

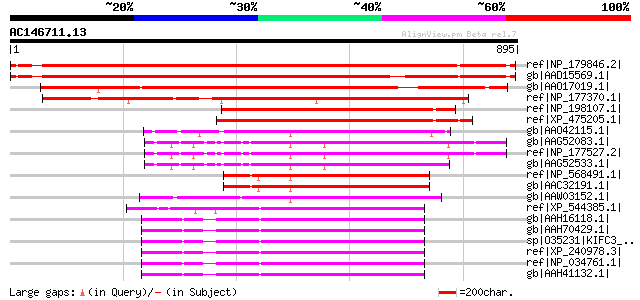
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146711.13 - phase: 0 /pseudo
(895 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_179846.2| kinesin motor protein-related [Arabidopsis thal... 1061 0.0
gb|AAD15569.1| putative kinesin heavy chain [Arabidopsis thalian... 999 0.0
gb|AAO17019.1| Hypothetical protein [Oryza sativa (japonica cult... 929 0.0
ref|NP_177370.1| kinesin motor protein-related [Arabidopsis thal... 655 0.0
ref|NP_198107.1| kinesin motor protein-related [Arabidopsis thal... 489 e-136
ref|XP_475205.1| putative kinesin-related protein [Oryza sativa ... 449 e-124
gb|AAO42115.1| putative kinesin [Arabidopsis thaliana] 348 3e-94
gb|AAG52083.1| kinesin-related protein; 103921-99132 [Arabidopsi... 339 3e-91
ref|NP_177527.2| kinesin motor protein-related [Arabidopsis thal... 339 3e-91
gb|AAG52533.1| putative kinesin; 97201-101676 [Arabidopsis thali... 338 4e-91
ref|NP_568491.1| kinesin motor protein-related [Arabidopsis thal... 338 4e-91
gb|AAC32191.1| kinesin-like heavy chain [Arabidopsis thaliana] 338 4e-91
gb|AAW03152.1| kinesin [Gossypium hirsutum] 338 6e-91
ref|XP_544385.1| PREDICTED: similar to Kifc3 protein [Canis fami... 337 1e-90
gb|AAH16118.1| Kifc3 protein [Mus musculus] 335 4e-90
gb|AAH70429.1| Kifc3 protein [Mus musculus] 335 4e-90
sp|O35231|KIFC3_MOUSE Kinesin-like protein KIFC3 335 4e-90
ref|XP_240978.3| PREDICTED: similar to Kifc3 protein [Rattus nor... 335 5e-90
ref|NP_034761.1| kinesin family member C3 [Mus musculus] gi|1258... 335 5e-90
gb|AAH41132.1| KIFC3 protein [Homo sapiens] 333 1e-89
>ref|NP_179846.2| kinesin motor protein-related [Arabidopsis thaliana]
Length = 1093
Score = 1061 bits (2744), Expect = 0.0
Identities = 550/898 (61%), Positives = 684/898 (75%), Gaps = 32/898 (3%)
Query: 1 MEDANDSLFDSMLVDS---SSKLI--QNGFARSQSSGLKINIMY*FVKLSVHF*RIHFEA 55
++D +D+L DSM DS S +LI ++GF + ++ I Y
Sbjct: 45 IDDCDDALGDSM--DSGFYSFRLILGEDGFNLMLTRRVRFLIQY---------------- 86
Query: 56 EECVMFVNVGGEATNEGADGVKFLSDTFFDGGDVFLTNEAIVEGGDYPSIYQSARVGSFS 115
E +MF+N GG+ + + D +F+GGDV T E+IVE GD+P IYQSARVG+F
Sbjct: 87 -ETIMFINAGGDDSKVLDSELNISRDDYFEGGDVLRTEESIVEAGDFPFIYQSARVGNFC 145
Query: 116 YRIDNLPPGQYLVDLHFVEIINVNGPKGMRVFNVYIQEEKVLSELDIYAAVGVNKPLQLI 175
Y+++NL PG+YL+D HF EIIN NGPKG+RVFNVY+Q+EK +E DI++ VG N+PL L+
Sbjct: 146 YQLNNLLPGEYLIDFHFAEIINTNGPKGIRVFNVYVQDEKA-TEFDIFSVVGANRPLLLV 204
Query: 176 DCRATVKDDGVILIRFESLNGRPIVSGICIRRASKESVPPVPSDFIECNYCAAQIEIPSS 235
D R V DDG+I +RFE +NG P+V GIC+R+A + SVP DFI+C CA +IEI +
Sbjct: 205 DLRVMVMDDGLIRVRFEGINGSPVVCGICLRKAPQVSVPRTSQDFIKCENCATEIEISPT 264
Query: 236 QIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSF 295
+ ++M+ K+ KYE KI EL+ + E K EC+EAW SLT + ++EKV MEL+ +++
Sbjct: 265 RKRLMRAKAHDKYEKKIAELSERYEHKTNECHEAWMSLTSANEQLEKVMMELNNKIYQAR 324
Query: 296 TTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDSIP 355
+ + T QA+ L+SI+ +YE DK+ WA AI SLQEK+++MK EQS+LS +AHECV+ IP
Sbjct: 325 SLDQTVITQADCLKSITRKYENDKRHWATAIDSLQEKIEIMKREQSQLSQEAHECVEGIP 384
Query: 356 ELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSS 415
EL KMV VQ LV QCEDLK KY EE +RK+L+N +QE KGNIRVFCRCRPLN E S+
Sbjct: 385 ELYKMVGGVQALVSQCEDLKQKYSEEQAKRKELYNHIQETKGNIRVFCRCRPLNTEETST 444
Query: 416 GCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNV 475
T+VDFD AKDG LG++ +SKK F+FDRVYTPKD QVDVFADAS MV+SVLDGYNV
Sbjct: 445 KSATIVDFDGAKDGELGVITGNNSKKSFKFDRVYTPKDGQVDVFADASPMVVSVLDGYNV 504
Query: 476 CIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQI 535
CIFAYGQTGTGKTFTMEGT QNRGVNYRT+E LF V++ER ET SY+ISVSVLEVYNEQI
Sbjct: 505 CIFAYGQTGTGKTFTMEGTPQNRGVNYRTVEQLFEVARERRETISYNISVSVLEVYNEQI 564
Query: 536 RDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEH 595
RDLLAT P SK+LEIKQ+ +G HHVPG+VEA V+NI++VW VLQAGSNAR+VGSNNVNEH
Sbjct: 565 RDLLATSPGSKKLEIKQSSDGSHHVPGLVEANVENINEVWNVLQAGSNARSVGSNNVNEH 624
Query: 596 SSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSA 655
SSRSHCML IMVK KNLMNG+CTKSKLWLVDL+GSERLAKTDVQGERLKEAQNINRSLSA
Sbjct: 625 SSRSHCMLSIMVKAKNLMNGDCTKSKLWLVDLAGSERLAKTDVQGERLKEAQNINRSLSA 684
Query: 656 LGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFAT 715
LGDVI ALA KSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPS+ DV ETLSSLNFAT
Sbjct: 685 LGDVIYALATKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSEHDVSETLSSLNFAT 744
Query: 716 RVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNI 775
RVRGVEL P +KQ+DTGE+QK KAM++KAR E R K+ES++K+EE++QN+E K KG+DN
Sbjct: 745 RVRGVELGPARKQVDTGEIQKLKAMVEKARQESRSKDESIKKMEENIQNLEGKNKGRDNS 804
Query: 776 HKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEETCCTLQHKVKELERKIKEQ 835
+++LQEK K+L+ Q+ S+ NQSEKQ +QL ERLK ++E C LQ KVKELE K++E+
Sbjct: 805 YRSLQEKNKDLQNQL---DSVHNQSEKQYAQLQERLKSRDEICSNLQQKVKELECKLRER 861
Query: 836 LQTETANFQQKVCNTFAKLLPTLFSNSLFLPKVWDLEKKLKDQLQGSESESSFLKDKV 893
Q+++A QKV + L + S+ ++ KV D E KLK+ SE S + K+
Sbjct: 862 HQSDSAANNQKVKDLENNLKESEGSSLVWQQKVKDYENKLKE----SEGNSLVWQQKI 915
>gb|AAD15569.1| putative kinesin heavy chain [Arabidopsis thaliana]
gi|25412125|pir||F84614 probable kinesin heavy chain
[imported] - Arabidopsis thaliana
Length = 1068
Score = 999 bits (2584), Expect = 0.0
Identities = 525/898 (58%), Positives = 659/898 (72%), Gaps = 57/898 (6%)
Query: 1 MEDANDSLFDSMLVDS---SSKLI--QNGFARSQSSGLKINIMY*FVKLSVHF*RIHFEA 55
++D +D+L DSM DS S +LI ++GF + ++ I Y
Sbjct: 45 IDDCDDALGDSM--DSGFYSFRLILGEDGFNLMLTRRVRFLIQY---------------- 86
Query: 56 EECVMFVNVGGEATNEGADGVKFLSDTFFDGGDVFLTNEAIVEGGDYPSIYQSARVGSFS 115
E +MF+N GG+ + + D +F+GGDV T E+IVE GD+P IYQSARVG+F
Sbjct: 87 -ETIMFINAGGDDSKVLDSELNISRDDYFEGGDVLRTEESIVEAGDFPFIYQSARVGNFC 145
Query: 116 YRIDNLPPGQYLVDLHFVEIINVNGPKGMRVFNVYIQEEKVLSELDIYAAVGVNKPLQLI 175
Y+++NL PG+YL+D HF EIIN NGPKG+RVFNVY+Q+EK +E DI++ VG N+PL L+
Sbjct: 146 YQLNNLLPGEYLIDFHFAEIINTNGPKGIRVFNVYVQDEKA-TEFDIFSVVGANRPLLLV 204
Query: 176 DCRATVKDDGVILIRFESLNGRPIVSGICIRRASKESVPPVPSDFIECNYCAAQIEIPSS 235
D R V DDG+I +RFE +NG P+V GIC+R+A + SVP DFI+C CA +IEI +
Sbjct: 205 DLRVMVMDDGLIRVRFEGINGSPVVCGICLRKAPQVSVPRTSQDFIKCENCATEIEISPT 264
