

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.7 + phase: 0
(376 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
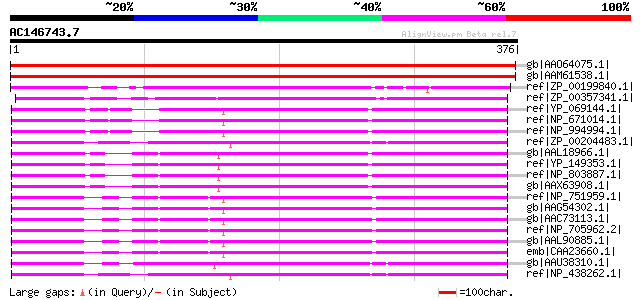
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO64075.1| putative homoserine dehydrogenase [Arabidopsis th... 515 e-145
gb|AAM61538.1| homoserine dehydrogenase-like protein [Arabidopsi... 512 e-144
ref|ZP_00199840.1| COG0460: Homoserine dehydrogenase [Rubrobacte... 214 3e-54
ref|ZP_00357341.1| COG0460: Homoserine dehydrogenase [Chloroflex... 213 6e-54
ref|YP_069144.1| bifunctional ThrA; aspartokinase I and homoseri... 209 1e-52
ref|NP_671014.1| aspartokinase I, homoserine dehydrogenase I [Ye... 209 1e-52
ref|NP_994994.1| bifunctional aspartokinase/homoserine dehydroge... 209 1e-52
ref|ZP_00204483.1| COG0527: Aspartokinases [Actinobacillus pleur... 208 2e-52
gb|AAL18966.1| aspartokinase I; homoserine dehydrogenase I [Salm... 200 5e-50
ref|YP_149353.1| aspartokinase I/homoserine dehydrogenase I [Sal... 200 7e-50
ref|NP_803887.1| aspartokinase I [Salmonella enterica subsp. ent... 200 7e-50
gb|AAX63908.1| aspartokinase I [Salmonella enterica subsp. enter... 200 7e-50
ref|NP_751959.1| Aspartokinase I; Homoserine dehydrogenase I [Es... 199 9e-50
gb|AAG54302.1| aspartokinase I, homoserine dehydrogenase I [Esch... 199 1e-49
gb|AAC73113.1| bifunctional: aspartokinase I (N-terminal); homos... 198 2e-49
ref|NP_705962.2| aspartokinase I, homoserine dehydrogenase I [Sh... 198 2e-49
gb|AAL90885.1| aspartokinase I-homoserine dehydrogenase I [Esche... 198 2e-49
emb|CAA23660.1| unnamed protein product [Escherichia coli] gi|21... 197 3e-49
gb|AAU38310.1| LysC protein [Mannheimia succiniciproducens MBEL5... 197 4e-49
ref|NP_438262.1| aspartokinase I/homoserine dehydrogenase I [Hae... 196 1e-48
>gb|AAO64075.1| putative homoserine dehydrogenase [Arabidopsis thaliana]
gi|28393222|gb|AAO42041.1| putative homoserine
dehydrogenase [Arabidopsis thaliana]
gi|15242147|ref|NP_197605.1| homoserine dehydrogenase
family protein [Arabidopsis thaliana]
Length = 376
Score = 515 bits (1327), Expect = e-145
Identities = 259/375 (69%), Positives = 313/375 (83%)
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDD 60
MK IP++LMGCGGVG HLLQHIVS RSLH+ G+ +RV+G+ DSKSLV D+L + +D
Sbjct: 1 MKKIPVLLMGCGGVGRHLLQHIVSCRSLHAKMGVHIRVIGVCDSKSLVAPMDVLKEELND 60
Query: 61 SFLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSD 120
L E+C +K G +LSKLG LG +V E + EIA LGK TGLA VDC+AS +
Sbjct: 61 ELLSEVCLIKSTGSALSKLGALGGYRVVHDSELSTETEEIAKLLGKSTGLAVVDCSASME 120
Query: 121 TIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRII 180
TI +L + VDLGCC+V+ANKKP+TST+ ++KL +PR IRHESTVGAGLPVI+SLNRII
Sbjct: 121 TIEILMKAVDLGCCIVLANKKPVTSTLEHYDKLALHPRFIRHESTVGAGLPVIASLNRII 180
Query: 181 SSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKA 240
SSGDPV+RI+GSLSGTLGYVMSE+EDGKPLSQVV+AAK LGYTEPDPRDDLGGMDVARK
Sbjct: 181 SSGDPVHRIVGSLSGTLGYVMSELEDGKPLSQVVQAAKKLGYTEPDPRDDLGGMDVARKG 240
Query: 241 LILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGN 300
