

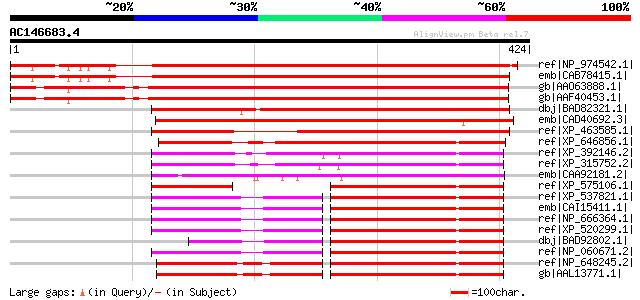
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.4 + phase: 0 /pseudo
(424 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_974542.1| RabGAP/TBC domain-containing protein [Arabidops... 508 e-142
emb|CAB78415.1| putative protein [Arabidopsis thaliana] gi|44553... 505 e-142
gb|AAO63888.1| unknown protein [Arabidopsis thaliana] gi|2839350... 477 e-133
gb|AAF40453.1| Similar to gi|3217452 F45E6.3 gene product from C... 473 e-132
dbj|BAD82321.1| RabGAP/TBC domain-containing protein-like [Oryza... 430 e-119
emb|CAD40692.3| OSJNBa0083D01.9 [Oryza sativa (japonica cultivar... 422 e-116
ref|XP_463585.1| P0497A05.14 [Oryza sativa (japonica cultivar-gr... 366 e-100
ref|XP_646856.1| hypothetical protein DDB0216614 [Dictyostelium ... 260 5e-68
ref|XP_392146.2| PREDICTED: similar to CG5978-PA [Apis mellifera] 214 4e-54
ref|XP_315752.2| ENSANGP00000018717 [Anopheles gambiae str. PEST... 213 7e-54
emb|CAA92181.2| Hypothetical protein F45E6.3 [Caenorhabditis ele... 196 9e-49
ref|XP_575106.1| PREDICTED: similar to TBC1 domain family, membe... 147 5e-34
ref|XP_537821.1| PREDICTED: similar to TBC1 domain family, membe... 147 8e-34
emb|CAI15411.1| RP11-545E17.5 [Homo sapiens] 146 1e-33
ref|NP_666364.1| TBC1 domain family, member 13 [Mus musculus] gi... 146 1e-33
ref|XP_520299.1| PREDICTED: similar to TBC1 domain family, membe... 146 1e-33
dbj|BAD92802.1| TBC1 domain family, member 13 variant [Homo sapi... 146 1e-33
ref|NP_060671.2| TBC1 domain family, member 13 [Homo sapiens] 146 1e-33
ref|NP_648245.2| CG5978-PA [Drosophila melanogaster] gi|7295061|... 139 1e-31
gb|AAL13771.1| LD24460p [Drosophila melanogaster] 139 1e-31
>ref|NP_974542.1| RabGAP/TBC domain-containing protein [Arabidopsis thaliana]
Length = 408
Score = 508 bits (1307), Expect = e-142
Identities = 266/437 (60%), Positives = 323/437 (73%), Gaps = 55/437 (12%)
Query: 1 MAKKRVPDWLNSPMWSS--PTEQNRFSATSYPS-EPPSPPPVTVVEDPP--SIHNAITTS 55
MAKK +P+WLNS +WSS P + + S PPSPP +VE PP S +AI+T+
Sbjct: 1 MAKKGIPEWLNSSLWSSSPPIDDRHLRNPAATSITPPSPP---IVERPPFSSSPSAISTA 57
Query: 56 P--------PSSSSSS--ASTSSTDDMSISRLAQS-------QSQLLAEVPFTLKSKCL* 98
P PS S + + S D I A S ++Q++AE
Sbjct: 58 PAPPLPVRPPSKSEINDLQNGSGNDGAGIETPAVSSVEDVSRKAQVVAE----------- 106
Query: 99 LLFSIQLY*YC*LFVY**LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRS 158
LS+KVID++ELRKIASQG+PD G+RS +WKLLL YL PDRS
Sbjct: 107 ------------------LSKKVIDLKELRKIASQGLPDDAGIRSIVWKLLLDYLSPDRS 148
Query: 159 LWSSELAKKRSQYKRFKQDILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEH 218
LWSSELAKKRSQYK+FK+++L+NPSE+TR+M S D++D K E+ G LSRS+ITH +H
