

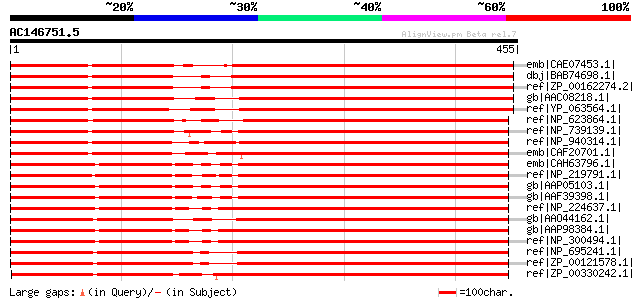
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.5 + phase: 0 /pseudo
(455 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAE07453.1| endopeptidase Clp ATP-binding chain C [Synechoco... 385 e-105
dbj|BAB74698.1| endopeptidase Clp ATP-binding chain [Nostoc sp. ... 379 e-104
ref|ZP_00162274.2| COG0542: ATPases with chaperone activity, ATP... 379 e-104
gb|AAC08218.1| Clp protease ATP binding subunit [Porphyra purpur... 364 3e-99
ref|YP_063564.1| Clp protease ATP binding subunit [Gracilaria te... 363 5e-99
ref|NP_623864.1| ATPases with chaperone activity, ATP-binding su... 362 1e-98
ref|NP_739139.1| putative endopeptidase Clp ATP-binding chain C ... 350 5e-95
ref|NP_940314.1| ATP-dependent Clp protease ATP-binding subunit ... 348 2e-94
emb|CAF20701.1| PROBABLE ATP-DEPENDENT PROTEASE (HEAT SHOCK PROT... 347 4e-94
emb|CAH63796.1| negative regulator of genetic competence clpc/me... 347 6e-94
ref|NP_219791.1| ClpC Protease ATPase [Chlamydia trachomatis D/U... 347 6e-94
gb|AAP05103.1| ATP-dependent Clp protease, ATP-binding subunit [... 345 2e-93
gb|AAF39398.1| ATP-dependent Clp protease, ATP-binding subunit C... 345 2e-93
ref|NP_224637.1| ClpC Protease [Chlamydophila pneumoniae CWL029]... 342 1e-92
gb|AAO44162.1| ATP-dependent Clp protease ATP-binding subunit [T... 341 2e-92
gb|AAP98384.1| class III stress response-related ATPase [Chlamyd... 341 2e-92
ref|NP_300494.1| ClpC protease [Chlamydophila pneumoniae J138] g... 341 2e-92
ref|NP_695241.1| protease [Bifidobacterium longum NCC2705] gi|23... 340 4e-92
ref|ZP_00121578.1| COG0542: ATPases with chaperone activity, ATP... 340 4e-92
ref|ZP_00330242.1| COG0542: ATPases with chaperone activity, ATP... 339 1e-91
>emb|CAE07453.1| endopeptidase Clp ATP-binding chain C [Synechococcus sp. WH 8102]
gi|33865472|ref|NP_897031.1| endopeptidase Clp
ATP-binding chain C [Synechococcus sp. WH 8102]
Length = 846
Score = 385 bits (988), Expect = e-105
Identities = 215/453 (47%), Positives = 295/453 (64%), Gaps = 42/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS G++ +R
Sbjct: 1 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGVAAKVLKSMGVNLKDAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLLLGL++
Sbjct: 61 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N G D+ K+R QVIR + E AE S GG
Sbjct: 118 GVAARVLENLGVDLAKVRTQVIRMLGET-------AEVSGGGGGG--------------- 155
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
GA+ + + L+ FG+NLT++A E KL P VGR ++ERVIQI+
Sbjct: 156 ------------GAK----GSTKTPTLDEFGSNLTQMANEAKLDPVVGRHNEIERVIQIL 199
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLAQRI G +P+ L+ K+V+ LD+ + +G
Sbjct: 200 GRRTKNNPVLIGEPGVGKTAIAEGLAQRIQQGEIPDILEDKRVLTLDIGLLVAGTKYRGE 259
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ +++EI+ GNVIL + EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 260 FEERLKKIMEEIKAAGNVILVIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 319
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPV V EPS+++TIEIL+GLR YE H++L TD+ALVAAA
Sbjct: 320 LDEYRKHIERDAALERRFQPVNVGEPSIDDTIEILRGLRERYEQHHRLKITDDALVAAAT 379
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
L +Y+S+RFLPDKAIDLIDEAGS V+L ++K+
Sbjct: 380 LGDRYISDRFLPDKAIDLIDEAGSRVRLLNSKL 412
>dbj|BAB74698.1| endopeptidase Clp ATP-binding chain [Nostoc sp. PCC 7120]
gi|17230491|ref|NP_487039.1| endopeptidase Clp
ATP-binding chain [Nostoc sp. PCC 7120]
gi|25289939|pir||AH2180 endopeptidase Clp ATP-binding
chain [imported] - Nostoc sp. (strain PCC 7120)
Length = 839
Score = 379 bits (974), Expect = e-104
Identities = 213/453 (47%), Positives = 292/453 (64%), Gaps = 46/453 (10%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS G++ +R
Sbjct: 17 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGVAAKVLKSMGVNLKDAR 76
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLLLGL++
Sbjct: 77 IEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLLGLIREGE 133
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N G D++K+R QVIR + E TAE S + Q
Sbjct: 134 GVAARVLENLGVDLSKVRTQVIRMLGE------------------------TAEVSATGQ 169
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + L+ FG+NLT++A + KL P VGR +++ERVIQI+
