckvl Peptide Derived From Rhob
gi|56553951|pdb|1TNU|F Chain F, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Gcincckvl Peptide Derived From Rhob
gi|56553949|pdb|1TNU|D Chain D, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Gcincckvl Peptide Derived From Rhob
gi|56553947|pdb|1TNU|B Chain B, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Gcincckvl Peptide Derived From Rhob
gi|56553939|pdb|1TNO|L Chain L, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvim Peptide Derived From K- Ras4b
gi|56553937|pdb|1TNO|J Chain J, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvim Peptide Derived From K- Ras4b
gi|56553935|pdb|1TNO|H Chain H, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvim Peptide Derived From K- Ras4b
gi|56553933|pdb|1TNO|F Chain F, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvim Peptide Derived From K- Ras4b
gi|56553931|pdb|1TNO|D Chain D, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvim Peptide Derived From K- Ras4b
gi|56553929|pdb|1TNO|B Chain B, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Kkksktkcvim Peptide Derived From K- Ras4b
gi|56553921|pdb|1TNB|L Chain L, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Substrate Kksktkcvif Peptide Derived From
Tc21 gi|56553919|pdb|1TNB|J Chain J, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Substrate Kksktkcvif Peptide Derived From
Tc21 gi|56553917|pdb|1TNB|H Chain H, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Substrate Kksktkcvif Peptide Derived From
Tc21 gi|56553915|pdb|1TNB|F Chain F, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Substrate Kksktkcvif Peptide Derived From
Tc21 gi|56553913|pdb|1TNB|D Chain D, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Substrate Kksktkcvif Peptide Derived From
Tc21 gi|56553911|pdb|1TNB|B Chain B, Rat Protein
Geranylgeranyltransferase Type-I Complexed With A Ggpp
Analog And A Substrate Kksktkcvif Peptide Derived From
Tc21 gi|474887|gb|AAA17756.1| geranylgeranyltransferase
type I
Length = 377
Score = 116 bits (291), Expect = 9e-25
Identities = 74/186 (39%), Positives = 106/186 (56%), Gaps = 11/186 (5%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MAATEDDRLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HN 111
L+SL +V K+ + W+ S QV T D +N GF GS P P +N G H ++
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLDRCGFRGSSYLGIPFNPSKNPGTAHPYD 118
Query: 112 NSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+ H+A TY L+ L I+G DLS +D E+ + ++ LQ DGSF + G E D+RFVYCA
Sbjct: 119 SGHIAMTYTGLSCLIILGDDLSRVDKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCA 178
Query: 172 GWITGM 177
I M
Sbjct: 179 SCICYM 184
>gb|AAH97794.1| Unknown (protein for MGC:115505) [Xenopus laevis]
Length = 372
Score = 115 bits (288), Expect = 2e-24
Identities = 70/186 (37%), Positives = 101/186 (53%), Gaps = 8/186 (4%)
Query: 1 MSSSEEASSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLH 60
M+ E+ S E D HV + Q LLP S E + LT+A+F +S LD+L+SL+
Sbjct: 1 MADDEDGSPEERLLFLPDRHVRYFQRSLQLLPEQCASLETSRLTIAFFALSGLDMLDSLN 60
Query: 61 LVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDENGVFHH--NNSHLA 116
++ K + W+ S QV T D +N GF GS P P + HH ++SH+A
Sbjct: 61 VINKSEIIEWIYSLQVL--PTEDQSNLHRCGFRGSSCLGLPFNPSKGHGLHHPYDSSHVA 118
Query: 117 STYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITG 176
TY A+A L I+G DLS ++ E+ + ++ LQ DGSF + E D+RFVYCA I
Sbjct: 119 MTYTAIASLLILGDDLSRVNKEACLAGLRALQLSDGSFCAVLEQSENDMRFVYCAACICY 178
Query: 177 M--EWT 180
M +W+
Sbjct: 179 MLNDWS 184
>ref|NP_766215.1| geranylgeranyltransferase type I [Mus musculus]
gi|26349349|dbj|BAC38314.1| unnamed protein product [Mus
musculus]
Length = 377
Score = 115 bits (287), Expect = 3e-24
Identities = 73/186 (39%), Positives = 105/186 (56%), Gaps = 11/186 (5%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MATTEDDRLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HN 111