Sbjct: 149 LWSSELAKKRSQYKQFKEELLMNPSEVTRKMDKSKGGDSNDPKIESPGALSRSEITHEDH 208
Query: 219 PLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAK 278
PLSLG TS+WN FF+DT+++EQI+RDV RTHPDMHFF GDS +AKSNQ+ALKNIL IFAK
Sbjct: 209 PLSLGTTSLWNNFFKDTEVLEQIERDVMRTHPDMHFFSGDSAVAKSNQDALKNILTIFAK 268
Query: 279 LNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNS 338
LNPGIRYVQGMNE+LAP+FY+FKNDPD+ NAA++E+D FFCFVEL+SGFRDNFCQQLDNS
Sbjct: 269 LNPGIRYVQGMNEILAPIFYIFKNDPDKGNAAYAESDAFFCFVELMSGFRDNFCQQLDNS 328
Query: 339 IVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWD 398
+VGIR TITRLS LLK HDEELWRHLEVTTK+NPQFYAFRWITLLLTQEF+F +SL IWD
Sbjct: 329 VVGIRYTITRLSLLLKHHDEELWRHLEVTTKINPQFYAFRWITLLLTQEFNFVESLHIWD 388
Query: 399 TLVSDPDGPQVQIYSFL 415
TL+SDP+GPQV ++S L
Sbjct: 389 TLLSDPEGPQV-VFSLL 404
>emb|CAB78415.1| putative protein [Arabidopsis thaliana] gi|4455302|emb|CAB36837.1|
putative protein [Arabidopsis thaliana]
gi|15236337|ref|NP_193109.1| RabGAP/TBC
domain-containing protein [Arabidopsis thaliana]
gi|7485689|pir||T05242 hypothetical protein F18A5.120 -
Arabidopsis thaliana
Length = 449
Score = 505 bits (1301), Expect = e-142
Identities = 263/430 (61%), Positives = 318/430 (73%), Gaps = 54/430 (12%)
Query: 1 MAKKRVPDWLNSPMWSS--PTEQNRFSATSYPS-EPPSPPPVTVVEDPP--SIHNAITTS 55
MAKK +P+WLNS +WSS P + + S PPSPP +VE PP S +AI+T+
Sbjct: 1 MAKKGIPEWLNSSLWSSSPPIDDRHLRNPAATSITPPSPP---IVERPPFSSSPSAISTA 57
Query: 56 P--------PSSSSSS--ASTSSTDDMSISRLAQS-------QSQLLAEVPFTLKSKCL* 98
P PS S + + S D I A S ++Q++AE
Sbjct: 58 PAPPLPVRPPSKSEINDLQNGSGNDGAGIETPAVSSVEDVSRKAQVVAE----------- 106
Query: 99 LLFSIQLY*YC*LFVY**LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRS 158
LS+KVID++ELRKIASQG+PD G+RS +WKLLL YL PDRS
Sbjct: 107 ------------------LSKKVIDLKELRKIASQGLPDDAGIRSIVWKLLLDYLSPDRS 148
Query: 159 LWSSELAKKRSQYKRFKQDILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEH 218
LWSSELAKKRSQYK+FK+++L+NPSE+TR+M S D++D K E+ G LSRS+ITH +H
Sbjct: 149 LWSSELAKKRSQYKQFKEELLMNPSEVTRKMDKSKGGDSNDPKIESPGALSRSEITHEDH 208
Query: 219 PLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAK 278
PLSLG TS+WN FF+DT+++EQI+RDV RTHPDMHFF GDS +AKSNQ+ALKNIL IFAK
Sbjct: 209 PLSLGTTSLWNNFFKDTEVLEQIERDVMRTHPDMHFFSGDSAVAKSNQDALKNILTIFAK 268
Query: 279 LNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNS 338
LNPGIRYVQGMNE+LAP+FY+FKNDPD+ NAA++E+D FFCFVEL+SGFRDNFCQQLDNS
Sbjct: 269 LNPGIRYVQGMNEILAPIFYIFKNDPDKGNAAYAESDAFFCFVELMSGFRDNFCQQLDNS 328
Query: 339 IVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWD 398
+VGIR TITRLS LLK HDEELWRHLEVTTK+NPQFYAFRWITLLLTQEF+F +SL IWD
Sbjct: 329 VVGIRYTITRLSLLLKHHDEELWRHLEVTTKINPQFYAFRWITLLLTQEFNFVESLHIWD 388
Query: 399 TLVSDPDGPQ 408
TL+SDP+GPQ