Sbjct: 170 ------------------SGRTKTPTLDEFGSNLTQMATDNKLDPVVGRAKEIERVIQIL 211
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLA RI + VP+ L+ K+VV LD+ + +G
Sbjct: 212 GRRTKNNPVLIGEPGVGKTAIAEGLASRIANKDVPDILEDKRVVTLDIGLLVAGTKYRGE 271
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++ EI GNVIL + EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 272 FEERLKKIMDEIRQAGNVILVIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 331
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPV V EPSV+ETIEIL GLR YE H+KL +DEALVAAA
Sbjct: 332 LDEYRKHIERDAALERRFQPVMVGEPSVDETIEILYGLRDRYEQHHKLKISDEALVAAAK 391
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS +Y+S+R+LPDKAIDL+DEAGS V+L ++++
Sbjct: 392 LSDRYISDRYLPDKAIDLVDEAGSRVRLMNSQL 424
>ref|ZP_00162274.2| COG0542: ATPases with chaperone activity, ATP-binding subunit
[Anabaena variabilis ATCC 29413]
Length = 839
Score = 379 bits (974), Expect = e-104
Identities = 213/453 (47%), Positives = 292/453 (64%), Gaps = 46/453 (10%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS G++ +R
Sbjct: 17 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGVAAKVLKSMGVNLKDAR 76
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLLLGL++
Sbjct: 77 IEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLLGLIREGE 133
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N G D++K+R QVIR + E TAE S + Q
Sbjct: 134 GVAARVLENLGVDLSKVRTQVIRMLGE------------------------TAEVSATGQ 169
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + L+ FG+NLT++A + KL P VGR +++ERVIQI+
Sbjct: 170 ------------------SGRTKTPTLDEFGSNLTQMATDNKLDPVVGRAKEIERVIQIL 211
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLA RI + VP+ L+ K+VV LD+ + +G
Sbjct: 212 GRRTKNNPVLIGEPGVGKTAIAEGLASRIANKDVPDILEDKRVVTLDIGLLVAGTKYRGE 271
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++ EI GNVIL + EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 272 FEERLKKIMDEIRQAGNVILVIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 331
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPV V EPSV+ETIEIL GLR YE H+KL +DEALVAAA
Sbjct: 332 LDEYRKHIERDAALERRFQPVMVGEPSVDETIEILYGLRDRYEQHHKLKISDEALVAAAK 391
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS +Y+S+R+LPDKAIDL+DEAGS V+L ++++
Sbjct: 392 LSDRYISDRYLPDKAIDLVDEAGSRVRLMNSQL 424
>gb|AAC08218.1| Clp protease ATP binding subunit [Porphyra purpurea]
gi|11465798|ref|NP_053942.1| Clp ATP binding subunit
[Porphyra purpurea] gi|2147187|pir||S73253 endopeptidase
Clp (EC 3.4.21.-) ATP-binding chain clpC [similarity] -
red alga (Porphyra purpurea) chloroplast
gi|1705925|sp|P51332|CLPC_PORPU ATP-dependent clp
protease ATP-binding subunit clpA homolog
Length = 821
Score = 364 bits (935), Expect = 3e-99
Identities = 208/453 (45%), Positives = 286/453 (62%), Gaps = 43/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AA++LKS ++ +R
Sbjct: 1 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLVGEGTGIAAQVLKSMNVNLKDAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLL+GL++
Sbjct: 61 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLMGLVREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N DV+ IR +VI+ + EN E NV+ ++ +
Sbjct: 118 GVAARVLENLAVDVSSIRAEVIQMLGEN------------------AEANVSGSNATQAR 159
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
K P LE FG+NLT++A EG L P VGR++++ERVIQI+
Sbjct: 160 SKTP---------------------TLEEFGSNLTQMAIEGGLDPVVGRQKEIERVIQIL 198
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLAQRI + VP L+ K V+ LDV + +G
Sbjct: 199 GRRTKNNPVLIGEPGVGKTAIAEGLAQRIANRDVPSILEDKLVITLDVGLLVAGTKYRGE 258
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++ EI+ NVIL + EVH + A A GA A +LK AL RG +QCI ATT
Sbjct: 259 FEERLKRIMDEIKSADNVILVIDEVHTLIGAGAAEGAIDAANLLKPALARGELQCIGATT 318
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+ E+R H+E D L+R F PV V EPSVEETIEIL GLR YE H++L +D AL AAA
Sbjct: 319 LEEYRKHIEKDPALERRFHPVVVGEPSVEETIEILFGLRDRYEKHHQLTMSDGALAAAAK 378
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
+ QY+S+RFLPDKAIDLIDEAGS V+L ++++
Sbjct: 379 YANQYISDRFLPDKAIDLIDEAGSRVRLLNSQL 411
>ref|YP_063564.1| Clp protease ATP binding subunit [Gracilaria tenuistipitata var.
