

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.3 - phase: 0
(282 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
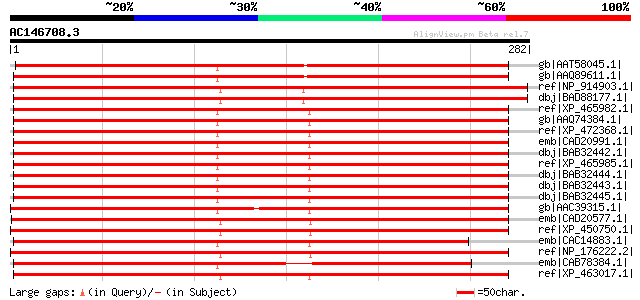
Score E
Sequences producing significant alignments: (bits) Value
gb|AAT58045.1| high-affinity K+ transporter [Capsicum annuum] 373 e-102
gb|AAQ89611.1| At4g13420 [Arabidopsis thaliana] gi|7108597|gb|AA... 367 e-100
ref|NP_914903.1| putative high-affinity potassium transporter [O... 358 1e-97
dbj|BAD88177.1| putative potassium transporter [Oryza sativa (ja... 358 1e-97
ref|XP_465982.1| putative high-affinity potassium transporter [O... 356 4e-97
gb|AAQ74384.1| KUP1 [Oryza sativa] 350 3e-95
ref|XP_472368.1| OSJNBb0012E08.7 [Oryza sativa (japonica cultiva... 350 3e-95
emb|CAD20991.1| putative potassium transporter [Oryza sativa (ja... 350 3e-95
dbj|BAB32442.1| high-affinity potassium transporter [Phragmites ... 350 3e-95
ref|XP_465985.1| putative high-affinity potassium transporter [O... 349 4e-95
dbj|BAB32444.1| high-affinity potassium transporter [Phragmites ... 349 6e-95
dbj|BAB32443.1| high-affinity potassium transporter [Phragmites ... 349 6e-95
dbj|BAB32445.1| high-affinity potassium transporter [Phragmites ... 348 1e-94
gb|AAC39315.1| putative high-affinity potassium transporter [Hor... 338 1e-91
emb|CAD20577.1| putative potassium transporter [Vicia faba] 318 8e-86
ref|XP_450750.1| putative HAK2 [Oryza sativa (japonica cultivar-... 315 1e-84
emb|CAC14883.1| putative potassium transporter [Hordeum vulgare] 313 3e-84
ref|NP_176222.2| potassium transporter family protein [Arabidops... 313 3e-84
emb|CAB78384.1| potassium transporter-like protein [Arabidopsis ... 312 8e-84
ref|XP_463017.1| putative potassium transporter [Oryza sativa (j... 311 2e-83
>gb|AAT58045.1| high-affinity K+ transporter [Capsicum annuum]
Length = 804
Score = 373 bits (958), Expect = e-102
Identities = 184/272 (67%), Positives = 225/272 (82%), Gaps = 5/272 (1%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFIV 63
L+LAFQS+GV+YGDIGTSPLYV ASTF I H DDILGVLSLI YTI+ +P+ KYVFIV
Sbjct: 60 LSLAFQSVGVIYGDIGTSPLYVFASTFTDKIGHKDDILGVLSLIIYTIILVPMTKYVFIV 119
Query: 64 LKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKLE 119
L AN+NG+GG FALYSLLCR+A VSLIPNQ+PED ELS+Y L+ PS++ Q+++ LE
Sbjct: 120 LWANNNGDGGAFALYSLLCRYAKVSLIPNQEPEDRELSHYSLDIPSNHIRRAQRIRHSLE 179
Query: 120 NSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDYVVSITIAILVILF 179
S FA+ L+F+ ILGT+MVIGDGV TP +SV+SAV+GI LGQ+ VV I++AILV LF
Sbjct: 180 KSKFAKFFLVFLAILGTSMVIGDGVLTPCISVLSAVSGIKP-LGQEAVVGISVAILVALF 238
Query: 180 CAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGKNA 239
CAQRFGT KVG++FAP + IWF+ I G+YN+FKYDV VL A NPKY+++YFQRNGK
Sbjct: 239 CAQRFGTDKVGYTFAPAICIWFMFISGIGLYNLFKYDVSVLRAFNPKYLINYFQRNGKKG 298
Query: 240 WMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
W+SLGGVFLCI+G EAMFADLGHF+VR+IQ+S
Sbjct: 299 WISLGGVFLCITGSEAMFADLGHFSVRSIQIS 330
>gb|AAQ89611.1| At4g13420 [Arabidopsis thaliana] gi|7108597|gb|AAF36490.1| K+
transporter HAK5 [Arabidopsis thaliana]
gi|38503206|sp|Q9M7K4|POT5_ARATH Potassium transporter 5
(AtPOT5) (AtHAK1) (AtHAK5) gi|18414004|ref|NP_567404.1|
