

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.11 + phase: 0
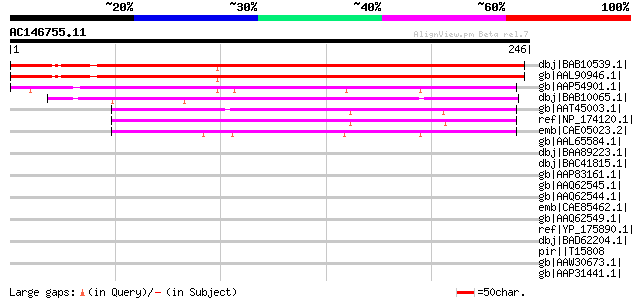
(246 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB10539.1| unnamed protein product [Arabidopsis thaliana] g... 252 7e-66
gb|AAL90946.1| AT5g52420/K24M7_17 [Arabidopsis thaliana] gi|1514... 250 3e-65
gb|AAP54901.1| unknown protein [Oryza sativa (japonica cultivar-... 171 2e-41
dbj|BAB10065.1| unnamed protein product [Arabidopsis thaliana] g... 157 2e-37
gb|AAT45003.1| unknown [Xerophyta humilis] 145 1e-33
ref|NP_174120.1| expressed protein [Arabidopsis thaliana] gi|123... 138 2e-31
emb|CAE05023.2| OSJNBa0044M19.10 [Oryza sativa (japonica cultiva... 131 2e-29
gb|AAL65584.1| macrolide-efflux protein [Streptococcus sp. 6] gi... 42 0.015
dbj|BAA89223.1| macrolide-efflux pump [Streptococcus pneumoniae] 42 0.015
dbj|BAC41815.1| unknown protein [Arabidopsis thaliana] 41 0.034
gb|AAP83161.1| macrolide-efflux protein A [Neisseria gonorrhoeae] 38 0.29
gb|AAQ62545.1| macrolide-efflux protein [Streptococcus sp. 'grou... 37 0.49
gb|AAQ62544.1| macrolide-efflux protein [Streptococcus sp. 'grou... 37 0.49
emb|CAE85462.1| macrolide-efflux protein [Streptococcus sp. 'gro... 37 0.49
gb|AAQ62549.1| macrolide-efflux protein [Streptococcus sp. 'grou... 36 0.84
ref|YP_175890.1| cytochrome caa3 oxidase controlling protein [Ba... 36 1.1
dbj|BAD62204.1| hypothetical protein [Oryza sativa (japonica cul... 35 1.4
pir||T15808 hypothetical protein C44E12.3 - Caenorhabditis elegans 34 3.2
gb|AAW30673.1| Twik family of potassium channels protein 17, iso... 34 3.2
gb|AAP31441.1| Twik family of potassium channels protein 17, iso... 34 3.2
>dbj|BAB10539.1| unnamed protein product [Arabidopsis thaliana]
gi|18423389|ref|NP_568771.1| expressed protein
[Arabidopsis thaliana]
Length = 242
Score = 252 bits (643), Expect = 7e-66
Identities = 130/245 (53%), Positives = 167/245 (68%), Gaps = 6/245 (2%)
Query: 1 MSGGVGPTGCDISLPKEEQEIEHKEEQDKSLNKSTSTPQRKTSFLSFRQLNCLAVVVVLS 60
MSGGVGPT DI+LPKEE+E EH + Q S ST P F SFRQLN LA+++VLS
Sbjct: 1 MSGGVGPTYNDITLPKEEEE-EH-QTQSTSTVSSTGKP---AGFFSFRQLNILAIIIVLS 55
Query: 61 ASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQP-PIFTQQIKILKIYVFIGAIIGL 119
ASG+V+ +DF F + ++IY F+SK+ FP H + + P+ + KI +IYV I+GL
Sbjct: 56 ASGLVTIQDFIFTILTLIYFFFLSKLIFPPHNNPNRDAPLTSSTNKIFRIYVTAAGIVGL 115
Query: 120 YAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFSNGFSSPIRAFVPVIYNSRR 179
PI YI GI E DK G+ AA PHVFLLASQ+FMEG+A GFS+P R VP++YN+RR
Sbjct: 116 IIPICYIFEGIVEDDKNGVSAAAPHVFLLASQIFMEGLATMFGFSAPARILVPIVYNARR 175