Sbjct: 389 TLLSDPEGPQ 398
>gb|AAO63888.1| unknown protein [Arabidopsis thaliana] gi|28393508|gb|AAO42175.1|
unknown protein [Arabidopsis thaliana]
gi|42561701|ref|NP_171975.2| RabGAP/TBC
domain-containing protein [Arabidopsis thaliana]
Length = 448
Score = 477 bits (1228), Expect = e-133
Identities = 240/410 (58%), Positives = 309/410 (74%), Gaps = 20/410 (4%)
Query: 1 MAKKRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPP---SIHNAITTSPP 57
M +K+VP+WLNS MWS+P + SY PVT +++ S+ + ++PP
Sbjct: 1 MVRKKVPEWLNSTMWSTPPPPS-----SYDDGLLRHSPVTKMKEEAESISVAPRLNSAPP 55
Query: 58 SSSSSSASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQLY*YC*LFVY**L 117
SS++S + S + + ++ + V + + FS Q + V L
Sbjct: 56 PSSNTSVPSPSHRPRNGNSISGGSGEYGHSVGPSAED------FSRQAH------VSAEL 103
Query: 118 SRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQD 177
S+KVI+M+ELR +A Q +PDSPG+RST+WKLLLGYLPP+RSLWS+EL +KRSQYK +K +
Sbjct: 104 SKKVINMKELRSLALQSLPDSPGIRSTVWKLLLGYLPPERSLWSTELKQKRSQYKHYKDE 163
Query: 178 ILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDI 237
+L +PSEIT +M S +D D+K E+R ML+RS+IT +HPLSLGK SIWN +FQDT+
Sbjct: 164 LLTSPSEITWKMVRSKGFDNYDLKSESRCMLARSRITDEDHPLSLGKASIWNTYFQDTET 223
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
IEQIDRDVKRTHPD+ FF G+S A+SNQE++KNIL++FAKLN GIRYVQGMNE+LAP+F
Sbjct: 224 IEQIDRDVKRTHPDIPFFSGESSFARSNQESMKNILLVFAKLNQGIRYVQGMNEILAPIF 283
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
YVF+NDPDE++++ +EAD FFCFVELLSGFRD +CQQLDNS+VGIRS ITRLSQL+++HD
Sbjct: 284 YVFRNDPDEDSSSHAEADAFFCFVELLSGFRDFYCQQLDNSVVGIRSAITRLSQLVRKHD 343
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGP 407
EELWRHLE+TTKVNPQFYAFRWITLLLTQEF F DSL IWD L+SDP+GP
Sbjct: 344 EELWRHLEITTKVNPQFYAFRWITLLLTQEFSFFDSLHIWDALLSDPEGP 393
>gb|AAF40453.1| Similar to gi|3217452 F45E6.3 gene product from C. elegans cosmid
gb|Z68117. [Arabidopsis thaliana]
gi|25354057|pir||E86181 hypothetical protein [imported]
- Arabidopsis thaliana
Length = 438
Score = 473 bits (1218), Expect = e-132
Identities = 238/410 (58%), Positives = 308/410 (75%), Gaps = 20/410 (4%)
Query: 1 MAKKRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPP---SIHNAITTSPP 57
M +K+VP+WLNS MWS+P + SY PVT +++ S+ + ++PP
Sbjct: 1 MVRKKVPEWLNSTMWSTPPPPS-----SYDDGLLRHSPVTKMKEEAESISVAPRLNSAPP 55
Query: 58 SSSSSSASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQLY*YC*LFVY**L 117
SS++S + S + + ++ + V + + FS Q + V L
Sbjct: 56 PSSNTSVPSPSHRPRNGNSISGGSGEYGHSVGPSAED------FSRQAH------VSAEL 103
Query: 118 SRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQD 177
S+KVI+M+ELR +A Q +PDSPG+RST+WKLLLGYLPP+RSLWS+EL +KRSQYK +K +
Sbjct: 104 SKKVINMKELRSLALQSLPDSPGIRSTVWKLLLGYLPPERSLWSTELKQKRSQYKHYKDE 163