potassium transporter (HAK5) [Arabidopsis thaliana]
Length = 785
Score = 367 bits (943), Expect = e-100
Identities = 177/273 (64%), Positives = 226/273 (81%), Gaps = 5/273 (1%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
T++LAFQSLGVVYGDIGTSPLYV ASTF GI+ DD++GVLSLI YTI + LLKYVFI
Sbjct: 58 TMSLAFQSLGVVYGDIGTSPLYVYASTFTDGINDKDDVVGVLSLIIYTITLVALLKYVFI 117
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL+ANDNG GGTFALYSL+CR+A + LIPNQ+PED+ELSNY LE P++ +K+KL
Sbjct: 118 VLQANDNGEGGTFALYSLICRYAKMGLIPNQEPEDVELSNYTLELPTTQLRRAHMIKEKL 177
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDYVVSITIAILVIL 178
ENS FA+++L +TI+GT+MVIGDG+ TP +SV+SAV+GI S LGQ+ VV +++AIL++L
Sbjct: 178 ENSKFAKIILFLVTIMGTSMVIGDGILTPSISVLSAVSGIKS-LGQNTVVGVSVAILIVL 236
Query: 179 FCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGKN 238
F QRFGT KVGFSFAPI+ +WF + G++N+FK+D+ VL A+NP YI+ YF+R G+
Sbjct: 237 FAFQRFGTDKVGFSFAPIILVWFTFLIGIGLFNLFKHDITVLKALNPLYIIYYFRRTGRQ 296
Query: 239 AWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
W+SLGGVFLCI+G EAMFADLGHF+VRA+Q+S
Sbjct: 297 GWISLGGVFLCITGTEAMFADLGHFSVRAVQIS 329
>ref|NP_914903.1| putative high-affinity potassium transporter [Oryza sativa
(japonica cultivar-group)]
Length = 745
Score = 358 bits (918), Expect = 1e-97
Identities = 180/286 (62%), Positives = 222/286 (76%), Gaps = 7/286 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+GVVYGD+GTSPLYV +STF GI T+DILGV+SLI YT++ LPL+KY FI
Sbjct: 61 TLHLAFQSIGVVYGDMGTSPLYVFSSTFTNGIKDTNDILGVMSLIIYTVVLLPLIKYCFI 120
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LKKKL 118
VL+ANDNG+GGTFALYSL+ R+A +SLIPNQQ ED +S+YKLE+PS+ + +K+K+
Sbjct: 121 VLRANDNGDGGTFALYSLISRYARISLIPNQQAEDAMVSHYKLESPSNRVKRAHWIKEKM 180
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGI---SSKLGQDYVVSITIAIL 175
ENS +++L +TIL T+MVIGDGV TP +SV+SAV GI + L Q + I IAIL
Sbjct: 181 ENSPNFKIILFLVTILATSMVIGDGVLTPCISVLSAVGGIKESAKSLTQGQIAGIAIAIL 240
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
++LF QRFGT KVG+SF PI+ WFI I TG+YN+FK+D VL A NPKYIVDYF+RN
Sbjct: 241 IVLFLVQRFGTDKVGYSFGPIILTWFIFIAGTGVYNLFKHDTGVLKAFNPKYIVDYFERN 300
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSIIKLELTFVL 281
GK W+SLGGV LCI+G EAMFADLGHFNVRAIQ+ + L VL
Sbjct: 301 GKQGWISLGGVILCITGTEAMFADLGHFNVRAIQIGFSVVLLPSVL 346
>dbj|BAD88177.1| putative potassium transporter [Oryza sativa (japonica
cultivar-group)] gi|57899652|dbj|BAD87321.1| putative
potassium transporter [Oryza sativa (japonica
cultivar-group)]
Length = 770
Score = 358 bits (918), Expect = 1e-97
Identities = 180/286 (62%), Positives = 222/286 (76%), Gaps = 7/286 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+GVVYGD+GTSPLYV +STF GI T+DILGV+SLI YT++ LPL+KY FI
Sbjct: 61 TLHLAFQSIGVVYGDMGTSPLYVFSSTFTNGIKDTNDILGVMSLIIYTVVLLPLIKYCFI 120
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LKKKL 118
VL+ANDNG+GGTFALYSL+ R+A +SLIPNQQ ED +S+YKLE+PS+ + +K+K+
Sbjct: 121 VLRANDNGDGGTFALYSLISRYARISLIPNQQAEDAMVSHYKLESPSNRVKRAHWIKEKM 180
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGI---SSKLGQDYVVSITIAIL 175
ENS +++L +TIL T+MVIGDGV TP +SV+SAV GI + L Q + I IAIL
Sbjct: 181 ENSPNFKIILFLVTILATSMVIGDGVLTPCISVLSAVGGIKESAKSLTQGQIAGIAIAIL 240