Query: 236 QIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSF 295
+ ++M+ K+ KYE KI EL+ + E K EC+EAW SLT + ++EKV MEL+ +++
Sbjct: 265 RKRLMRAKAHDKYEKKIAELSERYEHKTNECHEAWMSLTSANEQLEKVMMELNNKIYQAR 324
Query: 296 TTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDSIP 355
+ + T QA+ L+SI+ +YE DK+ WA AI SLQEK+++MK EQS+LS +AHECV+ IP
Sbjct: 325 SLDQTVITQADCLKSITRKYENDKRHWATAIDSLQEKIEIMKREQSQLSQEAHECVEGIP 384
Query: 356 ELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSS 415
EL KMV VQ LV QCEDLK KY EE +RK+L+N +QE KGNIRVFCRCRPLN E S+
Sbjct: 385 ELYKMVGGVQALVSQCEDLKQKYSEEQAKRKELYNHIQETKGNIRVFCRCRPLNTEETST 444
Query: 416 GCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNV 475
T+VDFD AKDG LG++ +SKK F+FDRVYTPKD QVDVFADAS MV+SVLDGYNV
Sbjct: 445 KSATIVDFDGAKDGELGVITGNNSKKSFKFDRVYTPKDGQVDVFADASPMVVSVLDGYNV 504
Query: 476 CIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQI 535
CIFAYGQTGTGKTFTMEGT QNRGVNYRT+E LF V++ER ET SY+ISVSVLEVYNEQI
Sbjct: 505 CIFAYGQTGTGKTFTMEGTPQNRGVNYRTVEQLFEVARERRETISYNISVSVLEVYNEQI 564
Query: 536 RDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEH 595
RDLLAT P SK+LEIKQ+ +G HHVPG+VEA V+NI++VW VLQAGSNAR+VGSNNVNEH
Sbjct: 565 RDLLATSPGSKKLEIKQSSDGSHHVPGLVEANVENINEVWNVLQAGSNARSVGSNNVNEH 624
Query: 596 SSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSA 655
SSRSHCML IMVK KNLMNG+CTKSKLWLVDL+GSERLAKTDVQGERLKEAQNINRSLSA
Sbjct: 625 SSRSHCMLSIMVKAKNLMNGDCTKSKLWLVDLAGSERLAKTDVQGERLKEAQNINRSLSA 684
Query: 656 LGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFAT 715
LGDVI ALA KSSHIPY SPS+ DV ETLSSLNFAT
Sbjct: 685 LGDVIYALATKSSHIPY-------------------------SPSEHDVSETLSSLNFAT 719
Query: 716 RVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNI 775
RVRGVEL P +KQ+DTGE+QK KAM++KAR E R K+ES++K+EE++QN+E K KG+DN
Sbjct: 720 RVRGVELGPARKQVDTGEIQKLKAMVEKARQESRSKDESIKKMEENIQNLEGKNKGRDNS 779
Query: 776 HKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEETCCTLQHKVKELERKIKEQ 835
+++LQEK K+L+ Q+ S+ NQSEKQ +QL ERLK ++E C LQ KVKELE K++E+
Sbjct: 780 YRSLQEKNKDLQNQLD---SVHNQSEKQYAQLQERLKSRDEICSNLQQKVKELECKLRER 836
Query: 836 LQTETANFQQKVCNTFAKLLPTLFSNSLFLPKVWDLEKKLKDQLQGSESESSFLKDKV 893
Q+++A QKV + L + S+ ++ KV D E KLK+ SE S + K+
Sbjct: 837 HQSDSAANNQKVKDLENNLKESEGSSLVWQQKVKDYENKLKE----SEGNSLVWQQKI 890
>gb|AAO17019.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 1045
Score = 929 bits (2400), Expect = 0.0
Identities = 480/832 (57%), Positives = 607/832 (72%), Gaps = 50/832 (6%)
Query: 55 AEECVMFVNVGGEATNEGADGVKFLSDTFFDGGDVFLTNEAIVEGGDYPSIYQSARVGSF 114
A++ V+F+N GG AT K D+FF+GGD T+E IVEGGDYPS+Y SAR G+F
Sbjct: 59 ADDSVLFINAGGSATEGCEPSSKLSEDSFFEGGDAIETSEDIVEGGDYPSLYHSARYGNF 118
Query: 115 SYRIDNLPPGQYLVDLHFVEIINVNGPKGMRVFNVYIQEEK------VLSELDIYAAVGV 168
SY+ID L PG Y +DLHF EI+N GPKG+R F+V +QEEK +LSELD+YA VG
Sbjct: 119 SYKIDGLAPGDYFLDLHFAEIVNTYGPKGIRAFDVLVQEEKANTLTHILSELDVYAVVGG 178
Query: 169 NKPLQLIDCRATVKDDGVILIRFESLNGRPIVSGICIRRASKESVPPVPSDF-IECNYCA 227
N+PLQ+ D R TV+ D I+I F+ + G P+V GICIR+ +V + ++ + C C+
Sbjct: 179 NRPLQVRDIRVTVESDSAIVINFKGVRGSPMVCGICIRKRVAMAVTDMVTEGNVLCKRCS 238
Query: 228 AQIEIPSSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMEL 287
A +S ++ +K +KYE +I+ELT QC +K+ ECY AW+S+ ++E+E++++EL
Sbjct: 239 AHTG--NSPLQTRTSKLISKYEKQIEELTNQCNMKSDECYMAWSSVESTNQELERLKIEL 296
Query: 288 DQVTFKSFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKA 347
Q +S E ++QA+ LRS+S +YE KK WA AI +L+ K++ MK EQ+ LS +A
Sbjct: 297 HQKVMQSDNIEQVVDRQADQLRSVSQKYENAKKLWAAAISNLENKIKAMKQEQTLLSLEA 356
Query: 348 HECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRP 407
H+C +++P+L+KM+ AVQ LV QCEDLK+KYYEEM +RKKL N V+E KGNIRVFCRCRP
Sbjct: 357 HDCANAVPDLSKMIGAVQTLVAQCEDLKLKYYEEMAKRKKLHNIVEETKGNIRVFCRCRP 416
Query: 408 LNKVEMSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVI 467
L+K E SSG VDFD AKDG + I+ G++KK F+FDRVY P D+Q DV+ADAS +V
Sbjct: 417 LSKDETSSGYKCAVDFDGAKDGDIAIVNGGAAKKTFKFDRVYMPTDNQADVYADASPLVT 476
Query: 468 SVLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSV 527
SVLDGYNVCIFAYGQTGTGKTFTMEGTE+NRGVNYRTLE LF++++ER ET +Y ISVSV
Sbjct: 477 SVLDGYNVCIFAYGQTGTGKTFTMEGTERNRGVNYRTLEELFKIAEERKETVTYSISVSV 536
Query: 528 LEVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAV 587
LEVYNEQIRDLLA+ P+SK+LEIKQ EG HHVPG+VEAKV+NI +VW VLQAGSNARAV
Sbjct: 537 LEVYNEQIRDLLASSPSSKKLEIKQASEGSHHVPGIVEAKVENIKEVWDVLQAGSNARAV 596
Query: 588 GSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQ 647
GSNNVNEHSSRSHCMLCIMV+ +NLMNGECT+SKLWLVDL+GSERLAKTDVQGERLKEAQ
Sbjct: 597 GSNNVNEHSSRSHCMLCIMVRAENLMNGECTRSKLWLVDLAGSERLAKTDVQGERLKEAQ 656
Query: 648 NINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGET 707
NINRSLSALGDVISALA K+SHIPYRNSKLTHLLQDSL
Sbjct: 657 NINRSLSALGDVISALATKNSHIPYRNSKLTHLLQDSL---------------------- 694
Query: 708 LSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIES 767
EL P KKQ+DT ELQK K ML++A+ + R K++SLRKLE++ QN+E+
Sbjct: 695 -------------ELGPAKKQVDTAELQKVKQMLERAKQDIRLKDDSLRKLEDNCQNLEN 741
Query: 768 KAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEETCCTLQHKV-K 826
KAKGK+ +KNLQEK+KELE Q+ K Q SEKQ ++L +LK KEE C TLQ K+ +
Sbjct: 742 KAKGKEQFYKNLQEKVKELESQLDSKMHSQITSEKQQNELFGKLKEKEEMCTTLQQKIAE 801
Query: 827 ELERKIKEQLQTETANFQQKVCNTFAKLLPTLFSNSLFLPKVWDLEKKLKDQ 878
E E K++ Q Q+E+ ++ KL S+ K+ +LE KLK+Q
Sbjct: 802 ESEHKLRLQQQSES-----EIKELELKLKEQEHHRSVAESKIKELELKLKEQ 848
>ref|NP_177370.1| kinesin motor protein-related [Arabidopsis thaliana]
gi|12323661|gb|AAG51794.1| kinesin, putative;
56847-62063 [Arabidopsis thaliana]
gi|25406160|pir||B96746 probable kinesin T9N14.6
[imported] - Arabidopsis thaliana
Length = 1195
Score = 655 bits (1689), Expect = 0.0
Identities = 367/780 (47%), Positives = 493/780 (63%), Gaps = 68/780 (8%)
Query: 59 VMFVNVGGEATNEGADGVKFLSDTFFDGGDVFLTNEAIVEGGDYPSIYQSARVGSFSYRI 118
V+ +N G +T+ + V FL D FF GG+ +T +A+V D +YQ+AR+G+F+Y+
Sbjct: 172 VISINSGSISTDVTVEDVTFLKDEFFSGGES-ITTDAVVGNEDEILLYQTARLGNFAYKF 230
Query: 119 DNLPPGQYLVDLHFVEIINVNGPKGMRVFNVYIQEEKVLSELDIYAAVGVNKPLQLIDCR 178
+L PG Y +DLHF EI GP G V+S LD+++ VG N PL + D R