L+SL +V K+ + W+ S QV T D +N GF GS P P +N G H ++
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLSRCGFRGSSYLGIPFNPSKNPGAAHPYD 118
Query: 112 NSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+ H+A TY L+ L I+G DL +D E+ + ++ LQ DGSF + G E D+RFVYCA
Sbjct: 119 SGHIAMTYTGLSCLIILGDDLGRVDKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCA 178
Query: 172 GWITGM 177
I M
Sbjct: 179 SCICYM 184
>ref|XP_538560.1| PREDICTED: similar to protein geranylgeranyltransferase type I,
beta subunit [Canis familiaris]
Length = 418
Score = 114 bits (286), Expect = 3e-24
Identities = 71/177 (40%), Positives = 100/177 (56%), Gaps = 8/177 (4%)
Query: 7 ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEK 64
A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+L+SL +V K
Sbjct: 51 AGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDMLDSLDVVNK 110
Query: 65 EAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HNNSHLASTYC 120
+ + W+ S QV T D +N GF GS P P +N G H +++ H+A TY
Sbjct: 111 DDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPFNPSKNPGTAHPYDSGHIAMTYT 168
Query: 121 ALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
L+ L I+G DLS ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M
Sbjct: 169 GLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYM 225
>ref|NP_001015560.1| protein geranylgeranyltransferase type I, beta subunit [Bos taurus]
gi|59857633|gb|AAX08651.1| protein
geranylgeranyltransferase type I, beta subunit [Bos
taurus]
Length = 377
Score = 114 bits (285), Expect = 4e-24
Identities = 67/165 (40%), Positives = 95/165 (56%), Gaps = 6/165 (3%)
Query: 17 KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQV 76
+D HV F Q +LP Y S E + LT+A+F +S LD+L+SL +V K+ + W+ S QV
Sbjct: 22 RDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDMLDSLDVVNKDDIIEWIYSLQV 81
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HNNSHLASTYCALAILKIVGYDL 132
T D +N GF GS P P +N G H +++ H+A TY L+ L I+G DL
Sbjct: 82 L--PTEDRSNLNRCGFRGSSYLGIPFNPSKNPGTAHPYDSGHIAMTYTGLSCLVILGDDL 139
Query: 133 SSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
S ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M
Sbjct: 140 SRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYM 184
>sp|P53609|PGT1_HUMAN Geranylgeranyl transferase type I beta subunit (Type I protein
geranyl-geranyltransferase beta subunit) (GGTase-I-beta)
gi|466491|gb|AAA35888.1| geranylgeranyltransferase type
I beta-subunit gi|4826900|ref|NP_005014.1| protein
geranylgeranyltransferase type I, beta subunit [Homo
sapiens]
Length = 377
Score = 113 bits (283), Expect = 7e-24
Identities = 74/192 (38%), Positives = 105/192 (54%), Gaps = 12/192 (6%)
Query: 7 ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEK 64
A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+L+SL +V K
Sbjct: 10 AGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDMLDSLDVVNK 69
Query: 65 EAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HNNSHLASTYC 120
+ + W+ S QV T D +N GF GS P + + G H +++ H+A TY
Sbjct: 70 DDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPFNPSKAPGTAHPYDSGHIAMTYT 127
Query: 121 ALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--E 178
L+ L I+G DLS ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M
Sbjct: 128 GLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYMLNN 187
Query: 179 WT--RKKSRITY 188
W+ K ITY
Sbjct: 188 WSGMDMKKAITY 199
>gb|AAV98360.1| geranylgeranyltransferase type I beta subunit [Homo sapiens]
Length = 300
Score = 110 bits (276), Expect = 5e-23
Identities = 74/199 (37%), Positives = 108/199 (54%), Gaps = 12/199 (6%)
Query: 7 ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEK 64
A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+L+SL +V K
Sbjct: 10 AGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDMLDSLDVVNK 69