liui] gi|50657654|gb|AAT79639.1| Clp protease ATP
binding subunit [Gracilaria tenuistipitata var. liui]
Length = 823
Score = 363 bits (933), Expect = 5e-99
Identities = 206/453 (45%), Positives = 284/453 (62%), Gaps = 43/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AA++LKS ++ +R
Sbjct: 1 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAQVLKSMNVNLKDAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLL+GL++
Sbjct: 61 IEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLMGLVREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N +V IR +VI+ +G E N ++ +
Sbjct: 118 GVAARVLENLAVNVASIRTEVIQ------------------MLGDTTEANTNGNNNTQTR 159
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
K P LE FG+NLT+LA EG L P VGR++++ERVIQI+
Sbjct: 160 SKTP---------------------TLEEFGSNLTELAIEGILDPVVGRQKEIERVIQIL 198
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLAQRI + +P L+ K VV LDV + +G
Sbjct: 199 GRRTKNNPVLIGEPGVGKTAIAEGLAQRIANKDIPSILEDKLVVTLDVGLLVAGTKYRGE 258
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++ EI+ N++L + EVH + A A GA A +LK AL RG +QCI ATT
Sbjct: 259 FEERLKRIMDEIKSANNIVLVIDEVHTLIGAGAAEGAIDAANLLKPALARGELQCIGATT 318
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+ E+R H+E D L+R FQPV V EPSVEETIEIL GLR YE H++L +DEAL AAA
Sbjct: 319 LEEYRKHIEKDAALERRFQPVLVGEPSVEETIEILFGLRDRYEKHHQLKMSDEALAAAAK 378
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
+ QY+S+RFLPDKAIDLIDEAGS V+L ++++
Sbjct: 379 YANQYISDRFLPDKAIDLIDEAGSRVRLLNSQL 411
>ref|NP_623864.1| ATPases with chaperone activity, ATP-binding subunit
[Thermoanaerobacter tengcongensis MB4]
gi|20517331|gb|AAM25468.1| ATPases with chaperone
activity, ATP-binding subunit [Thermoanaerobacter
tengcongensis MB4]
Length = 816
Score = 362 bits (930), Expect = 1e-98
Identities = 202/448 (45%), Positives = 279/448 (62%), Gaps = 45/448 (10%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M TE A+K + AQ+EAR H Y+ TEHILLG+L + G+AA++LK G+ + +R
Sbjct: 3 MFGRFTERAQKALYLAQEEARSFYHNYVGTEHILLGLLKEDEGIAARVLKKLGVTYEATR 62
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
E+V+ L+G G + +T AK VL+ SL A+ V T H+LLGLL+
Sbjct: 63 EKVLSLIGMG----NIPGDVVGYTPRAKRVLELSLSEARRFNTSYVGTEHILLGLLREGE 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G +++ QG D N++RE++++ + E PA G ++ SK+
Sbjct: 119 GVAVRILMEQGIDFNRVREEIVKMLSEE------PAAG-------------PSKVSKAKN 159
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
P L Q FG +LT LA++GKL P +GRE+++ERVIQI+
Sbjct: 160 TNTPTLNQ---------------------FGRDLTDLARDGKLDPVIGREKEIERVIQIL 198
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I++GLAQ+I+ G VPE LK K+VV LD+A + +G
Sbjct: 199 SRRTKNNPVLIGEPGVGKTAIVEGLAQKIVEGEVPEILKDKRVVTLDLASLVAGTKYRGE 258
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
E+R++ +I E+ GNVILFV E+H + A A GA + ILK AL RG IQ I ATT
Sbjct: 259 FEDRLKTVINEVIKAGNVILFVDEMHTLIGAGAAEGAIDASNILKPALARGEIQVIGATT 318
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R ++E D L+R FQP+ V EP+VEETIEILKGLR YE+H+++ TDEAL AAA
Sbjct: 319 LDEYRKYVERDPALERRFQPIMVEEPTVEETIEILKGLRDKYEAHHRVKITDEALEAAAK 378
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y+++RFLPDKAIDLIDEA S V+L
Sbjct: 379 LSHRYITDRFLPDKAIDLIDEAASRVRL 406
>ref|NP_739139.1| putative endopeptidase Clp ATP-binding chain C [Corynebacterium
efficiens YS-314] gi|23494372|dbj|BAC19339.1| putative
endopeptidase Clp ATP-binding chain C [Corynebacterium
efficiens YS-314]
Length = 927
Score = 350 bits (898), Expect = 5e-95
Identities = 197/453 (43%), Positives = 292/453 (63%), Gaps = 29/453 (6%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++VI+ AQ+EAR + H YI TEHILLG++ + G+AAK L+S GI + R
Sbjct: 1 MFERFTDRARRVIVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKALESMGISLDAVR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +++G G S + I FT AK VL+ SL+ +G+ + T LLLGL++
Sbjct: 61 QEVEEIIGHG---SKPTTGHIPFTPRAKKVLELSLREGLQMGHKYIGTEFLLLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSK-----NQMGGQVEENVTAES 175
G Q++ GAD+ ++R+QVI+ + EG + + GGQ + V A S
Sbjct: 118 GVAAQVLVKLGADLPRVRQQVIQLLSGY--------EGGQGGSPDSPQGGQGGDAVGAGS 169
Query: 176 SKSNQIKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVER 235
+ + GA + N+ L+ FG NLT+ A++GKL P VGR++++ER
Sbjct: 170 APGGRA--------TGSGAGERSNS----LVLDQFGRNLTQAAKDGKLDPVVGRDKEIER 217
Query: 236 VIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVI 295
++Q++ RR KNNP L+GEPGVGKT++++GLA I++G VPE LK K+V +LD+ +
Sbjct: 218 IMQVLSRRTKNNPVLIGEPGVGKTAVVEGLALDIVNGKVPETLKDKQVYSLDLGSLVAGS 277
Query: 296 SNQGSSEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQC 354
+G EER++ ++KEI G++ILF+ E+H + A A GA A +LK L RG +Q
Sbjct: 278 RYRGDFEERLKKVLKEINQRGDIILFIDEIHTLVGAGAAEGAIDAASLLKPKLARGELQT 337