LILAR+LG+RI MDSI+IESLYP+EMGP +M+ DDFL G++ LD++I+ERV+KA+S G
Sbjct: 241 LILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGIVKLDQNIEERVKKASSKGC 300
Query: 301 VLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTT 360
VLRYVCVIEG +VGI+E+ K+S LGRLRGSDN++E+Y+RCY QPLVIQGAGAGNDTT
Sbjct: 301 VLRYVCVIEGSSVQVGIREVSKDSPLGRLRGSDNIVEIYSRCYKEQPLVIQGAGAGNDTT 360
Query: 361 AAGVLADIVDIQDLF 375
AAGVLADI+D+QDLF
Sbjct: 361 AAGVLADIIDLQDLF 375
>gb|AAM61538.1| homoserine dehydrogenase-like protein [Arabidopsis thaliana]
Length = 376
Score = 512 bits (1318), Expect = e-144
Identities = 258/375 (68%), Positives = 312/375 (82%)
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDD 60
MK IP++LMGCGGVG HLLQHIVS RSLH+ G+ +RV+G+ DSKSLV D+L + +D
Sbjct: 1 MKKIPVLLMGCGGVGRHLLQHIVSCRSLHAKMGVHIRVIGVCDSKSLVAPMDVLKEELND 60
Query: 61 SFLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSD 120
L E+C +K G +LSKLG LG +V E + EIA LGK TGLA VDC+AS +
Sbjct: 61 ELLSEVCLIKSTGSALSKLGALGGYRVVHDSELSTETEEIAKLLGKSTGLAVVDCSASME 120
Query: 121 TIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRII 180
TI +L + VDLGCC+V+ANKKP+TST+ ++KL +PR IRHESTVGAGLPVI+SLNRII
Sbjct: 121 TIEILMKAVDLGCCIVLANKKPVTSTLEHYDKLALHPRFIRHESTVGAGLPVIASLNRII 180
Query: 181 SSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKA 240
SSGDPV+RI+GSLS TLGYVMSE+EDGKPLSQVV+AAK LGYTEPDPRDDLGGMDVARK
Sbjct: 181 SSGDPVHRIVGSLSVTLGYVMSELEDGKPLSQVVQAAKKLGYTEPDPRDDLGGMDVARKG 240
Query: 241 LILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGN 300
LILAR+LG+RI MDSI+IESLYP+EMGP +M+ DDFL G++ LD++I+ERV+KA+S G
Sbjct: 241 LILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGIVKLDQNIEERVKKASSKGC 300
Query: 301 VLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTT 360
VLRYVCVIEG +VGI+E+ K+S LGRLRGSDN++E+Y+RCY QPLVIQGAGAGNDTT
Sbjct: 301 VLRYVCVIEGSSVQVGIREVSKDSPLGRLRGSDNIVEIYSRCYKEQPLVIQGAGAGNDTT 360
Query: 361 AAGVLADIVDIQDLF 375
AAGVLADI+D+QDLF
Sbjct: 361 AAGVLADIIDLQDLF 375
>ref|ZP_00199840.1| COG0460: Homoserine dehydrogenase [Rubrobacter xylanophilus DSM
9941]
Length = 360
Score = 214 bits (546), Expect = 3e-54
Identities = 143/375 (38%), Positives = 210/375 (55%), Gaps = 30/375 (8%)
Query: 1 MKTIPLILMGCGGVGSHLLQHIVSSRSLHSSQ-GLCLRVVGIGDSKSLVVVDDLLNKGFD 59
MK + L+ +G G VG + Q ++ R + GL + + D+ + +DLL +
Sbjct: 1 MKRVELVQLGIGHVGRAVAQIVLEERKRWRERYGLDISYRAVADTSGALAGEDLLPQAI- 59
Query: 60 DSFLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASS 119
RLK G LS+LG P E +A G KT VD A
Sbjct: 60 --------RLKEAGGRLSELG--------AEPLEE----VLAEGPGPKTARVLVDAAAGE 99
Query: 120 DTIVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTY-PRRIRHESTVGAGLPVISSLNR 178
T + + V G +V+ NK P++ + +E L+ P R+R+E+TVGAG+PV+S++
Sbjct: 100 GTYDLDVRGVRRGSSLVLCNKGPISGSTERYEGLVGEGPERLRYEATVGAGVPVLSTIEA 159
Query: 179 IISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVAR 238
+ +SGD + I S SGTLG++MS VE+G+P S+VVR A L YTEPDPRDDL G+DVAR
Sbjct: 160 LQASGDDILEIQASPSGTLGFIMSGVEEGRPFSEVVREAAELHYTEPDPRDDLSGLDVAR 219
Query: 239 KALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASN 298