Sbjct: 231 QSLDPGDYFIDLHFAEIEFTKGPPG------------VISGLDLFSQVGANTPLVIEDLR 278
Query: 179 ATVKDDGVILIRFESLNGRPIVSGICIRR-------------ASKESVPPVPSDFIECNY 225
V +G + IR E + G I+ GI IR+ A K S V S + N
Sbjct: 279 MLVGREGELSIRLEGVTGAAILCGISIRKETTATYVEETGMLAVKGSTDTVLSQQTQENL 338
Query: 226 -CAAQIEIPSSQIKV-MQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKV 283
C A+ E + Q K + ++EL ++ + K +EC EA SL+E+ E+ +
Sbjct: 339 VCRAEEEAEGMRSDCEQQRKEMEDMKRMVEELKLENQQKTRECEEALNSLSEIQNELMRK 398
Query: 284 QMELDQVTFKSFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRL 343
M + S T E+ ++ + E+++ K E + + VQ
Sbjct: 399 SMHVG-----SLGTSQREEQMVLFIKRFDKKIEVEQIKLLEEATTYKHLVQ--------- 444
Query: 344 SFKAHECVDSIPELNKMVYAVQELVKQC----EDLKVKYYEEMTQRKKLFNEVQEAKGNI 399
++N+ +Q VKQ E+LKVK+ +RK+L+N++ E KGNI
Sbjct: 445 ------------DINEFSSHIQSRVKQDAELHENLKVKFVAGEKERKELYNKILELKGNI 492
Query: 400 RVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVF 459
RVFCRCRPLN E +G + +D ++ K+G + +++ G KK F+FD V+ P Q DVF
Sbjct: 493 RVFCRCRPLNFEETEAGVSMGIDVESTKNGEVIVMSNGFPKKSFKFDSVFGPNASQADVF 552
Query: 460 ADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETF 519
D + SV+DGYNVCIFAYGQTGTGKTFTMEGT+ +RGVNYRTLE+LFR+ K R +
Sbjct: 553 EDTAPFATSVIDGYNVCIFAYGQTGTGKTFTMEGTQHDRGVNYRTLENLFRIIKAREHRY 612
Query: 520 SYDISVSVLEVYNEQIRDLLA----TGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVW 575
+Y+ISVSVLEVYNEQIRDLL + A KR EI+Q EG+HHVPG+VEA V +I +VW
Sbjct: 613 NYEISVSVLEVYNEQIRDLLVPASQSASAPKRFEIRQLSEGNHHVPGLVEAPVKSIEEVW 672
Query: 576 TVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAK 635
VL+ GSNARAVG NEHSSRSHC+ C+MVK +NL+NGECTKSKLWLVDL+GSER+AK
Sbjct: 673 DVLKTGSNARAVGKTTANEHSSRSHCIHCVMVKGENLLNGECTKSKLWLVDLAGSERVAK 732
Query: 636 TDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFV 695
T+VQGERLKE QNIN+SLSALGDVI ALA KSSHIP+RNSKLTHLLQDSLGGDSKTLMFV
Sbjct: 733 TEVQGERLKETQNINKSLSALGDVIFALANKSSHIPFRNSKLTHLLQDSLGGDSKTLMFV 792
Query: 696 QISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESL 755
QISP++ D ETL SLNFA+RVRG+EL P KKQ+D EL K K M++K + + + K+E +
Sbjct: 793 QISPNENDQSETLCSLNFASRVRGIELGPAKKQLDNTELLKYKQMVEKWKQDMKGKDEQI 852
Query: 756 RKLEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQ------SEKQVSQLCE 809
RK+EE++ +E+K K +D +K LQ+K+KELE Q+ ++ + Q +E+Q Q E
Sbjct: 853 RKMEETMYGLEAKIKERDTKNKTLQDKVKELESQLLVERKLARQHVDTKIAEQQTKQQTE 912
>ref|NP_198107.1| kinesin motor protein-related [Arabidopsis thaliana]
Length = 425
Score = 489 bits (1259), Expect = e-136
Identities = 245/415 (59%), Positives = 320/415 (77%), Gaps = 6/415 (1%)
Query: 374 LKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGI 433
L+ +Y EE ++RK+L+NEV E KGNIRVFCRCRPLN+ E+++GC +V +FD ++ L I
Sbjct: 15 LEKQYLEESSERKRLYNEVIELKGNIRVFCRCRPLNQAEIANGCASVAEFDTTQENELQI 74
Query: 434 LATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG 493
L++ SSKK F+FD V+ P D Q VFA +V SVLDGYNVCIFAYGQTGTGKTFTMEG
Sbjct: 75 LSSDSSKKHFKFDHVFKPDDGQETVFAQTKPIVTSVLDGYNVCIFAYGQTGTGKTFTMEG 134
Query: 494 TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLA--TGPASKRLEIK 551
T +NRGVNYRTLE LFR S+ +S +++SVS+LEVYNE+IRDLL + K+LE+K
Sbjct: 135 TPENRGVNYRTLEELFRCSESKSHLMKFELSVSMLEVYNEKIRDLLVDNSNQPPKKLEVK 194
Query: 552 QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKN 611
Q+ EG VPG+VEA+V N VW +L+ G R+VGS NE SSRSHC+L + VK +N
Sbjct: 195 QSAEGTQEVPGLVEAQVYNTDGVWDLLKKGYAVRSVGSTAANEQSSRSHCLLRVTVKGEN 254
Query: 612 LMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIP 671
L+NG+ T+S LWLVDL+GSER+ K +V+GERLKE+Q IN+SLSALGDVISALA+K+SHIP
Sbjct: 255 LINGQRTRSHLWLVDLAGSERVGKVEVEGERLKESQFINKSLSALGDVISALASKTSHIP 314
Query: 672 YRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDT 731
YRNSKLTH+LQ+SLGGD KTLMFVQISPS D+GETL SLNFA+RVRG+E P +KQ D
Sbjct: 315 YRNSKLTHMLQNSLGGDCKTLMFVQISPSSADLGETLCSLNFASRVRGIESGPARKQADV 374
Query: 732 GELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNIHKNLQEKIKEL 786
EL K+K M +K + E E+ +KL++++Q+++ + +++I + LQ+K+ L
Sbjct: 375 SELLKSKQMAEKLKHE----EKETKKLQDNVQSLQLRLTAREHICRGLQDKVSPL 425
>ref|XP_475205.1| putative kinesin-related protein [Oryza sativa (japonica
cultivar-group)] gi|51854417|gb|AAU10796.1| putative
kinesin [Oryza sativa (japonica cultivar-group)]
gi|46981329|gb|AAT07647.1| putative kinesin-related
protein [Oryza sativa (japonica cultivar-group)]
Length = 840
Score = 449 bits (1156), Expect = e-124
Identities = 233/455 (51%), Positives = 323/455 (70%), Gaps = 9/455 (1%)
Query: 365 QELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFD 424
+E + + LK KY +E +R++L+NE+ E +GNIRVFCRCRPL+ E+S+GC+++V D
Sbjct: 144 EECAPRYDGLKKKYADECAERRRLYNELIELRGNIRVFCRCRPLSTAEISNGCSSIVQID 203
Query: 425 AAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTG 484
+ + L + + +K F+FD V+ P D+Q VFA++ +V SV+DG+NVCIFAYGQTG
Sbjct: 204 PSHETELQFVPSDKDRKAFKFDHVFGPSDNQETVFAESLPVVRSVMDGFNVCIFAYGQTG 263
Query: 485 TGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLL--ATG 542
TGKTFTMEG ++RGVNYR LE LFR+S+ERS + +Y +VS+LEVYNE+IRDLL ++
Sbjct: 264 TGKTFTMEGIPEDRGVNYRALEELFRLSEERSSSVAYTFAVSILEVYNEKIRDLLDESSE 323
Query: 543 PASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCM 602
++L+IKQ +G V G++EA + I VW L+ G+ R+VG+ + NE SSRSH +
Sbjct: 324 QTGRKLDIKQTADGTQEVAGLIEAPIYTIDGVWEKLKVGAKNRSVGATSANELSSRSHSL 383
Query: 603 LCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISA 662
+ + V++++L+ G+ +S +WLVDL+GSER+ KT+V+G+RLKE+Q IN+SLSALGDVISA
Sbjct: 384 VKVTVRSEHLVTGQKWRSHIWLVDLAGSERVNKTEVEGDRLKESQFINKSLSALGDVISA 443
Query: 663 LAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVEL 722
LA+K++HIPYRNSKLTHLLQ SLGGD KTLMFVQISPS D GETL SLNFA+RVR ++
Sbjct: 444 LASKNAHIPYRNSKLTHLLQSSLGGDCKTLMFVQISPSSADSGETLCSLNFASRVRAIDH 503
Query: 723 DPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNIHKNLQEK 782
P +KQ D E K K M +K R E E+ KL ESLQ + K ++N+ K LQEK
Sbjct: 504 GPARKQADPAETFKLKQMTEKIRHE----EKENAKLLESLQLTQLKYASRENVIKTLQEK 559
Query: 783 IKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEET 817
I+E E K + Q + + ++L K +T
Sbjct: 560 IREAEQTSK---TYQQRVRELENELANEKKAARDT 591