Query: 355 IFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEAL 414
I ATT++E+R H+E D L+R FQPV+V EPSVE +IEILKGLR YE+H+++ TD AL
Sbjct: 338 IGATTLDEYRKHIEKDAALERRFQPVQVPEPSVELSIEILKGLRDRYEAHHRVSITDGAL 397
Query: 415 VAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
VAAA LS +Y+++RFLPDKA+DLIDEAG+ +++
Sbjct: 398 VAAAQLSDRYINDRFLPDKAVDLIDEAGARMRI 430
>ref|NP_940314.1| ATP-dependent Clp protease ATP-binding subunit [Corynebacterium
diphtheriae NCTC 13129] gi|38200810|emb|CAE50514.1|
ATP-dependent Clp protease ATP-binding subunit
[Corynebacterium diphtheriae]
Length = 883
Score = 348 bits (894), Expect = 2e-94
Identities = 193/448 (43%), Positives = 285/448 (63%), Gaps = 24/448 (5%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++VI+ AQ+EAR + H YI TEHILLG++ + G+AAK L+S GI R
Sbjct: 6 MFERFTDRARRVIVLAQEEARGLNHNYIGTEHILLGLIHEGEGVAAKALESMGISLEAVR 65
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +++G G S I FT AK VL+ SL+ +G+ + T LLLGL++ +
Sbjct: 66 QEVEEIIGHG---SEPPAGHIPFTPRAKKVLELSLREGLQMGHKYIGTEFLLLGLIREGD 122
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q++ GAD+ ++R+QVI+ + GG+ EN +++
Sbjct: 123 GVAAQVLVKLGADLPRVRQQVIQLLS--------------GYEGGESPEN----NNEGEA 164
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ G + +N L+ FG NLT+ A+EGKL P VGRE+++ER++Q++
Sbjct: 165 VGAGTGAGRAGRGGGAGERSNSL--VLDQFGRNLTQAAKEGKLDPVVGREQEIERIMQVL 222
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++++GLA I++G VPE LK K+V +LD+ + +G
Sbjct: 223 SRRTKNNPVLIGEPGVGKTAVVEGLALDIVNGKVPETLKDKQVYSLDLGSLVAGSRYRGD 282
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++KEI G++ILF+ E+H + A A GA A +LK L RG +Q I ATT
Sbjct: 283 FEERLKKVLKEINQRGDIILFIDEIHTLVGAGAAEGAIDAASLLKPKLARGELQTIGATT 342
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPV+V EPSV+ TIEILKGLR YE+H+++ TD AL AAA
Sbjct: 343 LDEYRKHIEKDAALERRFQPVQVPEPSVDMTIEILKGLRDRYEAHHRVSITDGALAAAAQ 402
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y+++RFLPDKA+DLIDEAG+ +++
Sbjct: 403 LSDRYINDRFLPDKAVDLIDEAGARMRI 430
>emb|CAF20701.1| PROBABLE ATP-DEPENDENT PROTEASE (HEAT SHOCK PROTEIN)
[Corynebacterium glutamicum ATCC 13032]
gi|21325450|dbj|BAC00072.1| ATPases with chaperone
activity, ATP-binding subunit [Corynebacterium
glutamicum ATCC 13032] gi|62391515|ref|YP_226917.1|
PROBABLE ATP-DEPENDENT PROTEASE (HEAT SHOCK PROTEIN)
[Corynebacterium glutamicum ATCC 13032]
gi|19553872|ref|NP_601874.1| ATPase with chaperone
activity, ATP-binding subunit [Corynebacterium
glutamicum ATCC 13032]
Length = 925
Score = 347 bits (890), Expect = 4e-94
Identities = 191/451 (42%), Positives = 288/451 (63%), Gaps = 24/451 (5%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++VI+ AQ+EAR + H YI TEHILLG++ + G+AAK L+S GI + R
Sbjct: 1 MFERFTDRARRVIVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKALESMGISLDAVR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +++G+G S + I FT AK VL+ SL+ +G+ + T LLLGL++
Sbjct: 61 QEVEEIIGQG---SQPTTGHIPFTPRAKKVLELSLREGLQMGHKYIGTEFLLLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q++ GAD+ ++R+QVI+ + G + GG E A +
Sbjct: 118 GVAAQVLVKLGADLPRVRQQVIQLLS-----------GYEGGQGGSPEGGQGAPTGGD-- 164
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSA---LEIFGTNLTKLAQEGKLHPFVGREEQVERVI 237
+ G R + ++S L+ FG NLT+ A++GKL P VGR++++ER++
Sbjct: 165 ----AVGAGAAPGGRPSSGSPGERSTSLVLDQFGRNLTQAAKDGKLDPVVGRDKEIERIM 220
Query: 238 QIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISN 297
Q++ RR KNNP L+GEPGVGKT++++GLA I++G VPE LK K+V +LD+ +
Sbjct: 221 QVLSRRTKNNPVLIGEPGVGKTAVVEGLALDIVNGKVPETLKDKQVYSLDLGSLVAGSRY 280
Query: 298 QGSSEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIF 356
+G EER++ ++KEI G++ILF+ E+H + A A GA A +LK L RG +Q I
Sbjct: 281 RGDFEERLKKVLKEINQRGDIILFIDEIHTLVGAGAAEGAIDAASLLKPKLARGELQTIG 340
Query: 357 ATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVA 416
ATT++E+R H+E D L+R FQPV+V EPSV+ T+EILKGLR YE+H+++ TD AL A
Sbjct: 341 ATTLDEYRKHIEKDAALERRFQPVQVPEPSVDLTVEILKGLRDRYEAHHRVSITDGALTA 400
Query: 417 AANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
AA L+ +Y+++RFLPDKA+DLIDEAG+ +++
Sbjct: 401 AAQLADRYINDRFLPDKAVDLIDEAGARMRI 431
>emb|CAH63796.1| negative regulator of genetic competence clpc/mecb [Chlamydophila
abortus S26/3] gi|62184977|ref|YP_219762.1| negative
regulator of genetic competence clpc/mecb [Chlamydophila
abortus S26/3]
Length = 845
Score = 347 bits (889), Expect = 6e-94
Identities = 192/448 (42%), Positives = 285/448 (62%), Gaps = 22/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ G+DF+ ++
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRNLGVDFDTAK 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +L+G G + D T K + + + A L ++ V T HLLLG+L ++