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
++LF QRFGT KVG+SF PI+ WFI I TG+YN+FK+D VL A NPKYIVDYF+RN
Sbjct: 241 IVLFLVQRFGTDKVGYSFGPIILTWFIFIAGTGVYNLFKHDTGVLKAFNPKYIVDYFERN 300
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSIIKLELTFVL 281
GK W+SLGGV LCI+G EAMFADLGHFNVRAIQ+ + L VL
Sbjct: 301 GKQGWISLGGVILCITGTEAMFADLGHFNVRAIQIGFSVVLLPSVL 346
>ref|XP_465982.1| putative high-affinity potassium transporter [Oryza sativa
(japonica cultivar-group)] gi|49387769|dbj|BAD26327.1|
putative high-affinity potassium transporter [Oryza
sativa (japonica cultivar-group)]
Length = 742
Score = 356 bits (914), Expect = 4e-97
Identities = 172/276 (62%), Positives = 222/276 (80%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+GVVYGD+GTSPLYV +STFP GI H DD++GVLSLI YT++ +P++KYVFI
Sbjct: 44 TLQLAFQSIGVVYGDVGTSPLYVYSSTFPNGIKHPDDLVGVLSLILYTLILIPMVKYVFI 103
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ RHA + +IPN Q ED +SNY +E PSS + +K+KL
Sbjct: 104 VLYANDNGDGGTFALYSLISRHAKIRMIPNDQTEDANVSNYSIEAPSSQLRRAEWVKQKL 163
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S+ A++ L +TILGT+MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 164 ESSNAAKIALFTITILGTSMVMGDGTLTPAISVLSAVSGIREKAPNLTQSQVVWISVAIL 223
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAP++++WF+LI G+YN+ +++ +L A NPKYIVDYF+RN
Sbjct: 224 FVLFSMQRFGTDKVGYTFAPVISVWFLLIAGIGMYNLTVHEITILRAFNPKYIVDYFRRN 283
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK AW+SLGGV LCI+G EAMFADLGHFN+RAIQ+S
Sbjct: 284 GKEAWVSLGGVVLCITGTEAMFADLGHFNIRAIQLS 319
>gb|AAQ74384.1| KUP1 [Oryza sativa]
Length = 801
Score = 350 bits (897), Expect = 3e-95
Identities = 169/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI H DD++GVLSLI YT++ +P+LKYVFI
Sbjct: 55 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPDGIGHRDDLVGVLSLILYTLIIIPMLKYVFI 114
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + +IPNQQ ED +SNY +E PSS Q +K KL
Sbjct: 115 VLYANDNGDGGTFALYSLISRYAKIRMIPNQQAEDAMVSNYSIEAPSSQLRRAQWVKHKL 174
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A++ L F+TILGT+MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 175 ESSRAAKMALFFLTILGTSMVMGDGTLTPAISVLSAVSGIREKAPNLTQTQVVLISVAIL 234
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAPI+++WF+LI G+YN+ +++ +L A NP YIV YF+RN
Sbjct: 235 FMLFSVQRFGTDKVGYTFAPIISVWFLLIAGIGLYNLVVHEITILKAFNPWYIVQYFRRN 294
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK W+SLGGV LC++G E MFADLGHFN+RA+Q+S
Sbjct: 295 GKKGWVSLGGVVLCVTGTEGMFADLGHFNIRAVQIS 330
>ref|XP_472368.1| OSJNBb0012E08.7 [Oryza sativa (japonica cultivar-group)]
gi|21740625|emb|CAD40783.1| OSJNBb0012E08.7 [Oryza
sativa (japonica cultivar-group)]
gi|62900231|sp|Q6VVA6|HAK1_ORYSA Potassium transporter 1
(OsHAK1)
Length = 801
Score = 350 bits (897), Expect = 3e-95
Identities = 169/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI H DD++GVLSLI YT++ +P+LKYVFI
Sbjct: 55 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPDGIGHRDDLVGVLSLILYTLIIIPMLKYVFI 114
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + +IPNQQ ED +SNY +E PSS Q +K KL
Sbjct: 115 VLYANDNGDGGTFALYSLISRYAKIRMIPNQQAEDAMVSNYSIEAPSSQLRRAQWVKHKL 174