Query: 178 ILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDI 237
+L +P +IT +M S +D D+K E+R ML+RS+IT +HPLSLGK SIWN +FQDT+
Sbjct: 164 LLTSPVKITWKMVRSKGFDNYDLKSESRCMLARSRITDEDHPLSLGKASIWNTYFQDTET 223
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
IEQIDRDVKRTHPD+ FF G+S A+SNQE++KNIL++FAKLN GIRYVQGMNE+LAP+F
Sbjct: 224 IEQIDRDVKRTHPDIPFFSGESSFARSNQESMKNILLVFAKLNQGIRYVQGMNEILAPIF 283
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
YVF+NDPDE++++ +EAD FFCFVELLSGFRD +CQQLDNS+VGIRS ITRLSQL+++HD
Sbjct: 284 YVFRNDPDEDSSSHAEADAFFCFVELLSGFRDFYCQQLDNSVVGIRSAITRLSQLVRKHD 343
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGP 407
EELWRHLE+TTKVNPQFYAFRWITLLLTQEF F DSL IWD L+SDP+GP
Sbjct: 344 EELWRHLEITTKVNPQFYAFRWITLLLTQEFSFFDSLHIWDALLSDPEGP 393
>dbj|BAD82321.1| RabGAP/TBC domain-containing protein-like [Oryza sativa (japonica
cultivar-group)] gi|56785115|dbj|BAD82753.1| RabGAP/TBC
domain-containing protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 368
Score = 430 bits (1106), Expect = e-119
Identities = 204/294 (69%), Positives = 250/294 (84%), Gaps = 5/294 (1%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
LS+KVID+ ELR +A+QG+PD +R T+WKLLLGYLP DR+LW ELAKKRSQY FK+
Sbjct: 29 LSKKVIDLDELRMLAAQGVPDGAAVRPTVWKLLLGYLPSDRALWEQELAKKRSQYAAFKE 88
Query: 177 DILINPSEITRR--MFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQD 234
+ L NP EI R+ + S +A + G+L RS++T EHPLSLGKT+ WNQFF+
Sbjct: 89 EFLSNPMEIARQRELEGQGSENAGSIY---NGLLHRSEVTQEEHPLSLGKTTAWNQFFEY 145
Query: 235 TDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLA 294
++IIEQIDRDVKRTHPDMHFFCGDS AKSNQE+LKNILIIFAKLN GIRYVQGMNE+LA
Sbjct: 146 SEIIEQIDRDVKRTHPDMHFFCGDSSFAKSNQESLKNILIIFAKLNAGIRYVQGMNEILA 205
Query: 295 PLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLK 354
PLF+VF+NDPD++NA F+EAD+FFCF+ELLSGFRDNFCQ+LDNS VGI+ T+++LSQL+
Sbjct: 206 PLFFVFRNDPDDKNANFAEADSFFCFMELLSGFRDNFCQKLDNSAVGIQGTLSKLSQLVA 265
Query: 355 EHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQ 408
++D EL R+LE+TT++NPQFYAFRWITLLLTQEF+FAD++ IWDTL+SDPDGPQ
Sbjct: 266 KYDGELQRYLEITTEINPQFYAFRWITLLLTQEFNFADTIHIWDTLLSDPDGPQ 319
>emb|CAD40692.3| OSJNBa0083D01.9 [Oryza sativa (japonica cultivar-group)]
gi|50923695|ref|XP_472208.1| OSJNBa0083D01.9 [Oryza
sativa (japonica cultivar-group)]
Length = 392
Score = 422 bits (1085), Expect = e-116
Identities = 205/306 (66%), Positives = 240/306 (77%), Gaps = 14/306 (4%)
Query: 120 KVIDMRELRKIASQGIPDSPGLRSTIWK-LLLGYLPPDRSLWSSELAKKRSQYKRFKQDI 178
KV+D+ ELR++A QG+PD+ G+R +WK LLLGYLP D +LW+ EL KKRSQY FK ++
Sbjct: 36 KVVDLAELRRLACQGVPDAAGVRPVVWKQLLLGYLPTDHALWAYELEKKRSQYSAFKDEL 95