Sbjct: 61 QEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEAGILEHNYVGTEHLLLGILNQAD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q+++N D ++R+++++++E + LP S N G T+ SSKS+
Sbjct: 119 GVALQVLENLHIDPREVRKEILKELE--TFNLQLPPASSSNPRGS------TSSSSKSSS 170
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + LG K D SAL+ +G +LT++ +E KL P +GR +VER+I I+
Sbjct: 171 L-------GHTLGGDKSDKL----SALKAYGYDLTEMFRESKLDPVIGRSTEVERLILIL 219
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+S VP+ L+ K+++ LD+A + +G
Sbjct: 220 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIISNEVPDTLRKKRLITLDLALMIAGTKYRGQ 279
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA + ILK AL RG IQCI ATT
Sbjct: 280 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASNILKPALARGEIQCIGATT 339
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ETIEIL+GL+ YE H+ + T+EAL AAA
Sbjct: 340 IDEYRKHIEKDAALERRFQKILVQPPSVDETIEILRGLKKKYEEHHNVSITEEALKAAAT 399
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 400 LSDQYVHGRFLPDKAIDLLDEAGARVRV 427
>ref|NP_219791.1| ClpC Protease ATPase [Chlamydia trachomatis D/UW-3/CX]
gi|3328700|gb|AAC67879.1| ClpC Protease ATPase
[Chlamydia trachomatis D/UW-3/CX] gi|7428217|pir||C71533
endopeptidase Clp (EC 3.4.21.-) ATP-binding chain clpC
[similarity] - Chlamydia trachomatis (serotype D, strain
UW3/Cx) gi|14194496|sp|O84288|CLPC_CHLTR Probable
ATP-dependent CLP protease ATP-binding subunit
Length = 854
Score = 347 bits (889), Expect = 6e-94
Identities = 194/448 (43%), Positives = 284/448 (63%), Gaps = 19/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ G+DF+ ++
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRTLGVDFDTAK 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V +L+G G C D T K + + + A L ++ V T HLLLG+L S+
Sbjct: 61 HEVERLIGYGPEIQ--VCGDPALTGRVKKSFESANEEAALLEHNYVGTEHLLLGILNQSD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q+++N D +IR+++++++E + LP S SS S++
Sbjct: 119 GVALQVLENLHVDPKEIRKEILKELE--TFNLQLPPSSSITPRN--------TNSSSSSK 168
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
P+ + LG K + SAL+ +G +LT++ +E +L P +GR +VER+I I+
Sbjct: 169 SSSPL--GGHTLGGDKPEKL----SALKAYGYDLTEMFKESRLDPVIGRSAEVERLILIL 222
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP LVGE GVGKT+I++GLAQ+I+SG VPE L+ K+++ LD+A + +G
Sbjct: 223 CRRRKNNPVLVGEAGVGKTAIVEGLAQKIVSGEVPEALRKKRLITLDLALMIAGTKYRGQ 282
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA ++ILK AL RG IQCI ATT
Sbjct: 283 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASHILKPALARGEIQCIGATT 342
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ET+EIL+GL+ YE H+ + TDEALVAAA
Sbjct: 343 LDEYRKHIEKDAALERRFQKIVVQPPSVDETVEILRGLKKKYEEHHNVFITDEALVAAAK 402
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 403 LSDQYVHGRFLPDKAIDLLDEAGARVRV 430
>gb|AAP05103.1| ATP-dependent Clp protease, ATP-binding subunit [Chlamydophila
caviae GPIC] gi|29840119|ref|NP_829225.1| ATP-dependent
Clp protease, ATP-binding subunit [Chlamydophila caviae
GPIC]
Length = 846
Score = 345 bits (884), Expect = 2e-93
Identities = 191/448 (42%), Positives = 285/448 (62%), Gaps = 22/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ G+DF+ ++
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRNLGVDFDTAK 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +L+G G + D T K + + + A L ++ V T HLLLG+L ++
Sbjct: 61 QEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEAGVLEHNYVGTEHLLLGILNQAD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q+++N D ++R+++++++E + LP S N G ++ SSKS+
Sbjct: 119 GVALQVLENLHIDPREVRKEILKELE--TFNLQLPPSSSSNPRGS------SSSSSKSSS 170
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + LG K D SAL+ +G +LT++ +E KL P +GR +VER+I I+
Sbjct: 171 L-------GHALGGEKSDKL----SALKAYGYDLTEMFRESKLDPVIGRSAEVERLILIL 219
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+S VP+ L+ K+++ LD+A + +G
Sbjct: 220 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIISNEVPDTLRKKRLITLDLALMIAGTKYRGQ 279
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA + ILK AL RG IQCI ATT
Sbjct: 280 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASNILKPALARGEIQCIGATT 339
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ETIEIL+GL+ YE H+ + T+EAL AAA
Sbjct: 340 IDEYRKHIEKDAALERRFQKIIVQPPSVDETIEILRGLKKKYEEHHNVAITEEALKAAAT 399
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 400 LSDQYVHGRFLPDKAIDLLDEAGARVRV 427
>gb|AAF39398.1| ATP-dependent Clp protease, ATP-binding subunit ClpC [Chlamydia
muridarum Nigg] gi|15835176|ref|NP_296935.1|
ATP-dependent Clp protease, ATP-binding subunit ClpC
[Chlamydia muridarum Nigg] gi|11265231|pir||B81689
ATP-dependent Clp proteinase, ATP-binding chain ClpC,
TC0559 [imported] - Chlamydia muridarum (strain Nigg)