>gb|AAO42115.1| putative kinesin [Arabidopsis thaliana]
Length = 983
Score = 348 bits (894), Expect = 3e-94
Identities = 213/555 (38%), Positives = 323/555 (57%), Gaps = 31/555 (5%)
Query: 237 IKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSFT 296
I+ + +K ++EN++ T Q EL E+ +S S + E ++ +FK+
Sbjct: 250 IESLLSKVVEEFENRV---TNQYELVRAAPRESTSSQNNRSFLKPLGEREREEKSFKAI- 305
Query: 297 TELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKV----QLMKSEQSRLSFKAHECVD 352
+K N + + + + + K QE + Q + + ++ + F + +
Sbjct: 306 -----KKDDHNSQILDEKMKTRQFKQLTIFNQQQEDIEGLRQTLYTTRAGMQFMQKKFQE 360
Query: 353 SIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVE 412
L V+ + Y+ + + +KL+N+VQ+ KG+IRV+CR RP +
Sbjct: 361 EFSSLGMHVHGLAHAASG-------YHRVLEENRKLYNQVQDLKGSIRVYCRVRPFLPGQ 413
Query: 413 MSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
SS +T+ + + G G S K F F++V+ P Q +VF+D ++ SVLDG
Sbjct: 414 -SSFSSTIGNMEDDTIGINTASRHGKSLKSFTFNKVFGPSATQEEVFSDMQPLIRSVLDG 472
Query: 473 YNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVL 528
YNVCIFAYGQTG+GKTFTM G TE+++GVNYR L LF ++++R +TF YDI+V ++
Sbjct: 473 YNVCIFAYGQTGSGKTFTMSGPRDLTEKSQGVNYRALGDLFLLAEQRKDTFRYDIAVQMI 532
Query: 529 EVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVG 588
E+YNEQ+RDLL T ++KRLEI+ + + VP V + DV +++ G RAVG
Sbjct: 533 EIYNEQVRDLLVTDGSNKRLEIRNSSQKGLSVPDASLVPVSSTFDVIDLMKTGHKNRAVG 592
Query: 589 SNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQN 648
S +N+ SSRSH L + V+ ++L +G + + LVDL+GSER+ K++V G+RLKEAQ+
Sbjct: 593 STALNDRSSRSHSCLTVHVQGRDLTSGAVLRGCMHLVDLAGSERVDKSEVTGDRLKEAQH 652
Query: 649 INRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETL 708
INRSLSALGDVI++LA K+ H+PYRNSKLT LLQDSLGG +KTLMFV ISP VGET+
Sbjct: 653 INRSLSALGDVIASLAHKNPHVPYRNSKLTQLLQDSLGGQAKTLMFVHISPEADAVGETI 712
Query: 709 SSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDK-----ARSECRCKEESLRKLEESLQ 763
S+L FA RV VEL + DT ++++ K + AR E ++ ++ K +
Sbjct: 713 STLKFAERVATVELGAARVNNDTSDVKELKEQIATLKAALARKEAESQQNNILKTPGGSE 772
Query: 764 NIESKAKGKDNIHKN 778
++K G+ IH N
Sbjct: 773 KYKAKT-GEVEIHNN 786
>gb|AAG52083.1| kinesin-related protein; 103921-99132 [Arabidopsis thaliana]
Length = 1050
Score = 339 bits (869), Expect = 3e-91
Identities = 236/682 (34%), Positives = 366/682 (53%), Gaps = 64/682 (9%)
Query: 238 KVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQ------------- 284
KV+ T A E ++KEL E KE + A +L E +++++++
Sbjct: 366 KVVNTAKNA-LEERVKEL----EQMGKEAHSAKNALEEKIKQLQQMEKETKTANTSLEGK 420
Query: 285 -MELDQ--VTFKSFTTELTAEKQAENLRSISNRYELDKKKW----AEAIISLQEKVQLMK 337
EL+Q V +K+ E+ EK++E+ ++ EL K + ++A++ L+ + +K
Sbjct: 421 IQELEQNLVMWKTKVREM--EKKSESNHQRWSQKELSYKSFIDNQSQALLELRSYSRSIK 478
Query: 338 SEQSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKG 397
E ++ D +L K + EL E+ Y+ +T+ +KLFNE+QE KG
Sbjct: 479 QEILKVQ---ENYTDQFSQLGKKLI---ELSNAAEN----YHAVLTENRKLFNELQELKG 528
Query: 398 NIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGIL-ATGSSK---KLFRFDRVYTPKD 453
NIRVFCR RP + TVV++ +DG L + T K + F+F++VY+P
Sbjct: 529 NIRVFCRVRPF--LPAQGAANTVVEY-VGEDGELVVTNPTRPGKDGLRQFKFNKVYSPTA 585
Query: 454 DQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLF 509
Q DVF+D +V SVLDGYNVCIFAYGQTG+GKT+TM G +E++ GVNYR L LF
Sbjct: 586 SQADVFSDIRPLVRSVLDGYNVCIFAYGQTGSGKTYTMTGPDGSSEEDWGVNYRALNDLF 645
Query: 510 RVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQN-------YEGHHHVPG 562
++S+ R SY++ V ++E+YNEQ+ DLL+ + K+ N + VP
Sbjct: 646 KISQSRKGNISYEVGVQMVEIYNEQVLDLLSDDNSQKKYPFVLNPGILSTTQQNGLAVPD 705
Query: 563 VVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKL 622
V + SDV T++ G RAVGS +NE SSRSH ++ + V+ K+L G L
Sbjct: 706 ASMYPVTSTSDVITLMDIGLQNRAVGSTALNERSSRSHSIVTVHVRGKDLKTGSVLYGNL 765
Query: 623 WLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQ 682
LVDL+GSER+ +++V G+RL+EAQ+IN+SLS+LGDVI +LA+KSSH+PYRNSKLT LLQ
Sbjct: 766 HLVDLAGSERVDRSEVTGDRLREAQHINKSLSSLGDVIFSLASKSSHVPYRNSKLTQLLQ 825
Query: 683 DSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLD 742
SLGG +KTLMFVQ++P E++S+L FA RV GVEL K + +++ L
Sbjct: 826 TSLGGRAKTLMFVQLNPDATSYSESMSTLKFAERVSGVELGAAKTSKEGKDVRDLMEQLA 885
Query: 743 KARSECRCKEESLRKLEESLQNIESKAKGK------DNIHKNLQEKIKELEGQIKLKTSM 796
+ K+E + +L+ Q ++ + D+I+ + E + + S+
Sbjct: 886 SLKDTIARKDEEIERLQHQPQRLQKSMMRRKSIGHTDDINSDTGEYSSQSRYSVTDGESL 945
Query: 797 QNQSEKQVSQLCERLKGKEETCCTLQHKVKELER--KIKEQLQTETANFQQKVCNTFAKL 854
+ +E + + + + T Q + +R +I ++ ++ TA V KL
Sbjct: 946 ASSAEAEYDERLSEITSDAASMGT-QGSIDVTKRPPRISDRAKSVTAKSSTSVTRPLDKL 1004
Query: 855 LPTLFSNSLFLPKVWDLEKKLK 876
+ + KV L K
Sbjct: 1005 RKVATRTTSTVAKVTGLTSSSK 1026
Score = 42.7 bits (99), Expect = 0.053
Identities = 27/95 (28%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query: 758 LEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEET 817
LE L+ +E + K + L+E++KELE K S +N E+++ QL + K +
Sbjct: 354 LESRLKELEQEGKVVNTAKNALEERVKELEQMGKEAHSAKNALEEKIKQLQQMEKETKTA 413
Query: 818 CCTLQHKVKELER-------KIKEQLQTETANFQQ 845
+L+ K++ELE+ K++E + +N Q+
Sbjct: 414 NTSLEGKIQELEQNLVMWKTKVREMEKKSESNHQR 448
>ref|NP_177527.2| kinesin motor protein-related [Arabidopsis thaliana]
Length = 1030
Score = 339 bits (869), Expect = 3e-91
Identities = 236/682 (34%), Positives = 366/682 (53%), Gaps = 64/682 (9%)
Query: 238 KVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQ------------- 284
KV+ T A E ++KEL E KE + A +L E +++++++
Sbjct: 346 KVVNTAKNA-LEERVKEL----EQMGKEAHSAKNALEEKIKQLQQMEKETKTANTSLEGK 400
Query: 285 -MELDQ--VTFKSFTTELTAEKQAENLRSISNRYELDKKKW----AEAIISLQEKVQLMK 337
EL+Q V +K+ E+ EK++E+ ++ EL K + ++A++ L+ + +K
Sbjct: 401 IQELEQNLVMWKTKVREM--EKKSESNHQRWSQKELSYKSFIDNQSQALLELRSYSRSIK 458
Query: 338 SEQSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKG 397
E ++ D +L K + EL E+ Y+ +T+ +KLFNE+QE KG
Sbjct: 459 QEILKVQ---ENYTDQFSQLGKKLI---ELSNAAEN----YHAVLTENRKLFNELQELKG 508
Query: 398 NIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGIL-ATGSSK---KLFRFDRVYTPKD 453
NIRVFCR RP + TVV++ +DG L + T K + F+F++VY+P
Sbjct: 509 NIRVFCRVRPF--LPAQGAANTVVEY-VGEDGELVVTNPTRPGKDGLRQFKFNKVYSPTA 565