Query: 180 IFTIVDWLRDEISKVGEEHSGSYKRIYAGRALAVANMAFWCFNLFGFLIPVYLPRVFKAY 239
+ T+V+W+ E S+ + S +R+YAG+ LA AN+ W FNLFG LIPVYLPR FK Y
Sbjct: 176 VLTLVEWIMSEFSREDVTGTVSARRMYAGKVLAAANLGIWSFNLFGVLIPVYLPRAFKRY 235
Query: 240 YSSQK 244
Y S K
Sbjct: 236 YGSDK 240
>gb|AAL90946.1| AT5g52420/K24M7_17 [Arabidopsis thaliana]
gi|15146256|gb|AAK83611.1| AT5g52420/K24M7_17
[Arabidopsis thaliana]
Length = 242
Score = 250 bits (638), Expect = 3e-65
Identities = 129/245 (52%), Positives = 166/245 (67%), Gaps = 6/245 (2%)
Query: 1 MSGGVGPTGCDISLPKEEQEIEHKEEQDKSLNKSTSTPQRKTSFLSFRQLNCLAVVVVLS 60
MSGGVGPT DI+LPKEE+E EH + Q S ST P F SFRQLN LA+++VLS
Sbjct: 1 MSGGVGPTYNDITLPKEEEE-EH-QTQSTSTVSSTGKP---AGFFSFRQLNILAIIIVLS 55
Query: 61 ASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQP-PIFTQQIKILKIYVFIGAIIGL 119
ASG+V+ +DF F + ++IY F+SK+ FP H + + P+ + KI +IYV I+GL
Sbjct: 56 ASGLVTIQDFIFTILTLIYFFFLSKLIFPPHNNPNRDAPLTSSTNKIFRIYVTAAGIVGL 115
Query: 120 YAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFSNGFSSPIRAFVPVIYNSRR 179
PI YI GI E DK G+ AA PHVFLLASQ+FMEG+A GFS+P R VP++YN+RR
Sbjct: 116 IIPICYIFEGIVEDDKNGVSAAAPHVFLLASQIFMEGLATMFGFSAPARILVPIVYNARR 175
Query: 180 IFTIVDWLRDEISKVGEEHSGSYKRIYAGRALAVANMAFWCFNLFGFLIPVYLPRVFKAY 239
+ +V+W+ E S+ + S +R+YAG+ LA AN+ W FNLFG LIPVYLPR FK Y
Sbjct: 176 VLALVEWIMSEFSREDVTGTVSARRMYAGKVLAAANLGIWSFNLFGVLIPVYLPRAFKRY 235
Query: 240 YSSQK 244
Y S K
Sbjct: 236 YGSDK 240
>gb|AAP54901.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|37536624|ref|NP_922614.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|13876529|gb|AAK43505.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 277
Score = 171 bits (432), Expect = 2e-41
Identities = 101/253 (39%), Positives = 140/253 (54%), Gaps = 16/253 (6%)
Query: 1 MSGGVGPT-GCDISLPKEEQEIEHKEEQDKSLNKSTSTPQRKTSFLSFRQLNCLAVVVVL 59
MSGGVGPT G I+LP + S T+ P S +QLN VL
Sbjct: 1 MSGGVGPTCGGGITLPSTGAPLPPLHPTPTS---PTARPHHHYYLFSIKQLNSFGAAAVL 57
Query: 60 SASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQP--PIFTQQIK-ILKIYVFIGAI 116
+ S V D F L + Y+ +S +AFP P K P P+F + + +L+ + +G +
Sbjct: 58 AFSTTVPLSDIAFALLVIPYLVVLSVLAFPQRPGKPNPGAPVFLGRGRFLLRAHDALGFL 117
Query: 117 IGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA--FSNGFSSPIRAFVPVI 174
+G P YIL G+ GD G+ AA+PH FLLA+Q+F EG+A + FS P+RA V V+
Sbjct: 118 VGAALPALYILDGLRSGDTAGVAAASPHAFLLAAQIFTEGLAAAWPGRFSLPVRAAVVVM 177
Query: 175 YNSRRIFTIVDWLRDEISK-------VGEEHSGSYKRIYAGRALAVANMAFWCFNLFGFL 227
Y +RR+F +WLR E+ K VG + + +R+ AGR LAVAN+A+W NLF FL
Sbjct: 178 YGARRMFAASEWLRQEMEKRDQFGGGVGGAPAVARRRVVAGRVLAVANLAYWGINLFAFL 237