KALILAR +GRR+ + + ESL P+E+ ++ ++F+ L D+ R+ A
Sbjct: 220 KALILARKIGRRLEPEEVPYESLVPEELRE--VSVEEFME-RLSEFDEGFAARL-SAVRP 275
Query: 299 GNVLRYVCVI--EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G+VLRY+ I EGP EVG++E+P G G +NV TR YS+ L + G GAG
Sbjct: 276 GHVLRYLARIPKEGP-VEVGLREVPAAGCFGARSGVENVFNFRTRRYSDVTLTVSGPGAG 334
Query: 357 NDTTAAGVLADIVDI 371
+ TA+GV+ D++D+
Sbjct: 335 PERTASGVVCDLLDL 349
>ref|ZP_00357341.1| COG0460: Homoserine dehydrogenase [Chloroflexus aurantiacus]
Length = 354
Score = 213 bits (543), Expect = 6e-54
Identities = 142/367 (38%), Positives = 203/367 (54%), Gaps = 23/367 (6%)
Query: 5 PLILMGCGGVGSHLLQHIVS-SRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDSFL 63
P+I +G GGVG L++ +++ + ++ G+ LR + DS+ + D L D +
Sbjct: 3 PIIQLGLGGVGRALVRQMLAVAPTIRRRYGIDLRYTALVDSRGALAGDPALT----DDQI 58
Query: 64 LELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDTIV 123
+ K G + L H ++ LE+ T VD TA+
Sbjct: 59 QTVLTAKAAGHHIDSLA---------HAITDRHWLELLPA----TMAIVVDVTAAGSHAA 105
Query: 124 VLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRHESTVGAGLPVISSLNRIISSG 183
L + G VV+ANK+PL + F L T R+E+TVGAGLPVI L +I SG
Sbjct: 106 PLAAAISAGHRVVLANKRPLCESYELFAAL-TERGATRYEATVGAGLPVIGVLQSLIDSG 164
Query: 184 DPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVARKALIL 243
D V I +LSGTLG++MSE+E G + VR A LGYTEPDPRDDL G DVARKALIL
Sbjct: 165 DEVRAIEAALSGTLGFLMSELERGVGFADAVRTAHQLGYTEPDPRDDLSGADVARKALIL 224
Query: 244 ARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAASNGNVLR 303
AR G + D++ E L+P + +T +FL L + ++ R++ A++NG VLR
Sbjct: 225 ARTCGMHLPPDAVIAEPLFPATLKEISVT--EFLQ-RLEEAEHEVMNRLQVASANGKVLR 281
Query: 304 YVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGNDTTAA 362
Y+ I EVG++ELP + L LRG DN++ T Y+++PLV++G GAG + TAA
Sbjct: 282 YITRISPDNGVEVGLRELPTDHPLASLRGPDNMISFTTNRYADRPLVVRGPGAGVEVTAA 341
Query: 363 GVLADIV 369
GVL+DI+
Sbjct: 342 GVLSDII 348
>ref|YP_069144.1| bifunctional ThrA; aspartokinase I and homoserine dehydrogenase I
(C-terminal) [Yersinia pseudotuberculosis IP 32953]
gi|51588235|emb|CAH19842.1| bifunctional ThrA;
aspartokinase I and homoserine dehydrogenase I
(C-terminal) [Yersinia pseudotuberculosis IP 32953]
gi|15978551|emb|CAC89315.1| bifunctional
aspartokinase/homoserine dehydrogenase I [Yersinia
pestis CO92] gi|16120788|ref|NP_404101.1| bifunctional
aspartokinase/homoserine dehydrogenase I [Yersinia
pestis CO92] gi|25288159|pir||AH0056 aspartate kinase
(EC 2.7.2.4) [imported] - Yersinia pestis (strain CO92)
Length = 819
Score = 209 bits (532), Expect = 1e-52
Identities = 135/372 (36%), Positives = 202/372 (54%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +SK+++ + D+
Sbjct: 464 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQRHIDLRVCGIANSKAMLTNVHGIAL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL ++ +LS+L L + ++P VDCT+S