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A++ L F+TILGT+MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 175 ESSRAAKMALFFLTILGTSMVMGDGTLTPAISVLSAVSGIREKAPNLTQTQVVLISVAIL 234
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAPI+++WF+LI G+YN+ +++ +L A NP YIV YF+RN
Sbjct: 235 FMLFSVQRFGTDKVGYTFAPIISVWFLLIAGIGLYNLVVHEITILKAFNPWYIVQYFRRN 294
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK W+SLGGV LC++G E MFADLGHFN+RA+Q+S
Sbjct: 295 GKKGWVSLGGVVLCVTGTEGMFADLGHFNIRAVQIS 330
>emb|CAD20991.1| putative potassium transporter [Oryza sativa (japonica
cultivar-group)]
Length = 767
Score = 350 bits (897), Expect = 3e-95
Identities = 169/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI H DD++GVLSLI YT++ +P+LKYVFI
Sbjct: 30 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPDGIGHRDDLVGVLSLILYTLIIIPMLKYVFI 89
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + +IPNQQ ED +SNY +E PSS Q +K KL
Sbjct: 90 VLYANDNGDGGTFALYSLISRYAKIRMIPNQQAEDAMVSNYSIEAPSSQLRRAQWVKHKL 149
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A++ L F+TILGT+MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 150 ESSRAAKMALFFLTILGTSMVMGDGTLTPAISVLSAVSGIREKAPNLTQTQVVLISVAIL 209
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAPI+++WF+LI G+YN+ +++ +L A NP YIV YF+RN
Sbjct: 210 FMLFSVQRFGTDKVGYTFAPIISVWFLLIAGIGLYNLVVHEITILKAFNPWYIVQYFRRN 269
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK W+SLGGV LC++G E MFADLGHFN+RA+Q+S
Sbjct: 270 GKKGWVSLGGVVLCVTGTEGMFADLGHFNIRAVQIS 305
>dbj|BAB32442.1| high-affinity potassium transporter [Phragmites australis]
Length = 777
Score = 350 bits (897), Expect = 3e-95
Identities = 170/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI + DD+LGVLSLI YT++ +P+LKYVFI
Sbjct: 47 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPSGIKNNDDLLGVLSLIIYTLIIIPMLKYVFI 106
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + LIPNQQ ED +SNY +E PSS Q +K+K+
Sbjct: 107 VLYANDNGDGGTFALYSLISRYAKIRLIPNQQAEDAMVSNYSIEAPSSQLRRAQWVKQKI 166
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A++ L +TILGT MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 167 ESSKAAKIALFTLTILGTAMVMGDGTLTPAISVLSAVSGIREKAPSLSQTQVVWISVAIL 226
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAP++++WF+LI G+YN+ +D+ VL A NP YIV YF+RN
Sbjct: 227 FMLFSVQRFGTDKVGYTFAPVISVWFLLIAGIGLYNLVVHDIGVLRAFNPMYIVHYFKRN 286
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK+ W+SLGGV LC++G E MFADLGHFNVRA+Q+S
Sbjct: 287 GKDGWVSLGGVILCVTGTEGMFADLGHFNVRAVQIS 322
>ref|XP_465985.1| putative high-affinity potassium transporter [Oryza sativa
(japonica cultivar-group)] gi|49387772|dbj|BAD26330.1|
putative high-affinity potassium transporter [Oryza
sativa (japonica cultivar-group)]
Length = 747
Score = 349 bits (896), Expect = 4e-95
Identities = 167/276 (60%), Positives = 219/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+GVVYGD+GTSPLYV +STFP G+ H DD++GVLSL+ YT++ +P++KYVFI
Sbjct: 45 TLQLAFQSIGVVYGDVGTSPLYVYSSTFPDGVKHPDDLVGVLSLMLYTLILIPMVKYVFI 104
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ RHA + +IPN Q ED +SNY +E PSS + +K+KL
Sbjct: 105 VLYANDNGDGGTFALYSLISRHAKIRMIPNDQTEDANVSNYSIEAPSSQLRRAEWVKQKL 164
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S+ A++ L +TILGT+MV+GDG TP +SV+SAV+GI K L Q VV I++ IL