Query: 228 IPVYLPRVFKAYY 240
+P YLP+ F YY
Sbjct: 238 LPFYLPKAFNRYY 250
>dbj|BAB10065.1| unnamed protein product [Arabidopsis thaliana]
gi|29028832|gb|AAO64795.1| At5g23920 [Arabidopsis
thaliana] gi|15237854|ref|NP_197780.1| expressed protein
[Arabidopsis thaliana]
Length = 229
Score = 157 bits (398), Expect = 2e-37
Identities = 87/225 (38%), Positives = 129/225 (56%), Gaps = 6/225 (2%)
Query: 19 QEIEHKEEQDKSLNKSTSTPQRKTSFLSF-RQLNCLAVVVVLSASGMVSPEDFGFVLFSV 77
+EI K+ D S P K +S RQL L+ +++L+A G+V + FV+
Sbjct: 3 EEIREKDLTDSK--PSPPQPPPKRGLISRKRQLVFLSFMILLAAKGLVGIGEIAFVILCY 60
Query: 78 IYMC-FISKVAFPSHPSKEQPPIFTQQIKILKIYVFIGAIIGLYAPIAYILHGIFEGDKE 136
IY+ F+S+ AFP ++++ + + K+ + Y AIIGL P+ YI GI+ GD
Sbjct: 61 IYLYEFLSRFAFPRKQTEQKKRLSNPKNKLFQAYFLATAIIGLLFPLCYIGDGIYRGDIH 120
Query: 137 GIKAATPHVFLLASQVFMEGVAFSNGFSSPIRAFVPVIYNSRRIFTIVDWLRDEISKVGE 196
G AA PH+FLL+ Q F E + FS+ +S PI PV YN+RRIF ++DW++ E S
Sbjct: 121 GAGAAAPHLFLLSGQAFTEPIGFSDKYSMPIGILGPVFYNARRIFALLDWVKAEFSDT-- 178
Query: 197 EHSGSYKRIYAGRALAVANMAFWCFNLFGFLIPVYLPRVFKAYYS 241
+ G R+Y GR +A N W +NLFG L+PV+LPR + Y+S
Sbjct: 179 QRPGGPLRLYGGRVIASVNTVMWFYNLFGLLLPVFLPRSCEIYFS 223
>gb|AAT45003.1| unknown [Xerophyta humilis]
Length = 239
Score = 145 bits (365), Expect = 1e-33
Identities = 77/196 (39%), Positives = 118/196 (59%), Gaps = 6/196 (3%)
Query: 49 QLNCLAVVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILK 108
+L +A ++V SASG+V D F +F+ Y +S+VAFP++ ++ + K +
Sbjct: 8 ELQLVAFIMVFSASGLVPLFDLVFPVFATAYFIILSRVAFPTYHTQAREVFHGS--KPFQ 65
Query: 109 IYVFIGAIIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFS-NGFSSPI 167
YV IG +GL+ P+AY+L G GD ++AATPH+FLL+ Q+ E V + FS P+
Sbjct: 66 AYVVIGTAVGLFLPLAYVLGGFARGDNMAVRAATPHLFLLSCQILTENVISGLSLFSPPV 125
Query: 168 RAFVPVIYNSRRIFTIVDWLRDEISKVGEEHSGSYKR---IYAGRALAVANMAFWCFNLF 224
RA VP++Y RRIF +DW+ + ++ + K ++ GR LAVANM ++ NLF
Sbjct: 126 RALVPLLYTVRRIFVDLDWVNEVVNNKTLPANAKLKDSTWLWFGRVLAVANMIYFSINLF 185
Query: 225 GFLIPVYLPRVFKAYY 240
FLIP +LPR F+ Y+
Sbjct: 186 AFLIPRFLPRAFERYF 201
>ref|NP_174120.1| expressed protein [Arabidopsis thaliana] gi|12322980|gb|AAG51470.1|
unknown protein [Arabidopsis thaliana]
gi|25403048|pir||D86405 unknown protein [imported] -
Arabidopsis thaliana
Length = 271
Score = 138 bits (347), Expect = 2e-31
Identities = 75/196 (38%), Positives = 115/196 (58%), Gaps = 4/196 (2%)
Query: 49 QLNCLAVVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILK 108
+L +A ++V SASG+V D F F+ IY+ +S++AFPSH P + K+ +
Sbjct: 47 ELQLVAFILVFSASGLVPILDMLFPAFASIYIIALSRLAFPSHGVSTASPEVFRGSKLFR 106