Sbjct: 521 WRQELAEVQEPF-NLSRLIRLVKEYHLLNP-------------------VIVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + ++ R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLTDGFHVVTPNKKANTSSMNYYRQMRAAATKSCRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + R G LSG+L ++ ++++G LS+ R AK+LGYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELMRFTGILSGSLSFIFGKLDEGMSLSEATRQAKALGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G ++ + I++ES+ P D D D L LD + V AA
Sbjct: 681 RKLLILAREAGYKLELADIEVESVLPASF--DASGDVDTFLARLPSLDAEFTRLVANAAE 738
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV VIE RC+V ++ + N L +++ +N L YTR Y PLV++G GAGN
Sbjct: 739 QGKVLRYVGVIEDGRCKVRMEAVDGNDPLYKVKNGENALAFYTRYYQPIPLVLRGYGAGN 798
Query: 358 DTTAAGVLADIV 369
D TAAGV AD++
Sbjct: 799 DVTAAGVFADLL 810
>ref|NP_671014.1| aspartokinase I, homoserine dehydrogenase I [Yersinia pestis KIM]
gi|21960699|gb|AAM87265.1| aspartokinase I, homoserine
dehydrogenase I [Yersinia pestis KIM]
Length = 845
Score = 209 bits (532), Expect = 1e-52
Identities = 135/372 (36%), Positives = 202/372 (54%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +SK+++ + D+
Sbjct: 490 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQRHIDLRVCGIANSKAMLTNVHGIAL---DN 546
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL ++ +LS+L L + ++P VDCT+S
Sbjct: 547 WRQELAEVQEPF-NLSRLIRLVKEYHLLNP-------------------VIVDCTSSQAV 586
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + ++ R+ +++ VGAGLPVI +L
Sbjct: 587 ADQYADFLTDGFHVVTPNKKANTSSMNYYRQMRAAATKSCRKFLYDTNVGAGLPVIENLQ 646
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + R G LSG+L ++ ++++G LS+ R AK+LGYTEPDPRDDL GMDVA
Sbjct: 647 NLLNAGDELMRFTGILSGSLSFIFGKLDEGMSLSEATRQAKALGYTEPDPRDDLSGMDVA 706
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G ++ + I++ES+ P D D D L LD + V AA
Sbjct: 707 RKLLILAREAGYKLELADIEVESVLPASF--DASGDVDTFLARLPSLDAEFTRLVANAAE 764
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV VIE RC+V ++ + N L +++ +N L YTR Y PLV++G GAGN
Sbjct: 765 QGKVLRYVGVIEDGRCKVRMEAVDGNDPLYKVKNGENALAFYTRYYQPIPLVLRGYGAGN 824
Query: 358 DTTAAGVLADIV 369
D TAAGV AD++
Sbjct: 825 DVTAAGVFADLL 836
>ref|NP_994994.1| bifunctional aspartokinase/homoserine dehydrogenase I [Yersinia
pestis biovar Medievalis str. 91001]
gi|45438324|gb|AAS63871.1| bifunctional
aspartokinase/homoserine dehydrogenase I [Yersinia
pestis biovar Medievalis str. 91001]
Length = 817
Score = 209 bits (532), Expect = 1e-52
Identities = 135/372 (36%), Positives = 202/372 (54%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG L++ I + + + LRV GI +SK+++ + D+
Sbjct: 462 QVIEVFVIGVGGVGGALIEQIYRQQPWLKQRHIDLRVCGIANSKAMLTNVHGIAL---DN 518
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL ++ +LS+L L + ++P VDCT+S
Sbjct: 519 WRQELAEVQEPF-NLSRLIRLVKEYHLLNP-------------------VIVDCTSSQAV 558
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP----RRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + ++ R+ +++ VGAGLPVI +L
Sbjct: 559 ADQYADFLTDGFHVVTPNKKANTSSMNYYRQMRAAATKSCRKFLYDTNVGAGLPVIENLQ 618
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + R G LSG+L ++ ++++G LS+ R AK+LGYTEPDPRDDL GMDVA
Sbjct: 619 NLLNAGDELMRFTGILSGSLSFIFGKLDEGMSLSEATRQAKALGYTEPDPRDDLSGMDVA 678