gi|14194530|sp|Q9PKA8|CLPC_CHLMU Probable ATP-dependent
Clp protease ATP-binding subunit
Length = 870
Score = 345 bits (884), Expect = 2e-93
Identities = 191/448 (42%), Positives = 287/448 (63%), Gaps = 18/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ G+DF+ ++
Sbjct: 17 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRTLGVDFDTAK 76
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V +L+G G + D T K + + + A L ++ V T HLLLG+L ++
Sbjct: 77 NEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEASILEHNYVGTEHLLLGILNQAD 134
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q+++N D +IR+++++++E + LP S + ++ SSKS+
Sbjct: 135 GVALQVLENLHIDPKEIRKEILKELE--TFNLQLPPSSSITPRN---TNSSSSSSSKSSS 189
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ ++LG K + SAL+ +G +LT++ +E +L P +GR +VER+I I+
Sbjct: 190 LG------GHSLGGDKPEKL----SALKAYGYDLTEMFKEARLDPVIGRSAEVERLILIL 239
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+SG VPE L+ K+++ LD+A + +G
Sbjct: 240 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIVSGEVPEALRKKRLITLDLALMIAGTKYRGQ 299
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA ++ILK AL RG IQCI ATT
Sbjct: 300 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASHILKPALARGEIQCIGATT 359
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ET+EIL+GL+ YE H+ + TDEALVAAA
Sbjct: 360 LDEYRKHIEKDAALERRFQKIVVQPPSVDETVEILRGLKKKYEEHHNVFITDEALVAAAK 419
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 420 LSDQYVHGRFLPDKAIDLLDEAGARVRV 447
>ref|NP_224637.1| ClpC Protease [Chlamydophila pneumoniae CWL029]
gi|4376719|gb|AAD18581.1| ClpC Protease [Chlamydophila
pneumoniae CWL029]
Length = 845
Score = 342 bits (878), Expect = 1e-92
Identities = 190/448 (42%), Positives = 283/448 (62%), Gaps = 21/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ GIDF+ +R
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRNLGIDFDTAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +L+G G + D T K + + + A L ++ V T HLLLG+L S+
Sbjct: 61 QEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEASLLEHNYVGTEHLLLGILHQSD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
Q+++N D ++R++++R++E + LP S + + SS+SN
Sbjct: 119 SVALQVLENLHIDPREVRKEILRELE--TFNLQLPPSSSSS-----------SSSSRSNP 165
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + LG + N++ SAL+ +G +LT++ +E KL P +GR +VER+I I+
Sbjct: 166 S-----SSKSPLGHSLGSDKNEKLSALKAYGYDLTEMVRESKLDPVIGRSSEVERLILIL 220
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+ VP+ L+ K+++ LD+A + +G
Sbjct: 221 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIILNEVPDALRKKRLITLDLALMIAGTKYRGQ 280
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA + ILK AL RG IQCI ATT
Sbjct: 281 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASNILKPALARGEIQCIGATT 340
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ETIEIL+GL+ YE H+ + T+EAL AAA
Sbjct: 341 IDEYRKHIEKDAALERRFQKIVVHPPSVDETIEILRGLKKKYEEHHNVFITEEALKAAAT 400
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 401 LSDQYVHGRFLPDKAIDLLDEAGARVRV 428
>gb|AAO44162.1| ATP-dependent Clp protease ATP-binding subunit [Tropheryma whipplei
str. Twist] gi|28572245|ref|NP_789025.1| putative
Clp-family ATP-binding protease/regulator [Tropheryma
whipplei TW08/27] gi|28410376|emb|CAD66762.1| putative
Clp-family ATP-binding protease/regulator [Tropheryma
whipplei TW08/27] gi|28493032|ref|NP_787193.1|
ATP-dependent Clp protease ATP-binding subunit
[Tropheryma whipplei str. Twist]
Length = 840
Score = 341 bits (875), Expect = 2e-92
Identities = 187/448 (41%), Positives = 283/448 (62%), Gaps = 41/448 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++VI+ AQ+EAR + H YI TEH+LLG++ + +G+AA+ L+S I +R
Sbjct: 5 MFERFTDKARRVIVLAQEEARTLSHNYIGTEHVLLGLISEGDGIAAQALESLDITLERAR 64
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
E V +L+GRG + I FT AK VL+ SL+ A LG++ + T H+LLG+L
Sbjct: 65 EGVAELIGRGQNATS---GHIPFTPRAKKVLELSLREALQLGHNYIGTEHILLGILHEGE 121
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q++ N GA++ I+++V+ +E+ G+ +E V+ S+S +
Sbjct: 122 GIAAQVLVNMGAELPAIQQRVMHLLED-----------------GREQEPVSVGPSESGK 164
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
I L+ FG +LT+ A+EGKL P +GRE+++ERV+Q++
Sbjct: 165 IS--------------------GSQILDQFGRHLTRAAKEGKLDPVIGREKEIERVMQVL 204
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++++ LAQ I++G VP L+ K+V +LD+ + +G
Sbjct: 205 SRRTKNNPILIGEPGVGKTAVVEALAQAIVNGDVPVNLRNKQVYSLDLGSLIAGSRYRGD 264
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDAATS-GARSFAYILKHALERGVIQCIFATT 359
EER++ + KEI G++I+F+ E+H + A ++ GA A ILK L RG +Q I ATT
Sbjct: 265 FEERLKKVTKEIRSRGDIIVFIDEIHSLVGAGSAEGAIDAATILKPLLARGELQTIGATT 324
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R ++E D+ L+R FQPV V EPS+ I+ILKGLR YE+H+K+ TDEA+ AA