Query: 179 LINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDII 238
L+NPSE+TRRM + E G+L R++I H EHPLSLGKTS+WNQFFQ+++ I
Sbjct: 96 LVNPSEVTRRMEEMTISKGNRHNSEGTGVLPRAEIVHDEHPLSLGKTSVWNQFFQESETI 155
Query: 239 EQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFY 298
EQIDRDVKRTHP+M FF GDS A SNQE+LK IL IFAKLNPGIRYVQGMNEVLAPL+Y
Sbjct: 156 EQIDRDVKRTHPEMQFFNGDSSDALSNQESLKRILTIFAKLNPGIRYVQGMNEVLAPLYY 215
Query: 299 VFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDE 358
VFKNDP+E NA +E D FFCFVELLSGFRDNFC+QLDNS+VGIRSTI++LSQLLK HDE
Sbjct: 216 VFKNDPEENNAESAEPDAFFCFVELLSGFRDNFCKQLDNSVVGIRSTISKLSQLLKRHDE 275
Query: 359 ELWRHLEVTTK-------------VNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPD 405
ELWRHLEV TK VNPQFYAFRWITLLLTQEF F D + IWD L+ DP+
Sbjct: 276 ELWRHLEVVTKCISAPTLMLCIFQVNPQFYAFRWITLLLTQEFKFRDCIHIWDALLGDPE 335
Query: 406 GPQVQI 411
GPQ +
Sbjct: 336 GPQATL 341
>ref|XP_463585.1| P0497A05.14 [Oryza sativa (japonica cultivar-group)]
gi|20804889|dbj|BAB92570.1| P0497A05.14 [Oryza sativa
(japonica cultivar-group)]
Length = 426
Score = 366 bits (940), Expect = e-100
Identities = 180/292 (61%), Positives = 217/292 (73%), Gaps = 51/292 (17%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
LS+KVID+ ELR +A+QG+PD +R T+WKLLLGYLP DR+LW ELAKKRSQY FK+
Sbjct: 74 LSKKVIDLDELRMLAAQGVPDGAAVRPTVWKLLLGYLPSDRALWEQELAKKRSQYAAFKE 133
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+ L NP ++
Sbjct: 134 EFLSNPY---------------------------------------------------SE 142
Query: 237 IIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPL 296
IIEQIDRDVKRTHPDMHFFCGDS AKSNQE+LKNILIIFAKLN GIRYVQGMNE+LAPL
Sbjct: 143 IIEQIDRDVKRTHPDMHFFCGDSSFAKSNQESLKNILIIFAKLNAGIRYVQGMNEILAPL 202
Query: 297 FYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEH 356
F+VF+NDPD++NA F+EAD+FFCF+ELLSGFRDNFCQ+LDNS VGI+ T+++LSQL+ ++
Sbjct: 203 FFVFRNDPDDKNANFAEADSFFCFMELLSGFRDNFCQKLDNSAVGIQGTLSKLSQLVAKY 262
Query: 357 DEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQ 408
D EL R+LE+TT++NPQFYAFRWITLLLTQEF+FAD++ IWDTL+SDPDGPQ
Sbjct: 263 DGELQRYLEITTEINPQFYAFRWITLLLTQEFNFADTIHIWDTLLSDPDGPQ 314
>ref|XP_646856.1| hypothetical protein DDB0216614 [Dictyostelium discoideum]
gi|60475179|gb|EAL73115.1| hypothetical protein
DDB0216614 [Dictyostelium discoideum]
Length = 604
Score = 260 bits (665), Expect = 5e-68
Identities = 133/285 (46%), Positives = 188/285 (65%), Gaps = 23/285 (8%)
Query: 122 IDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILIN 181
ID+ ++ +A QGIP+S GLR+ WK+LL YLP DR+ W L K R+ Y+ F +++I+
Sbjct: 23 IDISIIQHLAEQGIPESHGLRACYWKILLRYLPLDRTQWEIHLKKSRNGYQDFINELMID 82
Query: 182 PSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQI 241