Sbjct: 165 ESSNAAKIALFTITILGTSMVMGDGTLTPAISVLSAVSGIREKAPSLTQLQVVWISVPIL 224
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
++LF QRFGT KVG+SFAP++++WF+LI G YN+ +++ +L A NP YI+DYF+RN
Sbjct: 225 IVLFSVQRFGTDKVGYSFAPVISVWFVLIAGIGAYNLAVHEITILRAFNPMYIIDYFRRN 284
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK AW+SLGG LCI+G EAMFADLGHFN+RAIQ+S
Sbjct: 285 GKEAWVSLGGAVLCITGTEAMFADLGHFNIRAIQLS 320
>dbj|BAB32444.1| high-affinity potassium transporter [Phragmites australis]
Length = 776
Score = 349 bits (895), Expect = 6e-95
Identities = 170/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI + DD+LGVLSLI YT++ +P+LKYVFI
Sbjct: 47 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPSGIKNNDDLLGVLSLIIYTLIIIPMLKYVFI 106
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + LIPNQQ ED +SNY +E PSS Q +K+K+
Sbjct: 107 VLYANDNGDGGTFALYSLISRYAKIRLIPNQQAEDAMVSNYSIEAPSSQLRRAQWVKQKI 166
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A++ L +TILGT MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 167 ESSKAAKIALFTLTILGTAMVMGDGTLTPAISVLSAVSGIREKAPSLTQTQVVWISVAIL 226
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAP++++WF+LI G+YN+ +D+ VL A NP YIV YF+RN
Sbjct: 227 FMLFSVQRFGTDKVGYTFAPVISVWFLLIAGIGLYNLVVHDIGVLRAFNPMYIVHYFKRN 286
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK+ W+SLGGV LC++G E MFADLGHFNVRA+Q+S
Sbjct: 287 GKDGWVSLGGVILCVTGTEGMFADLGHFNVRAVQIS 322
>dbj|BAB32443.1| high-affinity potassium transporter [Phragmites australis]
Length = 777
Score = 349 bits (895), Expect = 6e-95
Identities = 170/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI + DD+LGVLSLI YT++ +P+LKYVFI
Sbjct: 47 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPSGIKNNDDLLGVLSLIIYTLIIIPMLKYVFI 106
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + LIPNQQ ED +SNY +E PSS Q +K+K+
Sbjct: 107 VLYANDNGDGGTFALYSLISRYAKIRLIPNQQAEDAMVSNYSIEAPSSQLRRAQWVKQKI 166
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A++ L +TILGT MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 167 ESSKAAKIALFTLTILGTAMVMGDGTLTPAISVLSAVSGIREKAPSLTQTQVVWISVAIL 226
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAP++++WF+LI G+YN+ +D+ VL A NP YIV YF+RN
Sbjct: 227 FMLFSVQRFGTDKVGYTFAPVISVWFLLIAGIGLYNLVVHDIGVLRAFNPMYIVHYFKRN 286
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK+ W+SLGGV LC++G E MFADLGHFNVRA+Q+S
Sbjct: 287 GKDGWVSLGGVILCVTGTEGMFADLGHFNVRAVQIS 322
>dbj|BAB32445.1| high-affinity potassium transporter [Phragmites australis]
Length = 777
Score = 348 bits (893), Expect = 1e-94
Identities = 170/276 (61%), Positives = 218/276 (78%), Gaps = 7/276 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL LAFQS+G++YGDIGTSPLYV +STFP GI + DD+LGVLSLI YT++ +P+LKYVFI
Sbjct: 47 TLHLAFQSVGIIYGDIGTSPLYVYSSTFPSGIKNNDDLLGVLSLIIYTLIIIPMLKYVFI 106
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + LIPNQQ ED +SNY +ETPSS Q +K+K+
Sbjct: 107 VLYANDNGDGGTFALYSLISRYAKIRLIPNQQAEDAMVSNYSIETPSSQLRRAQWVKQKI 166
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
+S A++ L +TILGT MV+GDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 167 VSSKAAKIALFTLTILGTAMVMGDGTLTPAISVLSAVSGIREKAPSLTQTQVVWISVAIL 226