Query: 109 IYVFIGAIIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFS-NGFSSPI 167
+YV G IGL+ P+AY+L G GD +++ATPH+FLL+ Q+ E V + FS P+
Sbjct: 107 LYVISGTTIGLFLPLAYVLGGFARGDDHAVRSATPHLFLLSCQILTENVISGLSLFSPPV 166
Query: 168 RAFVPVIYNSRRIFTIVDWLRDEISKVGEEHSGSYKRI---YAGRALAVANMAFWCFNLF 224
RA VP++Y RIF I+ W +D + + + + GR LA+AN+ ++ NL
Sbjct: 167 RALVPLLYTVWRIFVIIGWSKDVWFNKSLPINATPNVVTWFWFGRYLALANLGYFGVNLL 226
Query: 225 GFLIPVYLPRVFKAYY 240
FLIP +LPR F+ Y+
Sbjct: 227 CFLIPRFLPRAFEQYF 242
>emb|CAE05023.2| OSJNBa0044M19.10 [Oryza sativa (japonica cultivar-group)]
gi|50923831|ref|XP_472276.1| OSJNBa0044M19.10 [Oryza
sativa (japonica cultivar-group)]
Length = 274
Score = 131 bits (329), Expect = 2e-29
Identities = 72/201 (35%), Positives = 115/201 (56%), Gaps = 9/201 (4%)
Query: 49 QLNCLAVVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPS-HPSKEQPPIFTQQI--- 104
+L +A ++V SASG+V D F + + +Y+ +S+++FP H + +Q+I
Sbjct: 42 ELQLVAFIMVFSASGLVPLIDLAFPVATTLYLLLLSRLSFPPLHSTLPSSSSSSQEIFRG 101
Query: 105 -KILKIYVFIGAIIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGV--AFSN 161
+ YV +G +GL+ P+A++L G GD +++ATPH+FLL+ Q+ E V A
Sbjct: 102 STWFQAYVVLGTTVGLFLPLAHVLGGFARGDDGAVRSATPHLFLLSCQILTENVVGALGA 161
Query: 162 GFSSPIRAFVPVIYNSRRIFTIVDWLRDEISK--VGEEHSGSYKRIYAGRALAVANMAFW 219
FS P+RA VP++Y RR+F VDW+ D + ++ GR LAVAN+ ++
Sbjct: 162 AFSPPVRALVPLLYTVRRVFVAVDWVYDAWGNRAAAAAPQEAVAWMWFGRYLAVANLVYF 221
Query: 220 CFNLFGFLIPVYLPRVFKAYY 240
NL FLIP +LPR F+ Y+
Sbjct: 222 STNLLVFLIPKFLPRSFEKYF 242
>gb|AAL65584.1| macrolide-efflux protein [Streptococcus sp. 6]
gi|22121181|gb|AAL65583.1| macrolide-efflux protein
[Enterococcus sp. 130] gi|22121179|gb|AAL58636.1|
macrolide efflux protein [Streptococcus intermedius]
gi|22121177|gb|AAL58635.1| macrolide efflux protein
[Staphylococcus aureus] gi|21322646|emb|CAC87432.1| MefE
protein [Streptococcus salivarius]
gi|37725264|gb|AAR02322.1| macrolide-efflux protein
[Gemella haemolysans] gi|37725262|gb|AAR02321.1|
macrolide-efflux protein [Granulicatella adiacens]
gi|37725260|gb|AAR02320.1| macrolide-efflux protein
[Granulicatella adiacens] gi|37725258|gb|AAR02319.1|
macrolide-efflux protein [Granulicatella adiacens]
gi|19421856|gb|AAL87749.1| macrolide efflux protein E
[Streptococcus pneumoniae] gi|14578840|gb|AAK69027.1|
MefE [Streptococcus pneumoniae]
gi|1800301|gb|AAB41436.1| macrolide-efflux determinant
[Streptococcus pneumoniae]
Length = 405
Score = 42.0 bits (97), Expect = 0.015
Identities = 37/103 (35%), Positives = 57/103 (54%), Gaps = 14/103 (13%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F V C I ++ P + S Q +F ++IK + +++ IG+