Query: 454 DQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLF 509
Q DVF+D +V SVLDGYNVCIFAYGQTG+GKT+TM G +E++ GVNYR L LF
Sbjct: 566 SQADVFSDIRPLVRSVLDGYNVCIFAYGQTGSGKTYTMTGPDGSSEEDWGVNYRALNDLF 625
Query: 510 RVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQN-------YEGHHHVPG 562
++S+ R SY++ V ++E+YNEQ+ DLL+ + K+ N + VP
Sbjct: 626 KISQSRKGNISYEVGVQMVEIYNEQVLDLLSDDNSQKKYPFVLNPGILSTTQQNGLAVPD 685
Query: 563 VVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKL 622
V + SDV T++ G RAVGS +NE SSRSH ++ + V+ K+L G L
Sbjct: 686 ASMYPVTSTSDVITLMDIGLQNRAVGSTALNERSSRSHSIVTVHVRGKDLKTGSVLYGNL 745
Query: 623 WLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQ 682
LVDL+GSER+ +++V G+RL+EAQ+IN+SLS+LGDVI +LA+KSSH+PYRNSKLT LLQ
Sbjct: 746 HLVDLAGSERVDRSEVTGDRLREAQHINKSLSSLGDVIFSLASKSSHVPYRNSKLTQLLQ 805
Query: 683 DSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLD 742
SLGG +KTLMFVQ++P E++S+L FA RV GVEL K + +++ L
Sbjct: 806 TSLGGRAKTLMFVQLNPDATSYSESMSTLKFAERVSGVELGAAKTSKEGKDVRDLMEQLA 865
Query: 743 KARSECRCKEESLRKLEESLQNIESKAKGK------DNIHKNLQEKIKELEGQIKLKTSM 796
+ K+E + +L+ Q ++ + D+I+ + E + + S+
Sbjct: 866 SLKDTIARKDEEIERLQHQPQRLQKSMMRRKSIGHTDDINSDTGEYSSQSRYSVTDGESL 925
Query: 797 QNQSEKQVSQLCERLKGKEETCCTLQHKVKELER--KIKEQLQTETANFQQKVCNTFAKL 854
+ +E + + + + T Q + +R +I ++ ++ TA V KL
Sbjct: 926 ASSAEAEYDERLSEITSDAASMGT-QGSIDVTKRPPRISDRAKSVTAKSSTSVTRPLDKL 984
Query: 855 LPTLFSNSLFLPKVWDLEKKLK 876
+ + KV L K
Sbjct: 985 RKVATRTTSTVAKVTGLTSSSK 1006
Score = 42.7 bits (99), Expect = 0.053
Identities = 27/95 (28%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query: 758 LEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEET 817
LE L+ +E + K + L+E++KELE K S +N E+++ QL + K +
Sbjct: 334 LESRLKELEQEGKVVNTAKNALEERVKELEQMGKEAHSAKNALEEKIKQLQQMEKETKTA 393
Query: 818 CCTLQHKVKELER-------KIKEQLQTETANFQQ 845
+L+ K++ELE+ K++E + +N Q+
Sbjct: 394 NTSLEGKIQELEQNLVMWKTKVREMEKKSESNHQR 428
>gb|AAG52533.1| putative kinesin; 97201-101676 [Arabidopsis thaliana]
gi|25406299|pir||B96766 protein kinesin F2P9.27
[imported] - Arabidopsis thaliana
Length = 987
Score = 338 bits (868), Expect = 4e-91
Identities = 219/573 (38%), Positives = 329/573 (57%), Gaps = 55/573 (9%)
Query: 238 KVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQ------------- 284
KV+ T A E ++KEL E KE + A +L E +++++++
Sbjct: 336 KVVNTAKNA-LEERVKEL----EQMGKEAHSAKNALEEKIKQLQQMEKETKTANTSLEGK 390
Query: 285 -MELDQ--VTFKSFTTELTAEKQAENLRSISNRYELDKKKW----AEAIISLQEKVQLMK 337
EL+Q V +K+ E+ EK++E+ ++ EL K + ++A++ L+ + +K
Sbjct: 391 IQELEQNLVMWKTKVREM--EKKSESNHQRWSQKELSYKSFIDNQSQALLELRSYSRSIK 448
Query: 338 SEQSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKG 397
E ++ D +L K + EL E+ Y+ +T+ +KLFNE+QE KG
Sbjct: 449 QEILKVQ---ENYTDQFSQLGKKLI---ELSNAAEN----YHAVLTENRKLFNELQELKG 498
Query: 398 NIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGIL-ATGSSK---KLFRFDRVYTPKD 453
NIRVFCR RP + TVV++ +DG L + T K + F+F++VY+P
Sbjct: 499 NIRVFCRVRPF--LPAQGAANTVVEY-VGEDGELVVTNPTRPGKDGLRQFKFNKVYSPTA 555
Query: 454 DQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLF 509
Q DVF+D +V SVLDGYNVCIFAYGQTG+GKT+TM G +E++ GVNYR L LF
Sbjct: 556 SQADVFSDIRPLVRSVLDGYNVCIFAYGQTGSGKTYTMTGPDGSSEEDWGVNYRALNDLF 615
Query: 510 RVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQN-------YEGHHHVPG 562
++S+ R SY++ V ++E+YNEQ+ DLL+ + K+ N + VP
Sbjct: 616 KISQSRKGNISYEVGVQMVEIYNEQVLDLLSDDNSQKKYPFVLNPGILSTTQQNGLAVPD 675
Query: 563 VVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKL 622
V + SDV T++ G RAVGS +NE SSRSH ++ + V+ K+L G L
Sbjct: 676 ASMYPVTSTSDVITLMDIGLQNRAVGSTALNERSSRSHSIVTVHVRGKDLKTGSVLYGNL 735
Query: 623 WLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQ 682
LVDL+GSER+ +++V G+RL+EAQ+IN+SLS+LGDVI +LA+KSSH+PYRNSKLT LLQ
Sbjct: 736 HLVDLAGSERVDRSEVTGDRLREAQHINKSLSSLGDVIFSLASKSSHVPYRNSKLTQLLQ 795
Query: 683 DSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLD 742
SLGG +KTLMFVQ++P E++S+L FA RV GVEL K + +++ L
Sbjct: 796 TSLGGRAKTLMFVQLNPDATSYSESMSTLKFAERVSGVELGAAKTSKEGKDVRDLMEQLA 855
Query: 743 KARSECRCKEESLRKLEESLQNIESKAKGKDNI 775
+ K+E + +L+ Q ++ + +I
Sbjct: 856 SLKDTIARKDEEIERLQHQPQRLQKSMMRRKSI 888
Score = 42.7 bits (99), Expect = 0.053
Identities = 27/95 (28%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query: 758 LEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEET 817
LE L+ +E + K + L+E++KELE K S +N E+++ QL + K +
Sbjct: 324 LESRLKELEQEGKVVNTAKNALEERVKELEQMGKEAHSAKNALEEKIKQLQQMEKETKTA 383
Query: 818 CCTLQHKVKELER-------KIKEQLQTETANFQQ 845
+L+ K++ELE+ K++E + +N Q+
Sbjct: 384 NTSLEGKIQELEQNLVMWKTKVREMEKKSESNHQR 418
>ref|NP_568491.1| kinesin motor protein-related [Arabidopsis thaliana]
gi|34921410|sp|O81635|ATK4_ARATH Kinesin-4 (Kinesin-like
protein D)
Length = 987
Score = 338 bits (868), Expect = 4e-91
Identities = 184/372 (49%), Positives = 247/372 (65%), Gaps = 11/372 (2%)
Query: 378 YYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATG 437
Y + + +KL+N VQ+ KGNIRV+CR RP + S G + V D D +G + I
Sbjct: 374 YKRVLEENRKLYNLVQDLKGNIRVYCRVRPFLPGQESGGLSAVEDID---EGTITIRVPS 430
Query: 438 ----SSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG 493
+ +K F F++V+ P Q +VF+D +V SVLDGYNVCIFAYGQTG+GKTFTM G
Sbjct: 431 KYGKAGQKPFMFNKVFGPSATQEEVFSDMQPLVRSVLDGYNVCIFAYGQTGSGKTFTMTG 490
Query: 494 ----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLE 549
TE++ GVNYR L LF +S +R +T SY+ISV +LE+YNEQ+RDLLA +KRLE
Sbjct: 491 PKELTEESLGVNYRALADLFLLSNQRKDTTSYEISVQMLEIYNEQVRDLLAQDGQTKRLE 550
Query: 550 IKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKT 609
I+ N +VP V + DV ++ G RAV S +N+ SSRSH + + V+
Sbjct: 551 IRNNSHNGINVPEASLVPVSSTDDVIQLMDLGHMNRAVSSTAMNDRSSRSHSCVTVHVQG 610
Query: 610 KNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSH 669
++L +G + LVDL+GSER+ K++V G+RLKEAQ+IN+SLSALGDVIS+L+ K+SH
Sbjct: 611 RDLTSGSILHGSMHLVDLAGSERVDKSEVTGDRLKEAQHINKSLSALGDVISSLSQKTSH 670
Query: 670 IPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQI 729
+PYRNSKLT LLQDSLGG +KTLMFV ISP +GET+S+L FA RV VEL +
Sbjct: 671 VPYRNSKLTQLLQDSLGGSAKTLMFVHISPEPDTLGETISTLKFAERVGSVELGAARVNK 730