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAP++++WF+LI G+YN+ +D+ VL A NP YIV YF+RN
Sbjct: 227 FMLFSVQRFGTDKVGYTFAPVISVWFLLIAGIGLYNLVVHDIGVLRAFNPMYIVHYFKRN 286
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
GK+ W+SLGGV LC++G E MFADLGHFNVRA+Q+S
Sbjct: 287 GKDGWVSLGGVILCVTGTEGMFADLGHFNVRAVQIS 322
>gb|AAC39315.1| putative high-affinity potassium transporter [Hordeum vulgare]
gi|7489431|pir||T04379 probable potassium transport
protein - barley
Length = 773
Score = 338 bits (867), Expect = 1e-91
Identities = 167/278 (60%), Positives = 218/278 (78%), Gaps = 9/278 (3%)
Query: 1 MATLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYV 60
M TL+LAFQS+G++YGDIGTSPLYV +STFP GI + DD+LGVLSLI YT++ +P+LKYV
Sbjct: 43 MRTLSLAFQSVGIIYGDIGTSPLYVYSSTFPDGIKNRDDLLGVLSLILYTLIIIPMLKYV 102
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKK 116
FIVL ANDNG+GGTFALYSL+ R+A + LIP+QQ ED +SNY +E P+S Q LK+
Sbjct: 103 FIVLYANDNGDGGTFALYSLISRYAKIRLIPDQQAEDAAVSNYHIEAPNSQLKRAQWLKQ 162
Query: 117 KLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIA 173
KLE+S A+++L +TI +MVIGDG TP +SV+SAV+GI K L Q VV I++A
Sbjct: 163 KLESSKAAKIVLFTLTI--PSMVIGDGTLTPAISVLSAVSGIREKAPSLTQTQVVLISVA 220
Query: 174 ILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQ 233
IL +LF QRFGT KVG++FAP++++WF+LI G+YN+ +D+ VL A NP YIV YF
Sbjct: 221 ILFMLFSVQRFGTDKVGYTFAPVISVWFLLIAGIGMYNLVVHDIGVLRAFNPMYIVQYFI 280
Query: 234 RNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
RNGK+ W+SLGG+ LC++G E MFADLGHFN+RA+Q+S
Sbjct: 281 RNGKSGWVSLGGIILCVTGTEGMFADLGHFNIRAVQLS 318
>emb|CAD20577.1| putative potassium transporter [Vicia faba]
Length = 837
Score = 318 bits (816), Expect = 8e-86
Identities = 154/278 (55%), Positives = 212/278 (75%), Gaps = 8/278 (2%)
Query: 2 ATLALAFQSLGVVYGDIGTSPLYVLASTFPK-GIDHTDDILGVLSLIYYTILALPLLKYV 60
+T+ALAFQ+LGVVYGD+GTSPLYV A F K I+ +D+LG LSL+ YTI +PL KYV
Sbjct: 91 STIALAFQTLGVVYGDMGTSPLYVFADVFSKVPINSDNDVLGALSLVMYTIALIPLAKYV 150
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LKK 116
FIVLKANDNG GGTFALYSL+CR+ANV+L+PN+Q D ++S+++L+ P+ Q +K+
Sbjct: 151 FIVLKANDNGEGGTFALYSLICRYANVNLLPNRQQADEQISSFRLKLPTPELQRALKIKE 210
Query: 117 KLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKL---GQDYVVSITIA 173
LE + + +LL + ++GT+M+IGDG+ TP +SV+SA++G+ ++ G VVSI+I
Sbjct: 211 TLEKTSILKNVLLVLVLIGTSMIIGDGILTPAISVMSAISGLQDQIDGFGTSEVVSISIV 270
Query: 174 ILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQ 233
+LV LF QRFGT+KVGF FAP+L +WF +G+ G+YN+ KYD+ V+ A+NP YI +F
Sbjct: 271 VLVALFNIQRFGTAKVGFMFAPVLALWFFSLGSIGLYNMLKYDITVVRALNPAYIYYFFN 330
Query: 234 RNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
NGK+AW +LGG LCI+G EAMFADLGHF V +IQ++
Sbjct: 331 NNGKSAWSALGGCVLCITGAEAMFADLGHFTVPSIQIA 368
>ref|XP_450750.1| putative HAK2 [Oryza sativa (japonica cultivar-group)]
gi|49389043|dbj|BAD26283.1| putative HAK2 [Oryza sativa
(japonica cultivar-group)] gi|49387698|dbj|BAD26044.1|
putative HAK2 [Oryza sativa (japonica cultivar-group)]
Length = 877
Score = 315 bits (806), Expect = 1e-84
Identities = 158/279 (56%), Positives = 209/279 (74%), Gaps = 8/279 (2%)
Query: 1 MATLALAFQSLGVVYGDIGTSPLYVLASTFPK-GIDHTDDILGVLSLIYYTILALPLLKY 59