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G ++ + I++ES+ P D D D L LD + V AA
Sbjct: 679 RKLLILAREAGYKLELADIEVESVLPASF--DASGDVDTFLARLPSLDAEFTRLVANAAE 736
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV VIE RC+V ++ + N L +++ +N L YTR Y PLV++G GAGN
Sbjct: 737 QGKVLRYVGVIEDGRCKVRMEAVDGNDPLYKVKNGENALAFYTRYYQPIPLVLRGYGAGN 796
Query: 358 DTTAAGVLADIV 369
D TAAGV AD++
Sbjct: 797 DVTAAGVFADLL 808
>ref|ZP_00204483.1| COG0527: Aspartokinases [Actinobacillus pleuropneumoniae serovar 1
str. 4074]
Length = 818
Score = 208 bits (530), Expect = 2e-52
Identities = 130/372 (34%), Positives = 201/372 (53%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
K+I + L+G GGVGS L+ I + + + + +RV + ++ +++ ++ LN
Sbjct: 464 KSIEVFLVGVGGVGSELIDQIKQQKEFLAKKDIEIRVCALANADKMLLNENGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L K E+ ++ D F+ +L FVDCTA+
Sbjct: 518 ----LDNWKSDLETATQPSDFDVLLSFI-------------KLHHVVNPVFVDCTAAQSV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRH----ESTVGAGLPVISSLN 177
+ + + G VV NKK T + +++L R+ +H E+ VGAGLPVI +L
Sbjct: 561 ANLYTRALKEGFHVVTPNKKANTGSYRYYQELREAARQGQHKFLYETNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD V + G LSG+L ++ ++++G LS+ AK G+TEPDPRDDL G DVA
Sbjct: 621 NLLAAGDEVEKFEGILSGSLSFIFGKLDEGLSLSEATLIAKEKGFTEPDPRDDLSGADVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G + D I++E + PK D M+ ++FL L +D ERV+ A +
Sbjct: 681 RKLLILARETGLALEFDDIEVEGVLPKGFS-DGMSKEEFLKV-LPEIDAQFNERVQAAKA 738
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV I G +C V I+ + +N L +++ +N L TR YS PL+++G GAG
Sbjct: 739 EGKVLRYVGSITGDKCRVAIEAVDENHPLYKVKDGENALAFLTRYYSPIPLLLRGYGAGT 798
Query: 358 DTTAAGVLADIV 369
D TAAG+ ADI+
Sbjct: 799 DVTAAGIFADIL 810
>gb|AAL18966.1| aspartokinase I; homoserine dehydrogenase I [Salmonella typhimurium
LT2] gi|16763392|ref|NP_459007.1| aspartokinase
I/homoserine dehydrogenase I [Salmonella typhimurium
LT2]
Length = 820
Score = 200 bits (509), Expect = 5e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPNEF--DASGDVTAFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>ref|YP_149353.1| aspartokinase I/homoserine dehydrogenase I [Salmonella enterica
subsp. enterica serovar Paratyphi A str. ATCC 9150]
gi|56126535|gb|AAV76041.1| aspartokinase I/homoserine
dehydrogenase I [Salmonella enterica subsp. enterica
serovar Paratyphi A str. ATCC 9150]
Length = 820
Score = 200 bits (508), Expect = 7e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPDEF--DASGDVTAFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>ref|NP_803887.1| aspartokinase I [Salmonella enterica subsp. enterica serovar Typhi
Ty2] gi|16501285|emb|CAD01155.1| aspartokinase
I/homoserine dehydrogenase I [Salmonella enterica subsp.
enterica serovar Typhi] gi|29136169|gb|AAO67736.1|
aspartokinase I [Salmonella enterica subsp. enterica
serovar Typhi Ty2] gi|16758995|ref|NP_454612.1|
aspartokinase I/homoserine dehydrogenase I [Salmonella
enterica subsp. enterica serovar Typhi str. CT18]
gi|25288156|pir||AC0502 aspartokinase I/homoserine
dehydrogenase I [imported] - Salmonella enterica subsp.