Query: 65 EAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HNNSHLASTYC 120
+ + W+ S QV T D +N GF GS P + + G H +++ H+A T
Sbjct: 70 DDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPFNPSKAPGTAHPYDSGHIAMTCT 127
Query: 121 ALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--E 178
L+ L I+G DLS ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M
Sbjct: 128 GLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYMLNN 187
Query: 179 WT--RKKSRITY*IASLMM 195
W+ K ITY S+++
Sbjct: 188 WSGMDMKKAITYIRRSMLL 206
>emb|CAF90463.1| unnamed protein product [Tetraodon nigroviridis]
Length = 611
Score = 108 bits (270), Expect = 2e-22
Identities = 64/174 (36%), Positives = 94/174 (53%), Gaps = 7/174 (4%)
Query: 17 KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQV 76
KD HV F Q +LP Y S E LT+ +F +S LD+L++L V+K + W+ S QV
Sbjct: 18 KDRHVRFFQRTLQVLPERYASLETTRLTIVFFALSGLDVLDALDAVDKNVMIEWIYSLQV 77
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDEN--GVFH-HNNSHLASTYCALAILKIVGYDLS 133
T D +N GF GS P GV H +++ H+A TY L L I+G +LS
Sbjct: 78 L--PTEDQSNLSRCGFRGSSYMGIPYSTKGPGVSHPYDSGHVAMTYTGLCSLLILGDNLS 135
Query: 134 SLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWTRKKSR 185
++ ++ + ++ LQ DGSF + G E D+RF+YCA I M +W+ +R
Sbjct: 136 RVNKQACLAGLRALQMEDGSFYALPEGSENDIRFIYCAASICYMLDDWSGMDTR 189
>gb|AAH97132.1| Unknown (protein for MGC:114070) [Danio rerio]
Length = 355
Score = 106 bits (265), Expect = 9e-22
Identities = 63/179 (35%), Positives = 97/179 (53%), Gaps = 8/179 (4%)
Query: 8 SSMETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAV 67
+ E+ ++ HV F Q ++LP Y E + LT+A+F +S LD+L +L +V++ ++
Sbjct: 2 ADFESVDFLRERHVRFFQRCLYVLPERYAPYETSRLTIAFFSLSGLDVLGALDVVDRHSL 61
Query: 68 ANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HNNSHLASTYCALA 123
W+ S Q+ T D +N Q GF GS P + G H +++ H+ TY LA
Sbjct: 62 IEWIYSLQIL--PTADQSNLQRCGFRGSSHIGVPYSSSKGPGAPHPYDSGHVTMTYTGLA 119
Query: 124 ILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWT 180
L I+G DL +D + S ++ LQ DGSF + G E D+RFVYCA I M +W+
Sbjct: 120 CLLILGDDLGRVDRAACVSGLRALQLEDGSFYAVPEGSENDMRFVYCAACICFMLDDWS 178
>ref|XP_526978.1| PREDICTED: protein geranylgeranyltransferase type I, beta subunit
[Pan troglodytes]
Length = 418
Score = 99.8 bits (247), Expect = 1e-19
Identities = 77/240 (32%), Positives = 112/240 (46%), Gaps = 52/240 (21%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MAATEDERLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQV--------------------------------------- 76
L+SL +V K+ + W+ S QV
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVLPTEDSELTIAFFALSGLDMLDSLDVVNKDDIIEWIYSL 120
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HNNSHLASTYCALAILKIVGYDL 132
Q T D +N GF GS P + + G H +++ H+A TY L+ L I+G DL
Sbjct: 121 QVLPTEDRSNLNRCGFRGSSYLGIPFNPSKAPGTAHPYDSGHIAMTYTGLSCLVILGDDL 180
Query: 133 SSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWT--RKKSRITY 188
S ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M W+ K ITY
Sbjct: 181 SRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYMLNNWSGMDMKKAITY 240
>ref|XP_612542.1| PREDICTED: similar to protein geranylgeranyltransferase type I,
beta subunit, partial [Bos taurus]
Length = 166
Score = 96.3 bits (238), Expect = 1e-18
Identities = 57/139 (41%), Positives = 82/139 (58%), Gaps = 6/139 (4%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP- 101
LT+A+F +S LD+L+SL +V K+ + W+ S QV T D +N GF GS P
Sbjct: 10 LTIAFFALSGLDMLDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPF 67
Query: 102 -PDEN-GVFH-HNNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIH 158
P +N G H +++ H+A TY L+ L I+G DLS ++ E+ + ++ LQ DGSF +
Sbjct: 68 NPSKNPGTAHPYDSGHIAMTYTGLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVP 127
Query: 159 IGGETDLRFVYCAGWITGM 177
G E D+RFVYCA I M
Sbjct: 128 EGSENDMRFVYCASCICYM 146
>ref|XP_317045.2| ENSANGP00000019628 [Anopheles gambiae str. PEST]