++T+A+AFQ+LGVVYGD+GTSPLYV + F K I +ILG LSL+ YTI +P KY
Sbjct: 130 LSTVAMAFQTLGVVYGDMGTSPLYVFSDVFSKVPIKSEVEILGALSLVMYTIALIPFAKY 189
Query: 60 VFIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LK 115
VFIVLKANDNG GGTFALYSL+CR+A VSL+PNQQ D ++S+++L+ P+ + +K
Sbjct: 190 VFIVLKANDNGEGGTFALYSLICRYAKVSLLPNQQRVDEDISSFRLKLPTPELERALSVK 249
Query: 116 KKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKL---GQDYVVSITI 172
+ LE + + +LLF+ ++GT+MVIGDG+ TP MSV+SAV+G+ ++ G D VV ++I
Sbjct: 250 ESLEKNPVFKNILLFLVLMGTSMVIGDGILTPSMSVMSAVSGLQGRVPGFGTDAVVIVSI 309
Query: 173 AILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYF 232
LV+LF QRFGT KVGF FAPIL +WFI +G GIYN+ KYD+ V+ A NP YI +F
Sbjct: 310 LFLVLLFSVQRFGTGKVGFMFAPILALWFINLGTIGIYNLAKYDISVVRAFNPVYIYLFF 369
Query: 233 QRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
Q NG AW +LGG LCI+G EAMFADLGHF+V++IQV+
Sbjct: 370 QTNGIKAWSALGGCVLCITGAEAMFADLGHFSVKSIQVA 408
>emb|CAC14883.1| putative potassium transporter [Hordeum vulgare]
Length = 255
Score = 313 bits (802), Expect = 3e-84
Identities = 154/254 (60%), Positives = 201/254 (78%), Gaps = 7/254 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
TL+LAFQS+G++YGDIGTSPLYV +STFP GI + DD+LGVLSLI YT++ +P+LKYVFI
Sbjct: 2 TLSLAFQSVGIIYGDIGTSPLYVYSSTFPDGIRNRDDLLGVLSLILYTLIIIPMLKYVFI 61
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL ANDNG+GGTFALYSL+ R+A + LIP+QQ ED +SNY +E P+S Q LK+KL
Sbjct: 62 VLYANDNGDGGTFALYSLISRYAKIRLIPDQQAEDAAVSNYHIEAPNSQLKRAQWLKQKL 121
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAIL 175
E+S A+++L +TILGT+MVIGDG TP +SV+SAV+GI K L Q VV I++AIL
Sbjct: 122 ESSKAAKIVLFTLTILGTSMVIGDGTLTPAISVLSAVSGIREKAPSLTQTQVVLISVAIL 181
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
+LF QRFGT KVG++FAP++++WF+LI + G+YN+ +D+ VL A NP YIV YF RN
Sbjct: 182 FMLFSVQRFGTDKVGYTFAPVISVWFLLIASIGMYNLVVHDIGVLRAFNPMYIVQYFIRN 241
Query: 236 GKNAWMSLGGVFLC 249
GK+ W+SLGG+ LC
Sbjct: 242 GKSGWVSLGGIILC 255
>ref|NP_176222.2| potassium transporter family protein [Arabidopsis thaliana]
gi|38502862|sp|O80739|POT12_ARATH Putative potassium
transporter 12 (AtPOT12)
Length = 827
Score = 313 bits (802), Expect = 3e-84
Identities = 153/279 (54%), Positives = 207/279 (73%), Gaps = 8/279 (2%)
Query: 1 MATLALAFQSLGVVYGDIGTSPLYVLASTFPK-GIDHTDDILGVLSLIYYTILALPLLKY 59
+ TL +AFQ+LGVVYGD+GTSPLYV + F K I D+LG LSL+ YTI +PL KY
Sbjct: 86 LTTLGIAFQTLGVVYGDMGTSPLYVFSDVFSKVPIRSEVDVLGALSLVIYTIAVIPLAKY 145
Query: 60 VFIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LK 115
VF+VLKANDNG GGTFALYSL+CR+A V+ +PNQQP D ++S+++L+ P+ + +K
Sbjct: 146 VFVVLKANDNGEGGTFALYSLICRYAKVNKLPNQQPADEQISSFRLKLPTPELERALGIK 205
Query: 116 KKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKL---GQDYVVSITI 172
+ LE + + LLL + ++GT+M+IGDG+ TP MSV+SA++G+ ++ G + +V +I
Sbjct: 206 EALETKGYLKTLLLLLVLMGTSMIIGDGILTPAMSVMSAMSGLQGEVKGFGTNALVMSSI 265
Query: 173 AILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYF 232
ILV LF QRFGT KVGF FAP+L +WF +GA GIYN+ KYD V+ A+NP YIV +F
Sbjct: 266 VILVALFSIQRFGTGKVGFLFAPVLALWFFSLGAIGIYNLLKYDFTVIRALNPFYIVLFF 325
Query: 233 QRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
+N K AW +LGG LCI+G EAMFADLGHF+VR+IQ++