Sbjct: 299 LAVSGILPPN--GFVIFVVC--CAIMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLIGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA 158
I+ L PI IL G F DK G+ H FLL S + + G+A
Sbjct: 354 IMSLAMPIGLILSGFF-ADKIGVN----HWFLL-SGILIIGIA 390
>dbj|BAA89223.1| macrolide-efflux pump [Streptococcus pneumoniae]
Length = 402
Score = 42.0 bits (97), Expect = 0.015
Identities = 37/103 (35%), Positives = 57/103 (54%), Gaps = 14/103 (13%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F V C I ++ P + S Q +F ++IK + +++ IG+
Sbjct: 299 LAVSGILPPN--GFVIFVVC--CAIMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLIGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA 158
I+ L PI IL G F DK G+ H FLL S + + G+A
Sbjct: 354 IMSLAMPIGLILSGFF-ADKIGVN----HWFLL-SGILIIGIA 390
>dbj|BAC41815.1| unknown protein [Arabidopsis thaliana]
Length = 119
Score = 40.8 bits (94), Expect = 0.034
Identities = 22/53 (41%), Positives = 33/53 (61%), Gaps = 1/53 (1%)
Query: 49 QLNCLAVVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSH-PSKEQPPIF 100
+L +A ++V SASG+V D F F+ IY+ +S++AFPSH S P +F
Sbjct: 47 ELQLVAFILVFSASGLVPILDMLFPAFASIYIIALSRLAFPSHGVSTASPEVF 99
>gb|AAP83161.1| macrolide-efflux protein A [Neisseria gonorrhoeae]
Length = 405
Score = 37.7 bits (86), Expect = 0.29
Identities = 36/103 (34%), Positives = 56/103 (53%), Gaps = 14/103 (13%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F V C I ++ P + S Q +F ++IK + +++ IG+
Sbjct: 299 LAVSGILPPN--GFVIFVVC--CAIMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLIGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA 158
I+ L PI IL F DK G+ H FLL S + + G+A
Sbjct: 354 IMSLAMPIGLILSVSF-ADKIGVN----HWFLL-SGILIIGIA 390
>gb|AAQ62545.1| macrolide-efflux protein [Streptococcus sp. 'group G']
Length = 405
Score = 37.0 bits (84), Expect = 0.49
Identities = 32/103 (31%), Positives = 55/103 (53%), Gaps = 14/103 (13%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F + C + ++ P + S Q +F ++IK + +++ G+
Sbjct: 299 LAVSGLLPPS--GFVIF--VACCAVMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLTGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA 158
I+ PI IL G F D+ G+ H FLL S + + G+A
Sbjct: 354 IMSFAMPIGLILSGFF-ADRIGVN----HWFLL-SGILIIGIA 390
>gb|AAQ62544.1| macrolide-efflux protein [Streptococcus sp. 'group G']
Length = 405
Score = 37.0 bits (84), Expect = 0.49
Identities = 32/103 (31%), Positives = 55/103 (53%), Gaps = 14/103 (13%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F + C + ++ P + S Q +F ++IK + +++ G+