Sbjct: 325 LDEYRKNIEKDSALERRFQPVNVSEPSIPMCIQILKGLRDRYEAHHKVKITDEAIYAAVT 384
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y+++RFLPDKAIDLIDEAG+ ++L
Sbjct: 385 LSSRYINDRFLPDKAIDLIDEAGARLRL 412
>gb|AAP98384.1| class III stress response-related ATPase [Chlamydophila pneumoniae
TW-183] gi|7189244|gb|AAF38172.1| ATP-dependent Clp
protease, ATP-binding subunit [Chlamydophila pneumoniae
AR39] gi|33241786|ref|NP_876727.1| class III stress
response-related ATPase [Chlamydophila pneumoniae
TW-183] gi|14194545|sp|Q9Z8A6|CLPC_CHLPN Probable
ATP-dependent Clp protease ATP-binding subunit
gi|16752602|ref|NP_444865.1| ATP-dependent Clp protease,
ATP-binding subunit [Chlamydophila pneumoniae AR39]
Length = 845
Score = 341 bits (875), Expect = 2e-92
Identities = 189/448 (42%), Positives = 283/448 (62%), Gaps = 21/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ GIDF+ +R
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRNLGIDFDTAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +L+G G + D T K + + + A L ++ V T HLLLG+L S+
Sbjct: 61 QEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEASLLEHNYVGTEHLLLGILHQSD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
Q+++N D ++R+++++++E + LP S + + SS+SN
Sbjct: 119 SVALQVLENLHIDPREVRKEILKELE--TFNLQLPPSSSSS-----------SSSSRSNP 165
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + LG + N++ SAL+ +G +LT++ +E KL P +GR +VER+I I+
Sbjct: 166 S-----SSKSPLGHSLGSDKNEKLSALKAYGYDLTEMVRESKLDPVIGRSSEVERLILIL 220
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+ VP+ L+ K+++ LD+A + +G
Sbjct: 221 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIILNEVPDALRKKRLITLDLALMIAGTKYRGQ 280
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA + ILK AL RG IQCI ATT
Sbjct: 281 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASNILKPALARGEIQCIGATT 340
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ETIEIL+GL+ YE H+ + T+EAL AAA
Sbjct: 341 IDEYRKHIEKDAALERRFQKIVVHPPSVDETIEILRGLKKKYEEHHNVFITEEALKAAAT 400
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 401 LSDQYVHGRFLPDKAIDLLDEAGARVRV 428
>ref|NP_300494.1| ClpC protease [Chlamydophila pneumoniae J138]
gi|8978809|dbj|BAA98645.1| ClpC protease [Chlamydophila
pneumoniae J138]
Length = 845
Score = 341 bits (875), Expect = 2e-92
Identities = 189/448 (42%), Positives = 283/448 (62%), Gaps = 21/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ GIDF+ +R
Sbjct: 1 MFETFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRNLGIDFDTAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +L+G G + D T K + + + A L ++ V T HLLLG+L S+
Sbjct: 61 QEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEASLLEHNYVGTEHLLLGILHQSD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
Q+++N D ++R+++++++E + LP S + + SS+SN
Sbjct: 119 SVALQVLENLHIDPREVRKEILKELE--TFNLQLPPSSSSS-----------SSSSRSNP 165
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + LG + N++ SAL+ +G +LT++ +E KL P +GR +VER+I I+
Sbjct: 166 S-----SSKSPLGHSLGSDKNEKLSALKAYGYDLTEMVRESKLDPVIGRSSEVERLILIL 220
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+ VP+ L+ K+++ LD+A + +G
Sbjct: 221 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIILNEVPDALRKKRLITLDLALMIAGTKYRGQ 280
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA + ILK AL RG IQCI ATT
Sbjct: 281 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASNILKPALARGEIQCIGATT 340
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ETIEIL+GL+ YE H+ + T+EAL AAA
Sbjct: 341 IDEYRKHIEKDAALERRFQKIVVHPPSVDETIEILRGLKKKYEEHHNVFITEEALKAAAT 400
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 401 LSDQYVHGRFLPDKAIDLLDEAGARVRV 428
>ref|NP_695241.1| protease [Bifidobacterium longum NCC2705]
gi|23325195|gb|AAN23877.1| protease [Bifidobacterium
longum NCC2705]
Length = 869
Score = 340 bits (873), Expect = 4e-92
Identities = 188/448 (41%), Positives = 278/448 (61%), Gaps = 33/448 (7%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++VI+ AQ+EAR + H YI TEH+LLG++ + G+AAK L S G++ + +R
Sbjct: 1 MFERFTDRARRVIVLAQEEARALQHNYIGTEHLLLGLIREGEGVAAKALASKGVELDATR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+QV +++G+G I FT AK VL+FSL+ A LG+ + T H+LLGL++
Sbjct: 61 KQVEEMIGKGNAAPN---GHIPFTPHAKQVLEFSLREALQLGHSYIGTEHILLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G TQ++ D+ ++R I I N +G GG VT + +KS
Sbjct: 118 GVGTQVLIKMDVDLGELRSATIDMIRGNSGTGTGDGKGDLANAGG-----VTDKQNKSG- 171
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ L+ FG NLT A EGKL P +GR ++ERV+ ++
Sbjct: 172 -----------------------SAILDQFGRNLTAEAAEGKLDPVIGRTNEIERVMVVL 208
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++++GLAQ+I +G VPE LKGK+V +LD+ + +G
Sbjct: 209 SRRTKNNPVLIGEPGVGKTAVVEGLAQKIQAGDVPETLKGKQVYSLDLGSMVAGSRYRGD 268