P Y D + +HPLS+ S WN++++D +I+ I
Sbjct: 83 P------------YKGDSPPPKQEEF---------DHPLSVQTDSKWNEYWKDQNILIDI 121
Query: 242 DRDVKRTHPDMHFFCGDSQLAKS-NQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVF 300
++DV+RT P MHFF + KS + EAL+ IL I+AKLNPGI+YVQGMNE+L ++Y+F
Sbjct: 122 EKDVRRTFPSMHFFNYQQEDGKSIHYEALRRILFIYAKLNPGIKYVQGMNEILGHVYYIF 181
Query: 301 KNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEEL 360
DPD++ +EAD+F+CF L+S RDNFC+ LD S VGI S+I +L+++LK++D EL
Sbjct: 182 ATDPDQDCKKNAEADSFYCFTSLMSEIRDNFCKTLDRSDVGIISSIKKLNRILKDNDLEL 241
Query: 361 WRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPD 405
W LE K+NPQFY+FRWITLLL+QEF+ D LR+WD L SDP+
Sbjct: 242 WTDLE-DKKLNPQFYSFRWITLLLSQEFELPDVLRLWDALFSDPN 285
>ref|XP_392146.2| PREDICTED: similar to CG5978-PA [Apis mellifera]
Length = 396
Score = 214 bits (545), Expect = 4e-54
Identities = 122/342 (35%), Positives = 182/342 (52%), Gaps = 75/342 (21%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L+ + ID+ L+++ GIPD GLR WKLLL YLP R WS L +KR+ YK F +
Sbjct: 15 LNAEEIDLISLKRLCFHGIPDEGGLRPLCWKLLLNYLPSTRLSWSETLTRKRTLYKTFIE 74
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
D+++ P E A+ D + R +T +HPL+L S W +F+D +
Sbjct: 75 DLIVTPGE--------ANSDGE-----------RVDVTLHDHPLNLNPDSKWQTYFKDNE 115
Query: 237 IIEQIDRDVKRTHPDMHFF--------------------------------------CGD 258
++ QID+DV+R PD+ FF G
Sbjct: 116 VLLQIDKDVRRLCPDISFFQQGTDYPREEIVNASGQKRLHSRVQHTVLRSANVERKGLGV 175
Query: 259 SQLAKSNQEA-----------------LKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFK 301
+++A S ++A L+ IL ++AKLNPG YVQGMNE++ P+++ F
Sbjct: 176 TKIAVSIRKAAEDYAPLAEGGEAHWEVLERILFLYAKLNPGQGYVQGMNEIVGPIYHAFA 235
Query: 302 NDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELW 361
DPD++ +EADTFFCF L+S RD F + LD + GI S +++L+ +K +D E+W
Sbjct: 236 CDPDQKWREHAEADTFFCFTNLMSEIRDFFIKSLDEAEFGINSMMSKLTAQVKANDPEIW 295
Query: 362 RHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
L ++ PQ+Y+FRW+TLLL+QEF D +RIWD+L +D
Sbjct: 296 MRLH-QQELCPQYYSFRWLTLLLSQEFPLPDVMRIWDSLFAD 336
>ref|XP_315752.2| ENSANGP00000018717 [Anopheles gambiae str. PEST]
gi|55238540|gb|EAA11714.2| ENSANGP00000018717 [Anopheles
gambiae str. PEST]
Length = 381
Score = 213 bits (543), Expect = 7e-54
Identities = 123/340 (36%), Positives = 176/340 (51%), Gaps = 79/340 (23%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L + ID++ LR GIPD G R+ WKLLLGYL P +S W ++L K+R YK+F +
Sbjct: 15 LEQDEIDLKVLRSFCFNGIPDCNGFRALCWKLLLGYLSPKKSTWPAKLTKQRELYKQFVK 74
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I+P E D +C +HPLS G S W+ FF+D +
Sbjct: 75 EMVISPGE------------QDGPECI-------------DHPLSDGPESNWSTFFRDNE 109