Sbjct: 299 LAVSGLLPPS--GFVIF--VACCAVMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLTGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA 158
I+ PI IL G F D+ G+ H FLL S + + G+A
Sbjct: 354 IMSFAMPIGLILSGFF-ADRIGVN----HWFLL-SGILIIGIA 390
>emb|CAE85462.1| macrolide-efflux protein [Streptococcus sp. 'group G']
Length = 405
Score = 37.0 bits (84), Expect = 0.49
Identities = 32/103 (31%), Positives = 55/103 (53%), Gaps = 14/103 (13%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F + C + ++ P + S Q +F ++IK + +++ G+
Sbjct: 299 LAVSGLLPPS--GFVIF--VACCAVMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLTGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVA 158
I+ PI IL G F D+ G+ H FLL S + + G+A
Sbjct: 354 IMSFAMPIGLILSGFF-ADRIGVN----HWFLL-SGILIIGIA 390
>gb|AAQ62549.1| macrolide-efflux protein [Streptococcus sp. 'group G']
gi|34147938|gb|AAQ62547.1| macrolide-efflux protein
[Streptococcus sp. 'group G']
Length = 405
Score = 36.2 bits (82), Expect = 0.84
Identities = 33/113 (29%), Positives = 59/113 (52%), Gaps = 14/113 (12%)
Query: 59 LSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIK---ILKIYVFIGA 115
L+ SG++ P GFV+F V C I ++ P + S Q +F ++IK + +++ G+
Sbjct: 299 LAVSGILPPN--GFVIFVVC--CAIMGLSVPFY-SGVQTALFQEKIKPEYLGRVFSLTGS 353
Query: 116 IIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFSNGFSSPIR 168
I+ L PI IL G F ++ G+ + + L S V + G+A + + +R
Sbjct: 354 IMSLAMPIGLILSGFF-AERIGV-----NDWFLLSGVLIIGIAIACPMITEVR 400
>ref|YP_175890.1| cytochrome caa3 oxidase controlling protein [Bacillus clausii
KSM-K16] gi|56910402|dbj|BAD64929.1| cytochrome caa3
oxidase controlling protein [Bacillus clausii KSM-K16]
Length = 301
Score = 35.8 bits (81), Expect = 1.1
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 2/76 (2%)
Query: 54 AVVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFP-SHPSKEQPPIFTQQIKILKIYVF 112
A VV S ++ FGF S + ++++AF S+P K+ PI ++ K I+V
Sbjct: 108 AGAVVFGQSDLIMALHFGFSALSFASVVLLTRLAFEDSNPQKQYAPIVSKAYKGYVIFVA 167
Query: 113 IGAIIGLYAPIAYILH 128
I + + +Y AY+ H
Sbjct: 168 IYSYVAIYTG-AYVKH 182
>dbj|BAD62204.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 286
Score = 35.4 bits (80), Expect = 1.4
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 9/70 (12%)
Query: 24 KEEQDKSLNKSTSTPQRKTSFLSFRQLNCLAVVVVLSASGMVSPEDFGFVLFSVIYMCFI 83
K+ +DK+ +K T F S QL ++V++ G D ++ S+++ C
Sbjct: 24 KQSEDKATSKETDD-----CFTSRLQLTKISVLIATGGEGT----DLMVIVLSILFACAK 74
Query: 84 SKVAFPSHPS 93
SK+ FP HP+
Sbjct: 75 SKLRFPVHPN 84
>pir||T15808 hypothetical protein C44E12.3 - Caenorhabditis elegans
Length = 323