enterica serovar Typhi (strain CT18)
Length = 820
Score = 200 bits (508), Expect = 7e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPDEF--DASGDVTAFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>gb|AAX63908.1| aspartokinase I [Salmonella enterica subsp. enterica serovar
Choleraesuis str. SC-B67] gi|62178572|ref|YP_214989.1|
aspartokinase I [Salmonella enterica subsp. enterica
serovar Choleraesuis str. SC-B67]
Length = 820
Score = 200 bits (508), Expect = 7e-50
Identities = 133/373 (35%), Positives = 195/373 (51%), Gaps = 30/373 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + ++ ++ + LRV G+ +SK+L+ LN D+
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQTWLKNKHIDLRVCGVANSKALLTNVHGLNL---DN 520
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
+ EL + P + G+++ + + + VDCT+S
Sbjct: 521 WQAELAQANA-------------------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKL----LTYPRRIRHESTVGAGLPVISSLN 177
+ G VV NKK TS+M + +L R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRFAAAQSRRKFLYDTNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + + G LSG+L ++ ++E+G LSQ A+ +GYTEPDPRDDL GMDVA
Sbjct: 621 NLLNAGDELQKFSGILSGSLSFIFGKLEEGMSLSQATALAREMGYTEPDPRDDLSGMDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR GR + + I IE + P E D D L LD RV KA
Sbjct: 681 RKLLILARETGRELELSDIVIEPVLPDEF--DASGDVTTFMAHLPQLDDAFAARVAKARD 738
Query: 298 NGNVLRYVCVIEGPR-CEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAG 356
G VLRYV IE C V I E+ N L +++ +N L Y+ Y PLV++G GAG
Sbjct: 739 EGKVLRYVGNIEEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGAG 798
Query: 357 NDTTAAGVLADIV 369
ND TAAGV AD++
Sbjct: 799 NDVTAAGVFADLL 811
>ref|NP_751959.1| Aspartokinase I; Homoserine dehydrogenase I [Escherichia coli
CFT073] gi|26106317|gb|AAN78503.1| Aspartokinase I;
Homoserine dehydrogenase I [Escherichia coli CFT073]
Length = 841
Score = 199 bits (507), Expect = 9e-50
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 485 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 538
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 539 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 581
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 582 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 640
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 641 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 700
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 701 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDNLFAARVAKAR 758
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 759 DEGKVLRYVGNIDEDGVCRVKIAEVDSNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 818
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 819 GNDVTAAGVFADLL 832
>gb|AAG54302.1| aspartokinase I, homoserine dehydrogenase I [Escherichia coli
O157:H7 EDL933] gi|13359458|dbj|BAB33425.1|
aspartokinase I-homoserine dehydrogenase I [Escherichia
coli O157:H7] gi|15829256|ref|NP_308029.1| aspartokinase
I-homoserine dehydrogenase I [Escherichia coli O157:H7]
gi|25288151|pir||B90629 aspartokinase I-homoserine
dehydrogenase I [imported] - Escherichia coli (strain
O157:H7, substrain RIMD 0509952) gi|25288153|pir||B85480
aspartokinase I, homoserine dehydrogenase I [imported] -
Escherichia coli (strain O157:H7, substrain EDL933)
gi|15799682|ref|NP_285694.1| aspartokinase I, homoserine
dehydrogenase I [Escherichia coli O157:H7 EDL933]
Length = 820
Score = 199 bits (505), Expect = 1e-49
Identities = 133/374 (35%), Positives = 199/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ LL + R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHLLRHAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>gb|AAC73113.1| bifunctional: aspartokinase I (N-terminal); homoserine
dehydrogenase I (C-terminal) [Escherichia coli K12]
gi|537245|gb|AAA97301.1| aspartokinase I-homoserine
dehydrogenase I [Escherichia coli]
gi|16127996|ref|NP_414543.1| bifunctional: aspartokinase
I (N-terminal); homoserine dehydrogenase I (C-terminal)
[Escherichia coli K12] gi|34395933|sp|P00561|AK1H_ECOLI
Bifunctional aspartokinase/homoserine dehydrogenase I
(AKI-HDI) [Includes: Aspartokinase I ; Homoserine
dehydrogenase I ]
Length = 820
Score = 198 bits (504), Expect = 2e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>ref|NP_705962.2| aspartokinase I, homoserine dehydrogenase I [Shigella flexneri 2a
str. 301] gi|56383131|gb|AAN41669.2| aspartokinase I,
homoserine dehydrogenase I [Shigella flexneri 2a str.
301] gi|30061573|ref|NP_835744.1| aspartokinase I,
homoserine dehydrogenase I [Shigella flexneri 2a str.