Query: 730 DTGELQKTKAML 741
D E+++ K +
Sbjct: 731 DNSEVKELKEQI 742
>gb|AAC32191.1| kinesin-like heavy chain [Arabidopsis thaliana]
Length = 987
Score = 338 bits (868), Expect = 4e-91
Identities = 184/372 (49%), Positives = 247/372 (65%), Gaps = 11/372 (2%)
Query: 378 YYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATG 437
Y + + +KL+N VQ+ KGNIRV+CR RP + S G + V D D +G + I
Sbjct: 374 YKRVLEENRKLYNLVQDLKGNIRVYCRVRPFLPGQESGGLSAVEDID---EGTITIRVPS 430
Query: 438 ----SSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG 493
+ +K F F++V+ P Q +VF+D +V SVLDGYNVCIFAYGQTG+GKTFTM G
Sbjct: 431 KYGKAGQKPFMFNKVFGPSATQEEVFSDMQPLVRSVLDGYNVCIFAYGQTGSGKTFTMTG 490
Query: 494 ----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLE 549
TE++ GVNYR L LF +S +R +T SY+ISV +LE+YNEQ+RDLLA +KRLE
Sbjct: 491 PKELTEESLGVNYRALADLFLLSNQRKDTTSYEISVQMLEIYNEQVRDLLAQDGQTKRLE 550
Query: 550 IKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKT 609
I+ N +VP V + DV ++ G RAV S +N+ SSRSH + + V+
Sbjct: 551 IRNNSHNGINVPEASLVPVSSTDDVIQLMDLGHMNRAVSSTAMNDRSSRSHSCVTVHVQG 610
Query: 610 KNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSH 669
++L +G + LVDL+GSER+ K++V G+RLKEAQ+IN+SLSALGDVIS+L+ K+SH
Sbjct: 611 RDLTSGSILHGSMHLVDLAGSERVDKSEVTGDRLKEAQHINKSLSALGDVISSLSQKTSH 670
Query: 670 IPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQI 729
+PYRNSKLT LLQDSLGG +KTLMFV ISP +GET+S+L FA RV VEL +
Sbjct: 671 VPYRNSKLTQLLQDSLGGSAKTLMFVHISPEPDTLGETISTLKFAERVGSVELGAARVNK 730
Query: 730 DTGELQKTKAML 741
D E+++ K +
Sbjct: 731 DNSEVKELKEQI 742
>gb|AAW03152.1| kinesin [Gossypium hirsutum]
Length = 1018
Score = 338 bits (866), Expect = 6e-91
Identities = 221/551 (40%), Positives = 318/551 (57%), Gaps = 27/551 (4%)
Query: 229 QIEIPSSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELD 288
Q EIP ++ M K + +YE ++ T + K+ E+ +SR +E+D
Sbjct: 244 QDEIPMI-VESMIKKVSEEYERRLASHTELIKSSPKDTEESVPD-NSLSRTASCGNVEVD 301
Query: 289 QVTFKSFTTELTAEKQAENLRSI-SNRYELDKKKW---AEAIISLQEKVQLMKSEQSRLS 344
E AE+ ++ SI S + EL ++ EA L ++ L++ ++ L
Sbjct: 302 ------IEVEAPAEEPIDDESSIESEKKELPNEECNTDEEATRHLLKQKTLVEQQRQHLL 355
Query: 345 FKAHE--CVDSIPELNKMVYAVQ--ELVKQCEDLK---VKYYEEMTQRKKLFNEVQEAKG 397
H EL ++ Y + L K + + Y + + +KL+N+VQ+ KG
Sbjct: 356 ELKHSLHATKVGMELLQVTYREEFNNLGKHLHSIAYAAMGYQRVLEENRKLYNQVQDLKG 415
Query: 398 NIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATGSS-KKLFRFDRVYTPKDDQV 456
+IRV+CR RP + S+ + V D L TG +K F F+++++P Q
Sbjct: 416 SIRVYCRVRP-SLSGQSNNLSCVEHIDDTTITVLTPTKTGKEGRKSFTFNKIFSPSVTQA 474
Query: 457 DVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLFRVS 512
+VF+D ++ SVLDGYNVCIFAYGQTG+GKT+TM G TE+ GVNYR L LF +S
Sbjct: 475 EVFSDTQPLIRSVLDGYNVCIFAYGQTGSGKTYTMSGPTELTEEGLGVNYRALGDLFELS 534
Query: 513 KERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNIS 572
+R ET SY+ISV +LE+YNEQ+RDLLA+ +KRLEI+ + + +VP +V + S
Sbjct: 535 NQRKETISYEISVQMLEIYNEQVRDLLASDGLNKRLEIRNSSQNGINVPEAHLVRVSSTS 594
Query: 573 DVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSER 632
DV ++ G R V S +N+ SSRSH L + V+ K+L +G + LVDL+GSER
Sbjct: 595 DVINLMNLGQKNRTVFSTAMNDRSSRSHSCLTVHVQGKDLTSGNIIHGCMHLVDLAGSER 654
Query: 633 LAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTL 692
+ K++V G+RLKEAQ IN+SLSALGDVI+ALA+K SH+PYR SKLT LLQDSLGG +K L
Sbjct: 655 VDKSEVMGDRLKEAQYINKSLSALGDVIAALASKGSHVPYRTSKLTQLLQDSLGGQAKPL 714
Query: 693 MFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLD--KARSECRC 750
MFV I+P + GET+S+L FA RV VEL K D+GE+++ K + KA
Sbjct: 715 MFVHIAPEYEASGETISTLKFAERVATVELGAAKVNKDSGEVKELKGQISSLKAALTTAK 774
Query: 751 KEESLRKLEES 761
KE +L+ S
Sbjct: 775 KEGEPEQLQRS 785
>ref|XP_544385.1| PREDICTED: similar to Kifc3 protein [Canis familiaris]
Length = 927
Score = 337 bits (864), Expect = 1e-90
Identities = 206/547 (37%), Positives = 320/547 (57%), Gaps = 25/547 (4%)
Query: 207 RASKESVPPVPSDFIECNYCAAQIEIPSSQIKVMQTKSTAKYENKIKELTMQCELKAKEC 266
R S E++ + + Y +E+ SS+ K ++S A+ ++ +++ MQ ++ KE
Sbjct: 332 RDSHETIASLRAQSPPIKYVIKTVEVESSKTKQALSESQARNQHLQEQVAMQRQV-LKEM 390
Query: 267 YEAWTSLTEMSREVEKVQMELDQVTFKSFTTELTAEKQA---ENLRSISN---RYELDKK 320
+ S +++ ++ + Q+ + + + ++ E Q+ + R+I R +++ K
Sbjct: 391 EQQLQSSHQLTAQL-RAQIAMYESELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMK 449
Query: 321 KWAEAI-------ISLQEKVQLMKSEQSRLSFKAHECVDSIPELNKMV-----YAVQELV 368
E + ++LQ ++ + ++ + L + + E K V A++E+
Sbjct: 450 AVHENLAGVRTNLLTLQPALRTLTNDYNGLKRQVRGFPLLLQEALKSVKAEIGQAIEEVN 509
Query: 369 KQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSS-GCTTVVDFDAAK 427
++L KY E+ RKK NE+ KGNIRV R RP+ K + T V FDA
Sbjct: 510 SNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDGEGPDATNAVTFDADD 569
Query: 428 DGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGK 487
D + +L G F D+V++P+ Q DVF + +++ S +DG+NVCIFAYGQTG GK
Sbjct: 570 DSIIHLLHKGKPVS-FELDKVFSPRASQQDVFQEVQALITSCIDGFNVCIFAYGQTGAGK 628
Query: 488 TFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKR 547
T+TMEGT +N G+N R L+ LF +E++ + Y I+VS E+YNE +RDLL P K
Sbjct: 629 TYTMEGTPENPGINQRALQLLFSEVQEKASDWEYTITVSAAEIYNEVLRDLLGQEPQEK- 687
Query: 548 LEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCI 605
LEI+ + G +VPG+ E +V +++D+ V + G R N+NEHSSRSH +L +
Sbjct: 688 LEIRLCPDGSGQLYVPGLTEFQVQSVADINKVFEFGHTNRTTEFTNLNEHSSRSHALLIV 747
Query: 606 MVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAA 665
V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN+SLSALGDVI+AL +
Sbjct: 748 TVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGSRLREAQHINKSLSALGDVIAALRS 807
Query: 666 KSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPV 725
+ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL SL FA RVR VEL P
Sbjct: 808 RQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYSLKFAERVRSVELGPG 867
Query: 726 KKQIDTG 732
++ + G
Sbjct: 868 SRRAELG 874
>gb|AAH16118.1| Kifc3 protein [Mus musculus]
Length = 608
Score = 335 bits (859), Expect = 4e-90
Identities = 202/502 (40%), Positives = 289/502 (57%), Gaps = 27/502 (5%)
Query: 234 SSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFK 293