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDAATS-GARSFAYILKHALERGVIQCIFATT 359
EER++ ++KEI+ G+++LF+ E+H I A ++ GA + +LK L RG +Q I ATT
Sbjct: 269 FEERLKKVLKEIKTRGDIVLFIDEIHTIVGAGSADGALGASDMLKPMLARGELQTIGATT 328
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+E+R ++E D L+R FQP++V EP++ ETIEILKGLR YE+H+ + TD AL +AA
Sbjct: 329 TDEYRKYIEKDAALERRFQPIQVHEPTIAETIEILKGLRSRYENHHHVTITDGALQSAAE 388
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y+ +R LPDKAIDLIDEAG+ +++
Sbjct: 389 LSSRYIQDRNLPDKAIDLIDEAGARLRI 416
>ref|ZP_00121578.1| COG0542: ATPases with chaperone activity, ATP-binding subunit
[Bifidobacterium longum DJO10A]
Length = 869
Score = 340 bits (873), Expect = 4e-92
Identities = 188/448 (41%), Positives = 278/448 (61%), Gaps = 33/448 (7%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++VI+ AQ+EAR + H YI TEH+LLG++ + G+AAK L S G++ + +R
Sbjct: 1 MFERFTDRARRVIVLAQEEARALQHNYIGTEHLLLGLIREGEGVAAKALASKGVELDATR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+QV +++G+G I FT AK VL+FSL+ A LG+ + T H+LLGL++
Sbjct: 61 KQVEEMIGKGNAAPN---GHIPFTPHAKQVLEFSLREALQLGHSYIGTEHILLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G TQ++ D+ ++R I I N +G GG VT + +KS
Sbjct: 118 GVGTQVLIKMDVDLGELRSATIDMIRGNSGTGTGDGKGDLANAGG-----VTDKQNKSG- 171
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ L+ FG NLT A EGKL P +GR ++ERV+ ++
Sbjct: 172 -----------------------SAILDQFGRNLTAEAAEGKLDPVIGRTNEIERVMVVL 208
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++++GLAQ+I +G VPE LKGK+V +LD+ + +G
Sbjct: 209 SRRTKNNPVLIGEPGVGKTAVVEGLAQKIQAGDVPETLKGKQVYSLDLGSMVAGSRYRGD 268
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDAATS-GARSFAYILKHALERGVIQCIFATT 359
EER++ ++KEI+ G+++LF+ E+H I A ++ GA + +LK L RG +Q I ATT
Sbjct: 269 FEERLKKVLKEIKTRGDIVLFIDEIHTIVGAGSADGALGASDMLKPMLARGELQTIGATT 328
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+E+R ++E D L+R FQP++V EP++ ETIEILKGLR YE+H+ + TD AL +AA
Sbjct: 329 TDEYRKYIEKDAALERRFQPIQVHEPTIAETIEILKGLRSRYENHHHVTITDGALQSAAE 388
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y+ +R LPDKAIDLIDEAG+ +++
Sbjct: 389 LSSRYIQDRNLPDKAIDLIDEAGARLRI 416
>ref|ZP_00330242.1| COG0542: ATPases with chaperone activity, ATP-binding subunit
[Moorella thermoacetica ATCC 39073]
Length = 840
Score = 339 bits (869), Expect = 1e-91
Identities = 198/451 (43%), Positives = 276/451 (60%), Gaps = 21/451 (4%)
Query: 2 LENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSRE 61
+ TE A +V+ AQ+EA + H I TEHILLGIL + + +AA+ L + G++ RE
Sbjct: 1 MARFTERANRVLRLAQEEALALRHPAIGTEHILLGILREGDSVAARALTNLGVNIKAVRE 60
Query: 62 QVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNG 121
+V K++ G G ++ T AK VL+ + + A+ G + V T HLLLGLL+ G
Sbjct: 61 EVRKVIRPGEAVVG---GELGLTPRAKRVLELANEEARRQGVNYVGTEHLLLGLLEEGEG 117
Query: 122 TVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQI 181
Q++ G KIREQVI + P G + G + + A
Sbjct: 118 LAAQVLGGLGLSPEKIREQVIALLGG----AGQPQTGGWTVLFGNLPFGMAA-------- 165
Query: 182 KFP----VLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVI 237
FP Q GA+ + Q L+ F +LT+LA+E KL P VGRE+++ERV+
Sbjct: 166 -FPGGMGFQAQGMPAGAKMQERRPGQTPTLDQFSRDLTRLAREDKLDPVVGREKEIERVV 224
Query: 238 QIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISN 297
QI+ RR KNNP L+G+PGVGKT+I++GLAQ+I+ VPE L+ K+VVALD++ +
Sbjct: 225 QILSRRTKNNPVLIGDPGVGKTAIVEGLAQKIVQNQVPEVLRDKRVVALDMSGMVAGTKY 284
Query: 298 QGSSEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIF 356
+G EER + ++ E+ GNVILF+ E+H + A A GA A ILK AL RG +Q I
Sbjct: 285 RGEFEERFKKVMDEVRAAGNVILFIDELHTLIGAGAAEGAIDAANILKPALARGELQTIG 344
Query: 357 ATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVA 416
ATT++E+R H+E D L+R FQ V V EP+VEETI ILKGLR YE+H+++ TDEAL A
Sbjct: 345 ATTIDEYRKHIERDAALERRFQAVMVEEPTVEETIAILKGLRDRYEAHHRVKITDEALEA 404
Query: 417 AANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
AA LS +Y+++R+LPDKAIDL+DEAGS V+L
Sbjct: 405 AARLSDRYITDRYLPDKAIDLVDEAGSRVRL 435
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 719,751,550
Number of Sequences: 2540612
Number of extensions: 29814059
Number of successful extensions: 110267
Number of sequences better than 10.0: 1124
Number of HSP's better than 10.0 without gapping: 947
Number of HSP's successfully gapped in prelim test: 177
Number of HSP's that attempted gapping in prelim test: 106411
Number of HSP's gapped (non-prelim): 2103
length of query: 455
length of database: 863,360,394
effective HSP length: 131
effective length of query: 324
effective length of database: 530,540,222
effective search space: 171895031928
effective search space used: 171895031928
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146751.5