2457T] gi|30039815|gb|AAP15549.1| aspartokinase I,
homoserine dehydrogenase I [Shigella flexneri 2a str.
2457T]
Length = 820
Score = 198 bits (504), Expect = 2e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTSVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELVKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>gb|AAL90885.1| aspartokinase I-homoserine dehydrogenase I [Escherichia coli]
Length = 820
Score = 198 bits (504), Expect = 2e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + VDCT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVDCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ +++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>emb|CAA23660.1| unnamed protein product [Escherichia coli]
gi|21321892|dbj|BAB96579.1| ThrA bifunctional enzyme
[Escherichia coli] gi|147979|gb|AAA83914.1|
aspartokinase I-homoserine dehydrogenase I [Escherichia
coli]
Length = 820
Score = 197 bits (502), Expect = 3e-49
Identities = 133/374 (35%), Positives = 198/374 (52%), Gaps = 32/374 (8%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
+ I + ++G GGVG LL+ + +S ++ + LRV G+ +SK+L+ LN
Sbjct: 464 QVIEVFVIGVGGVGGALLEQLKRQQSWLKNKHIDLRVCGVANSKALLTNVHGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L++ E L++ + P + G+++ + + + V+CT+S
Sbjct: 518 -------LENWQEELAQAKE---------PFNLGRLIRLVKEYHLLNPV-IVNCTSSQAV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYP-----RRIRHESTVGAGLPVISSL 176
+ G VV NKK TS+M D+ L Y R+ ++ VGAGLPVI +L
Sbjct: 561 ADQYADFLREGFHVVTPNKKANTSSM-DYYHQLRYAAEKSRRKFLYDINVGAGLPVIENL 619
Query: 177 NRIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDV 236
++++GD + + G LSG+L Y+ ++++G S+ R A+ +GYTEPDPRDDL GMDV
Sbjct: 620 QNLLNAGDELMKFSGILSGSLSYIFGKLDEGMSFSEATRLAREMGYTEPDPRDDLSGMDV 679
Query: 237 ARKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAA 296
ARK LILAR GR + + I+IE + P E + D L LD RV KA
Sbjct: 680 ARKLLILARETGRELELADIEIEPVLPAEFNAE--GDVAAFMANLSQLDDLFAARVAKAR 737
Query: 297 SNGNVLRYVCVI-EGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGA 355
G VLRYV I E C V I E+ N L +++ +N L Y+ Y PLV++G GA
Sbjct: 738 DEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFKVKNGENALAFYSHYYQPLPLVLRGYGA 797
Query: 356 GNDTTAAGVLADIV 369
GND TAAGV AD++
Sbjct: 798 GNDVTAAGVFADLL 811
>gb|AAU38310.1| LysC protein [Mannheimia succiniciproducens MBEL55E]
gi|52425758|ref|YP_088895.1| LysC protein [Mannheimia
succiniciproducens MBEL55E]
Length = 816
Score = 197 bits (501), Expect = 4e-49
Identities = 125/372 (33%), Positives = 195/372 (51%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
K++ + L+G GGVG L++ I + + + + +RV + +S +++ ++ L+
Sbjct: 465 KSVDMFLVGVGGVGGELIEQIKQQKEYLAKKDIEIRVCALANSNKMLLNENGLS------ 518
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L + E LS S+ +L +L FVDCT +
Sbjct: 519 -------LDNWKEDLSNATQ----------PSDFDVLLSFIKLHHVVNPVFVDCTTAESV 561
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDF----EKLLTYPRRIRHESTVGAGLPVISSLN 177
+ + + G VV NKK T M + E R+ +++ VGAGLPVI +L
Sbjct: 562 SGLYARALSEGFHVVTPNKKANTREMAYYNLVRENARKNQRKFLYDTNVGAGLPVIENLQ 621
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD V R G LSG+L ++ ++E+G LSQ A+ G+TEPDPRDDL G DVA