Query: 237 IIEQIDRDVKRTHPD---------------------MHFFCGDSQLAKSNQE-------- 267
++ QID+DV+R PD +H + L+ +N E
Sbjct: 110 VLLQIDKDVRRLCPDISFFQQATEFPCEFVKERERKLHVRVAPTTLSSANVERKGVGMTK 169
Query: 268 ------------------------ALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND 303
++ IL ++AKLNPG YVQGMNE++ P++YVF +D
Sbjct: 170 INLITKRATESYEAMDAGQEAHWEVVERILFLYAKLNPGQGYVQGMNEIIGPIYYVFASD 229
Query: 304 PDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRH 363
P E ++EAD FFCF L+S RD F + LD S GI+ + +LS LL E D E+W
Sbjct: 230 PHLEYRRYAEADCFFCFTALMSEIRDFFIKTLDESEGGIKGMMAKLSNLLHEQDAEVWER 289
Query: 364 LEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
L ++ PQ+Y+FRW+TLLL+QEF D LRIWD++ +D
Sbjct: 290 LR-DQELYPQYYSFRWLTLLLSQEFPLPDVLRIWDSVFAD 328
>emb|CAA92181.2| Hypothetical protein F45E6.3 [Caenorhabditis elegans]
Length = 459
Score = 196 bits (499), Expect = 9e-49
Identities = 123/355 (34%), Positives = 188/355 (52%), Gaps = 69/355 (19%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
++ K ID+ ELR S G+P+S LR W+LLL YLP +R W + LA++R Y + +
Sbjct: 18 IANKKIDINELRAGCSYGVPES--LRPLAWRLLLHYLPLERHKWQTFLAEQRDNYDQMIE 75
Query: 177 DILINPSEITRRMFNSASYDAD--------------------------DVK-----CETR 205
I++ P + + + + D D DV+ +
Sbjct: 76 QIIVEPGTASLQQSAAQNQDNDHPLSDHPTSDWQAFFQDNKVLSQIDKDVRRLYPEIQFF 135
Query: 206 GMLSRSQITHG-EHPLS---LGKTSIWNQFFQ----------DTDIIEQIDRDVKRTHPD 251
+LSR HG ++PLS + + +Q F T+I +Q + + + +
Sbjct: 136 QLLSRFPHQHGMKYPLSRRVINHQELHSQEFGANRDGIVGCVKTNIAKQSQDENQAPNSE 195
Query: 252 MHFFCGDSQLAKSNQEAL----------------------KNILIIFAKLNPGIRYVQGM 289
H+ K +QE+L + IL I+AKLNPG++YVQGM
Sbjct: 196 FHWHQFFEYSYKFSQESLVFRKNCFNLMPNILNQFIIKIVERILFIYAKLNPGVQYVQGM 255
Query: 290 NEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRL 349
NE++AP++YVF ND D+E AA++EADTFFCF +L+S +DNF ++LD+S GI S+++
Sbjct: 256 NELVAPIYYVFANDADDEWAAYAEADTFFCFQQLMSEVKDNFIKKLDDSNCGIESSMSAF 315
Query: 350 SQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDP 404
L+ D EL +HL +T ++ PQFYAFRW++LLL+QEF D + +WD L SDP
Sbjct: 316 HNLISTFDTELHKHLTLTLEIKPQFYAFRWLSLLLSQEFPLPDVITLWDALFSDP 370
>ref|XP_575106.1| PREDICTED: similar to TBC1 domain family, member 13 [Rattus
norvegicus]
Length = 522
Score = 147 bits (372), Expect = 5e-34
Identities = 71/141 (50%), Positives = 98/141 (69%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 327 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 386
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LKE D EL+ L+ + PQF+AFRW+TL
Sbjct: 387 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKEKDVELYLKLQ-EQSIKPQFFAFRWLTL 445
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 446 LLSQEFLLPDVIRIWDSLFAD 466
Score = 60.1 bits (144), Expect = 1e-07