S Q+ V A YE +++ Q + + E E + +V+M+
Sbjct: 85 SHQLTVQLRAQIAMYEAELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMKAVHENLA 144
Query: 294 SFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDS 353
T L + A LR+++N Y K++ + LQE ++ +K+E +
Sbjct: 145 GVRTNLLTLQPA--LRTLTNDYNGLKRQVRGFPLLLQEALRSVKAEIGQ----------- 191
Query: 354 IPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEM 413
A++E+ ++L KY E+ RKK NE+ KGNIRV R RP+ K +
Sbjct: 192 ---------AIEEVNSNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDG 242
Query: 414 SSG-CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
T V FD D + +L G F D+V++P Q DVF + +++ S +DG
Sbjct: 243 EGPEATNAVTFDPDDDSIIHLLHKGKPVS-FELDKVFSPWASQQDVFQEVQALITSCIDG 301
Query: 473 YNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
+NVCIFAYGQTG GKT+TMEGT +N G+N R L+ LF +E++ + Y+I+VS E+YN
Sbjct: 302 FNVCIFAYGQTGAGKTYTMEGTPENPGINQRALQLLFSEVQEKASDWQYNITVSAAEIYN 361
Query: 533 EQIRDLLATGPASKRLEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
E +RDLL P K LEI+ + G +VPG+ E +V ++ D+ V + G N R
Sbjct: 362 EVLRDLLGKEPQEK-LEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGYNNRTTEFT 420
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
N+NEHSSRSH +L + V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN
Sbjct: 421 NLNEHSSRSHALLIVTVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGNRLREAQHIN 480
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
RSLSALGDVI+AL ++ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL S
Sbjct: 481 RSLSALGDVIAALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYS 540
Query: 711 LNFATRVRGVELDPVKKQIDTG 732
L FA RVR VEL P ++ + G
Sbjct: 541 LRFAERVRSVELGPGSRRTELG 562
>gb|AAH70429.1| Kifc3 protein [Mus musculus]
Length = 793
Score = 335 bits (859), Expect = 4e-90
Identities = 202/502 (40%), Positives = 289/502 (57%), Gaps = 27/502 (5%)
Query: 234 SSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFK 293
S Q+ V A YE +++ Q + + E E + +V+M+
Sbjct: 270 SHQLTVQLRAQIAMYEAELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMKAVHENLA 329
Query: 294 SFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDS 353
T L + A LR+++N Y K++ + LQE ++ +K+E +
Sbjct: 330 GVRTNLLTLQPA--LRTLTNDYNGLKRQVRGFPLLLQEALRSVKAEIGQ----------- 376
Query: 354 IPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEM 413
A++E+ ++L KY E+ RKK NE+ KGNIRV R RP+ K +
Sbjct: 377 ---------AIEEVNSNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDG 427
Query: 414 SSG-CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
T V FD D + +L G F D+V++P Q DVF + +++ S +DG
Sbjct: 428 EGPEATNAVTFDPDDDSIIHLLHKGKPVS-FELDKVFSPWASQQDVFQEVQALITSCIDG 486
Query: 473 YNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
+NVCIFAYGQTG GKT+TMEGT +N G+N R L+ LF +E++ + Y+I+VS E+YN
Sbjct: 487 FNVCIFAYGQTGAGKTYTMEGTPENPGINQRALQLLFSEVQEKASDWQYNITVSAAEIYN 546
Query: 533 EQIRDLLATGPASKRLEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
E +RDLL P K LEI+ + G +VPG+ E +V ++ D+ V + G N R
Sbjct: 547 EVLRDLLGKEPQEK-LEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGYNNRTTEFT 605
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
N+NEHSSRSH +L + V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN
Sbjct: 606 NLNEHSSRSHALLIVTVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGNRLREAQHIN 665
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
RSLSALGDVI+AL ++ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL S
Sbjct: 666 RSLSALGDVIAALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYS 725
Query: 711 LNFATRVRGVELDPVKKQIDTG 732
L FA RVR VEL P ++ + G
Sbjct: 726 LRFAERVRSVELGPGSRRTELG 747
>sp|O35231|KIFC3_MOUSE Kinesin-like protein KIFC3
Length = 687
Score = 335 bits (859), Expect = 4e-90
Identities = 202/502 (40%), Positives = 289/502 (57%), Gaps = 27/502 (5%)
Query: 234 SSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFK 293
S Q+ V A YE +++ Q + + E E + +V+M+
Sbjct: 164 SHQLTVQLRAQIAMYEAELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMKAVHENLA 223
Query: 294 SFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDS 353
T L + A LR+++N Y K++ + LQE ++ +K+E +
Sbjct: 224 GVRTNLLTLQPA--LRTLTNDYNGLKRQVRGFPLLLQEALRSVKAEIGQ----------- 270
Query: 354 IPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEM 413
A++E+ ++L KY E+ RKK NE+ KGNIRV R RP+ K +
Sbjct: 271 ---------AIEEVNSNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDG 321
Query: 414 SSG-CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
T V FD D + +L G F D+V++P Q DVF + +++ S +DG
Sbjct: 322 EGPEATNAVTFDPDDDSIIHLLHKGKPVS-FELDKVFSPWASQQDVFQEVQALITSCIDG 380
Query: 473 YNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
+NVCIFAYGQTG GKT+TMEGT +N G+N R L+ LF +E++ + Y+I+VS E+YN
Sbjct: 381 FNVCIFAYGQTGAGKTYTMEGTPENPGINQRALQLLFSEVQEKASDWQYNITVSAAEIYN 440
Query: 533 EQIRDLLATGPASKRLEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
E +RDLL P K LEI+ + G +VPG+ E +V ++ D+ V + G N R
Sbjct: 441 EVLRDLLGKEPQEK-LEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGYNNRTTEFT 499
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
N+NEHSSRSH +L + V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN
Sbjct: 500 NLNEHSSRSHALLIVTVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGNRLREAQHIN 559
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
RSLSALGDVI+AL ++ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL S
Sbjct: 560 RSLSALGDVIAALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYS 619
Query: 711 LNFATRVRGVELDPVKKQIDTG 732
L FA RVR VEL P ++ + G
Sbjct: 620 LRFAERVRSVELGPGSRRTELG 641
>ref|XP_240978.3| PREDICTED: similar to Kifc3 protein [Rattus norvegicus]
Length = 824
Score = 335 bits (858), Expect = 5e-90
Identities = 201/502 (40%), Positives = 289/502 (57%), Gaps = 27/502 (5%)
Query: 234 SSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFK 293
S Q+ + A YE +++ Q + + E E + +V+M+
Sbjct: 301 SHQLTIQLRAQIAMYEAELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMKAVHENLA 360
Query: 294 SFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDS 353
T L + A LR+++N Y K++ + LQE ++ +K+E +