Score = 34.3 bits (77), Expect = 3.2
Identities = 25/81 (30%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Query: 89 PSHPSKEQPPIFTQQIKILKI---YVFIGAIIGLYAPIAYILHGIFEGDKE--GIKAATP 143
P P + PI+ + +K+LKI +V + ++ Y + + E D E G KA
Sbjct: 62 PEQPIVVRVPIWRRILKLLKILLPHVGLNVLLLSYIAMGATVFIWLEADHELEGRKAKVK 121
Query: 144 HVFLLASQVFMEGVAFSNGFS 164
HVF + SQ+ E +A +N S
Sbjct: 122 HVFDIYSQIMNETIALTNNQS 142
>gb|AAW30673.1| Twik family of potassium channels protein 17, isoform f
[Caenorhabditis elegans] gi|3452393|gb|AAC32854.1|
putative potassium channel subunit n2P17m1-2
[Caenorhabditis elegans] gi|25151930|ref|NP_741843.1|
putative potassium channel subunit n2P17m1-2 family
member (58.5 kD) (XI220) [Caenorhabditis elegans]
Length = 512
Score = 34.3 bits (77), Expect = 3.2
Identities = 25/81 (30%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Query: 89 PSHPSKEQPPIFTQQIKILKI---YVFIGAIIGLYAPIAYILHGIFEGDKE--GIKAATP 143
P P + PI+ + +K+LKI +V + ++ Y + + E D E G KA
Sbjct: 55 PEQPIVVRVPIWRRILKLLKILLPHVGLNVLLLSYIAMGATVFIWLEADHELEGRKAKVK 114
Query: 144 HVFLLASQVFMEGVAFSNGFS 164
HVF + SQ+ E +A +N S
Sbjct: 115 HVFDIYSQIMNETIALTNNQS 135
>gb|AAP31441.1| Twik family of potassium channels protein 17, isoform e
[Caenorhabditis elegans] gi|3452403|gb|AAC32859.1|
putative potassium channel subunit n2P17m2-3
[Caenorhabditis elegans] gi|25151939|ref|NP_741846.1|
putative potassium channel subunit n2P17m2-3, possibly
N-myristoylated (62.1 kD) (XI220) [Caenorhabditis
elegans] gi|11359806|pir||T43364 potassium channel
subunit n2P17m2-3 homolog - Caenorhabditis elegans
Length = 544
Score = 34.3 bits (77), Expect = 3.2
Identities = 25/81 (30%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Query: 89 PSHPSKEQPPIFTQQIKILKI---YVFIGAIIGLYAPIAYILHGIFEGDKE--GIKAATP 143
P P + PI+ + +K+LKI +V + ++ Y + + E D E G KA
Sbjct: 119 PEQPIVVRVPIWRRILKLLKILLPHVGLNVLLLSYIAMGATVFIWLEADHELEGRKAKVK 178
Query: 144 HVFLLASQVFMEGVAFSNGFS 164
HVF + SQ+ E +A +N S
Sbjct: 179 HVFDIYSQIMNETIALTNNQS 199
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 402,875,612
Number of Sequences: 2540612
Number of extensions: 15915299
Number of successful extensions: 58196
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 58164
Number of HSP's gapped (non-prelim): 36
length of query: 246
length of database: 863,360,394
effective HSP length: 125
effective length of query: 121
effective length of database: 545,783,894
effective search space: 66039851174
effective search space used: 66039851174
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146755.11