Sbjct: 622 NLLAAGDEVERFNGILSGSLSFIFGKLEEGLTLSQATALAREKGFTEPDPRDDLSGQDVA 681
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G + + +++ES+ PK + + +F+ L LD + RVEKA +
Sbjct: 682 RKLLILARESGLELELSDVEVESVLPKGFS-EGKSAVEFMEI-LPQLDAEFAARVEKAGA 739
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
VLRYV I +C+V I E+ + L +++ +N L YTR Y PL+++G GAGN
Sbjct: 740 QNKVLRYVGQINDGKCKVSIVEVDADDPLYKVKNGENALAFYTRYYQPIPLLLRGYGAGN 799
Query: 358 DTTAAGVLADIV 369
TAAG+ ADI+
Sbjct: 800 AVTAAGIFADIL 811
>ref|NP_438262.1| aspartokinase I/homoserine dehydrogenase I [Haemophilus influenzae
Rd KW20] gi|1573040|gb|AAC21767.1| aspartokinase I /
homoserine dehydrogenase I (thrA) [Haemophilus
influenzae Rd KW20] gi|1073872|pir||A64048 aspartate
kinase (EC 2.7.2.4) / homoserine dehydrogenase (EC
1.1.1.3) I HI0089 [similarity] - Haemophilus influenzae
(strain Rd KW20) gi|1168401|sp|P44505|AKH_HAEIN
Bifunctional aspartokinase/homoserine dehydrogenase
(AK-HD) [Includes: Aspartokinase ; Homoserine
dehydrogenase ]
Length = 815
Score = 196 bits (497), Expect = 1e-48
Identities = 123/372 (33%), Positives = 199/372 (53%), Gaps = 29/372 (7%)
Query: 2 KTIPLILMGCGGVGSHLLQHIVSSRSLHSSQGLCLRVVGIGDSKSLVVVDDLLNKGFDDS 61
K + + L+G GGVG L++ + + + + + +RV I +S +++ ++ LN
Sbjct: 464 KVVDMFLVGVGGVGGELIEQVKRQKEYLAKKNVEIRVCAIANSNRMLLDENGLN------ 517
Query: 62 FLLELCRLKHGGESLSKLGDLGQCQVFVHPESEGKILEIASQLGKKTGLAFVDCTASSDT 121
L K+ E+ ++ D F+ +L FVDCT++
Sbjct: 518 ----LEDWKNDLENATQPSDFDVLLSFI-------------KLHHVVNPVFVDCTSAESV 560
Query: 122 IVVLKQVVDLGCCVVMANKKPLTSTMGDFEKLLTYPRRIRH----ESTVGAGLPVISSLN 177
+ + + G VV NKK T + + +L + +H E+ VGAGLPVI +L
Sbjct: 561 AGLYARALKEGFHVVTPNKKANTRELVYYNELRQNAQASQHKFLYETNVGAGLPVIENLQ 620
Query: 178 RIISSGDPVNRIIGSLSGTLGYVMSEVEDGKPLSQVVRAAKSLGYTEPDPRDDLGGMDVA 237
++++GD + G LSG+L ++ ++E+G LS+V A+ G+TEPDPRDDL G DVA
Sbjct: 621 NLLAAGDELEYFEGILSGSLSFIFGKLEEGLSLSEVTALAREKGFTEPDPRDDLSGQDVA 680
Query: 238 RKALILARILGRRINMDSIQIESLYPKEMGPDVMTDDDFLSCGLLLLDKDIQERVEKAAS 297
RK LILAR G + + +++E + PK D + D+F++ L LD++ + RV A +
Sbjct: 681 RKLLILAREAGIELELSDVEVEGVLPKGFS-DGKSADEFMAM-LPQLDEEFKTRVATAKA 738
Query: 298 NGNVLRYVCVIEGPRCEVGIQELPKNSALGRLRGSDNVLEVYTRCYSNQPLVIQGAGAGN 357
G VLRYV I +C+V I + N+ L +++ +N L YTR Y PL+++G GAGN
Sbjct: 739 EGKVLRYVGKISEGKCKVSIVAVDLNNPLYKVKDGENALAFYTRYYQPIPLLLRGYGAGN 798
Query: 358 DTTAAGVLADIV 369
TAAG+ ADI+
Sbjct: 799 AVTAAGIFADIL 810
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.139 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 622,125,441
Number of Sequences: 2540612
Number of extensions: 26226452
Number of successful extensions: 60948
Number of sequences better than 10.0: 346
Number of HSP's better than 10.0 without gapping: 284
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 59979
Number of HSP's gapped (non-prelim): 483
length of query: 376
length of database: 863,360,394
effective HSP length: 130
effective length of query: 246
effective length of database: 533,080,834
effective search space: 131137885164
effective search space used: 131137885164
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146743.7