Sbjct: 361 GVRTNLLTLQPA--LRTLTNDYNGLKRQVRGFPLLLQEALRSVKAEIGQ----------- 407
Query: 354 IPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEM 413
A++E+ ++L KY E+ RKK NE+ KGNIRV R RP+ K +
Sbjct: 408 ---------AIEEVNSNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDG 458
Query: 414 SSG-CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
T V FD D + +L G F D+V++P Q DVF + +++ S +DG
Sbjct: 459 EGPEATNAVTFDPDDDSIIHLLHKGKPVS-FELDKVFSPWASQQDVFQEVQALITSCIDG 517
Query: 473 YNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
+NVCIFAYGQTG GKT+TMEGT +N G+N R L+ LF +E++ + Y+I+VS E+YN
Sbjct: 518 FNVCIFAYGQTGAGKTYTMEGTPENPGINQRALQLLFSEVQEKASDWQYNITVSAAEIYN 577
Query: 533 EQIRDLLATGPASKRLEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
E +RDLL P K LEI+ + G +VPG+ E +V ++ D+ V + G N R
Sbjct: 578 EVLRDLLGKEPQEK-LEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGYNNRTTEFT 636
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
N+NEHSSRSH +L + V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN
Sbjct: 637 NLNEHSSRSHALLIVTVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGNRLREAQHIN 696
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
RSLSALGDVI+AL ++ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL S
Sbjct: 697 RSLSALGDVIAALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYS 756
Query: 711 LNFATRVRGVELDPVKKQIDTG 732
L FA RVR VEL P ++ + G
Sbjct: 757 LKFAERVRSVELGPGSRRTELG 778
>ref|NP_034761.1| kinesin family member C3 [Mus musculus] gi|12585614|gb|AAC39967.2|
kinesin motor protein KIFC3 [Mus musculus]
Length = 709
Score = 335 bits (858), Expect = 5e-90
Identities = 202/502 (40%), Positives = 289/502 (57%), Gaps = 27/502 (5%)
Query: 234 SSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFK 293
S Q+ V A YE +++ Q + + E E + +V+M+
Sbjct: 186 SHQLTVQLRAQIAMYEAELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMKAVHENLA 245
Query: 294 SFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDS 353
T L + A LR+++N Y K++ + LQE ++ +K+E +
Sbjct: 246 GVRTNLLTLQPA--LRTLTNDYNGLKRQVRGFPLLLQEALRSVKAEIGQ----------- 292
Query: 354 IPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEM 413
A++E+ ++L KY E+ RKK NE+ KGNIRV R RP+ K +
Sbjct: 293 ---------AIEEVNSNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDG 343
Query: 414 SSG-CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
T V FD D + +L G F D+V++P Q DVF + +++ S +DG
Sbjct: 344 EGPEATNAVTFDPDDDSIIHLLHKGKPVS-FELDKVFSPWASQQDVFQEVQALITSCIDG 402
Query: 473 YNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
+NVCIFAYGQTG GKT+TMEGT +N G+N R L+ LF +E++ + Y+I+VS E+YN
Sbjct: 403 FNVCIFAYGQTGAGKTYTMEGTPENPGINQRALQLLFSEVQEKASDWQYNITVSAREIYN 462
Query: 533 EQIRDLLATGPASKRLEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
E +RDLL P K LEI+ + G +VPG+ E +V ++ D+ V + G N R
Sbjct: 463 EVLRDLLGKEPQEK-LEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGYNNRTTEFT 521
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
N+NEHSSRSH +L + V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN
Sbjct: 522 NLNEHSSRSHALLIVTVRGVDCRTGLRTTGKLNLVDLAGSERVGKSGAEGNRLREAQHIN 581
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
RSLSALGDVI+AL ++ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL S
Sbjct: 582 RSLSALGDVIAALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYS 641
Query: 711 LNFATRVRGVELDPVKKQIDTG 732
L FA RVR VEL P ++ + G
Sbjct: 642 LRFAERVRSVELGPGSRRTELG 663
>gb|AAH41132.1| KIFC3 protein [Homo sapiens]
Length = 687
Score = 333 bits (855), Expect = 1e-89
Identities = 201/502 (40%), Positives = 289/502 (57%), Gaps = 27/502 (5%)
Query: 234 SSQIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFK 293
S Q+ A YE++++ Q + + E E + +V+M+
Sbjct: 164 SHQLTARLRAQIAMYESELERAHGQMLEEMQSLEEDKNRAIEEAFARAQVEMKAVHENLA 223
Query: 294 SFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDS 353
T L + A LR+++N Y K++ + LQE ++ +K+E +
Sbjct: 224 GVRTNLLTLQPA--LRTLTNDYNGLKRQVRVFPLLLQEALRSVKAEIGQ----------- 270
Query: 354 IPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEM 413
A++E+ ++L KY E+ RKK NE+ KGNIRV R RP+ K +
Sbjct: 271 ---------AIEEVNSNNQELLRKYRRELQLRKKCHNELVRLKGNIRVIARVRPVTKEDG 321
Query: 414 SSG-CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
T V FDA D + +L G F D+V++P+ Q DVF + ++V S +DG
Sbjct: 322 EGPEATNAVTFDADDDSIIHLLHKGKPVS-FELDKVFSPQASQQDVFQEVQALVTSCIDG 380
Query: 473 YNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
+NVCIFAYGQTG GKT+TMEGT +N G+N R L+ LF +E++ + Y I+VS E+YN
Sbjct: 381 FNVCIFAYGQTGAGKTYTMEGTAENPGINQRALQLLFSEVQEKASDWEYTITVSAAEIYN 440
Query: 533 EQIRDLLATGPASKRLEIK--QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
E +RDLL P K LEI+ + G +VPG+ E +V ++ D+ V + G R
Sbjct: 441 EVLRDLLGKEPQEK-LEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGHTNRTTEFT 499
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
N+NEHSSRSH +L + V+ + G T KL LVDL+GSER+ K+ +G RL+EAQ+IN
Sbjct: 500 NLNEHSSRSHALLIVTVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGSRLREAQHIN 559
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
+SLSALGDVI+AL ++ H+P+RNSKLT+LLQDSL GDSKTLM VQ+SP +++ ETL S
Sbjct: 560 KSLSALGDVIAALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVVQVSPVEKNTSETLYS 619
Query: 711 LNFATRVRGVELDPVKKQIDTG 732
L FA RVR VEL P ++ + G
Sbjct: 620 LKFAERVRSVELGPGLRRAELG 641
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,361,201,654
Number of Sequences: 2540612
Number of extensions: 55454672
Number of successful extensions: 285457
Number of sequences better than 10.0: 6283
Number of HSP's better than 10.0 without gapping: 2349
Number of HSP's successfully gapped in prelim test: 4166
Number of HSP's that attempted gapping in prelim test: 243618
Number of HSP's gapped (non-prelim): 30710
length of query: 895
length of database: 863,360,394
effective HSP length: 137
effective length of query: 758
effective length of database: 515,296,550
effective search space: 390594784900
effective search space used: 390594784900
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146711.13