Sbjct: 326 NKNSKQAWSALGGCVLCITGAEAMFADLGHFSVRSIQMA 364
>emb|CAB78384.1| potassium transporter-like protein [Arabidopsis thaliana]
gi|4584547|emb|CAB40777.1| potassium transporter-like
protein [Arabidopsis thaliana]
Length = 688
Score = 312 bits (799), Expect = 8e-84
Identities = 152/253 (60%), Positives = 195/253 (76%), Gaps = 18/253 (7%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
T++LAFQSLGVVYGDIGTSPLYV ASTF GI+ DD++GVLSLI YTI + LLKYVFI
Sbjct: 30 TMSLAFQSLGVVYGDIGTSPLYVYASTFTDGINDKDDVVGVLSLIIYTITLVALLKYVFI 89
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL+ANDNG GGTFALYSL+CR+A + LIPNQ+PED+ELSNY LE P++ +K+KL
Sbjct: 90 VLQANDNGEGGTFALYSLICRYAKMGLIPNQEPEDVELSNYTLELPTTQLRRAHMIKEKL 149
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDYVVSITIAILVIL 178
ENS FA+++L +TI+GT+MVIGDG+ TP +S D VV +++AIL++L
Sbjct: 150 ENSKFAKIILFLVTIMGTSMVIGDGILTPSIS--------------DTVVGVSVAILIVL 195
Query: 179 FCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGKN 238
F QRFGT KVGFSFAPI+ +WF + G++N+FK+D+ VL A+NP YI+ YF+R G+
Sbjct: 196 FAFQRFGTDKVGFSFAPIILVWFTFLIGIGLFNLFKHDITVLKALNPLYIIYYFRRTGRQ 255
Query: 239 AWMSLGGVFLCIS 251
W+SLGGVFLCI+
Sbjct: 256 GWISLGGVFLCIT 268
>ref|XP_463017.1| putative potassium transporter [Oryza sativa (japonica
cultivar-group)] gi|38093744|gb|AAR10860.1| putative
potassium transporter [Oryza sativa (japonica
cultivar-group)]
Length = 799
Score = 311 bits (796), Expect = 2e-83
Identities = 155/277 (55%), Positives = 205/277 (73%), Gaps = 8/277 (2%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
T++LAFQ +G++YGDIGTS LYV +STF GI H DD++GVLSLI Y+ + ++K VF+
Sbjct: 54 TMSLAFQCVGILYGDIGTSSLYVYSSTFEHGIGHPDDVVGVLSLIVYSFMLFTVIKIVFV 113
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLE-TPSSNQQ----LKKK 117
L AND+G+GGTFALYSL+ RHA VSLIPN Q ED +S Y PS+ + LK+
Sbjct: 114 ALHANDHGDGGTFALYSLISRHAKVSLIPNHQAEDELISGYSSSGKPSATLRRAHWLKQL 173
Query: 118 LENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAI 174
LE S A++ L +TIL MVI D V TPP+SV+SAV G+ K L D +V IT+AI
Sbjct: 174 LEASKAAKISLFLLTILAIAMVISDAVLTPPISVLSAVGGLREKVPHLTTDQIVWITVAI 233
Query: 175 LVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQR 234
LV+LF QR+GT KVG+SFAPI+ +W +LIGATG+YN+ K+D+ VL A NPKYI+DYF+R
Sbjct: 234 LVVLFAIQRYGTDKVGYSFAPIILLWLLLIGATGLYNLIKHDISVLRAFNPKYIIDYFRR 293
Query: 235 NGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
N K W+SLG + LC +G EA+FA+LG+F++R+IQ+S
Sbjct: 294 NKKEGWVSLGSILLCFTGSEALFANLGYFSIRSIQLS 330
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.143 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,137,668
Number of Sequences: 2540612
Number of extensions: 19097380
Number of successful extensions: 57087
Number of sequences better than 10.0: 278
Number of HSP's better than 10.0 without gapping: 251
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 56009
Number of HSP's gapped (non-prelim): 433
length of query: 282
length of database: 863,360,394
effective HSP length: 126
effective length of query: 156
effective length of database: 543,243,282
effective search space: 84745951992
effective search space used: 84745951992
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146708.3