gi|55237284|gb|EAA12255.2| ENSANGP00000019628 [Anopheles
gambiae str. PEST]
Length = 360
Score = 95.5 bits (236), Expect = 2e-18
Identities = 60/183 (32%), Positives = 91/183 (48%), Gaps = 13/183 (7%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVE---KEAVANWVLSFQV 76
H + H LP S + +T+A+F +S LD+L+SLH++ ++ + NW+ QV
Sbjct: 10 HAKYFIRFLHTLPGRLASHDSTRVTIAFFAVSGLDVLDSLHMLTDTFQQDICNWIYKLQV 69
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQF--PPDENGVFHHNNSHLASTYCALAILKIVGYDLSS 134
P + G GS T PD G+ + HLA TY +A+L +G DLS
Sbjct: 70 ----VPKPGERGYGGIQGSSTFDVIGTPDSCGLQLYRWGHLAITYTGIAVLVALGDDLSR 125
Query: 135 LDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--EWTR--KKSRITY*I 190
L+ ++ + +Q+ DGSF G E D+RFVYCA I M +W R +K Y +
Sbjct: 126 LNRRAIIEGVAAVQREDGSFSATIEGSEQDMRFVYCAAAICAMLNDWGRVDRKKMADYIL 185
Query: 191 ASL 193
S+
Sbjct: 186 KSI 188
>ref|XP_424421.1| PREDICTED: similar to tripartite motif protein 36; zinc-binding
protein Rbcc728 [Gallus gallus]
Length = 2099
Score = 94.0 bits (232), Expect = 6e-18
Identities = 54/139 (38%), Positives = 80/139 (56%), Gaps = 6/139 (4%)
Query: 43 LTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP- 101
LT+A+F +S LD+L+SL +V K+ + W+ S QV T D +N GF GS P
Sbjct: 280 LTIAFFALSGLDMLDSLDVVNKDDIIEWIYSLQVL--PTEDKSNLNRCGFRGSSYLGMPF 337
Query: 102 -PDENGVFHH--NNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIH 158
P + H ++ H+A TY L+ L I+G DLS ++ +++ + ++ LQ DGSF +
Sbjct: 338 NPSKGSDVSHPYDSGHIAMTYTGLSCLVILGDDLSRVNKDALLAGLRALQLEDGSFCAVL 397
Query: 159 IGGETDLRFVYCAGWITGM 177
G E D+RFVYCA I M
Sbjct: 398 EGSENDMRFVYCASCICYM 416
>ref|XP_396384.2| PREDICTED: similar to MGC89595 protein [Apis mellifera]
Length = 335
Score = 87.8 bits (216), Expect = 4e-16
Identities = 56/155 (36%), Positives = 78/155 (50%), Gaps = 15/155 (9%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEA---VANWVLSFQV 76
H + Q + ++PN + + L +A+F IS LDILN L+ + +E NW+ Q+
Sbjct: 10 HAKYFQRLLRIMPNCSAEFDCSRLAVAFFAISGLDILNCLNDLSEETKLEAINWIYRLQI 69
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLD 136
G GF S T P D + HLA TY L L I+G DLS +D
Sbjct: 70 TGA-------GPRSGFQPSTT--IPKDAP---KYQCGHLAMTYIGLVTLLILGDDLSRVD 117
Query: 137 SESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+S+ M+ Q PDGSF I G E+D+RF+YCA
Sbjct: 118 KKSIIEGMRACQNPDGSFTAIITGCESDMRFLYCA 152
>gb|EAK85648.1| hypothetical protein UM04373.1 [Ustilago maydis 521]
gi|49075918|ref|XP_401988.1| hypothetical protein
UM04373.1 [Ustilago maydis 521]
Length = 490
Score = 85.9 bits (211), Expect = 2e-15
Identities = 55/173 (31%), Positives = 85/173 (48%), Gaps = 18/173 (10%)
Query: 12 TTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLV---EKEAVA 68
T+ ++ H++F +LP PY S + +TL YF IS LD+LN+ + + EK +
Sbjct: 97 TSRLETKKHISFLLRCLRMLPQPYTSADDQRMTLGYFAISGLDLLNATNKIPTEEKVELI 156
Query: 69 NWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIV 128
+WV + Q+ G GF GS ++ P + +++A TY AL +L I+
Sbjct: 157 DWVYAQQLPTG-----------GFRGSPSTTSPCSSSTTSASGGANIAMTYAALLVLAIL 205
Query: 129 GYDLSSLDSESMSSSMKNLQQPDGSFMPIH--IGG--ETDLRFVYCAGWITGM 177
D + LD E + + +LQ DG F +GG + D RF YCA I M
Sbjct: 206 RDDFARLDREPLKRFISSLQHRDGGFAAEQAVVGGIVDRDPRFTYCAVAICSM 258
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.335 0.144 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 529,667,879
Number of Sequences: 2540612
Number of extensions: 19904647
Number of successful extensions: 65013
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 118
Number of HSP's that attempted gapping in prelim test: 64718
Number of HSP's gapped (non-prelim): 281
length of query: 348
length of database: 863,360,394
effective HSP length: 129
effective length of query: 219
effective length of database: 535,621,446
effective search space: 117301096674
effective search space used: 117301096674
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146757.11