Identities = 28/66 (42%), Positives = 41/66 (61%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R W S LAK+R Y +F +
Sbjct: 16 LKEPSIVLEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERDSWDSILAKQRGLYSQFLR 75
Query: 177 DILINP 182
+++I P
Sbjct: 76 EMIIQP 81
>ref|XP_537821.1| PREDICTED: similar to TBC1 domain family, member 13 [Canis
familiaris]
Length = 422
Score = 147 bits (370), Expect = 8e-34
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 227 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 286
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 287 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDRDMELYLKLQ-EQNIKPQFFAFRWLTL 345
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 346 LLSQEFLLPDVIRIWDSLFAD 366
Score = 97.1 bits (240), Expect = 9e-19
Identities = 50/139 (35%), Positives = 77/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 16 LKEPTIVLEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRELYSQFLR 75
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M +SR +T +HPL+ S WN +F+D +
Sbjct: 76 EMIIQPGIAKANM-----------------GVSREDVTFEDHPLNPNPDSRWNTYFKDNE 118
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 119 VLLQIDKDVRRLCPDISFF 137
>emb|CAI15411.1| RP11-545E17.5 [Homo sapiens]
Length = 400
Score = 146 bits (369), Expect = 1e-33
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 205 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 264
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 265 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQ-EQNIKPQFFAFRWLTL 323
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 324 LLSQEFLLPDVIRIWDSLFAD 344
Score = 96.3 bits (238), Expect = 2e-18
Identities = 50/139 (35%), Positives = 77/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 16 LKEPSIALEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRELYAQFLR 75
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M +SR +T +HPL+ S WN +F+D +
Sbjct: 76 EMIIQPGIAKANM-----------------GVSREDVTFEDHPLNPNPDSRWNTYFKDNE 118
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 119 VLLQIDKDVRRLCPDISFF 137
>ref|NP_666364.1| TBC1 domain family, member 13 [Mus musculus]
gi|19343763|gb|AAH25586.1| TBC1 domain family, member 13
[Mus musculus] gi|42559835|sp|Q8R3D1|TBC13_MOUSE TBC1
domain family member 13
Length = 400
Score = 146 bits (369), Expect = 1e-33
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 205 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 264
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 265 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQ-EQSIKPQFFAFRWLTL 323
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 324 LLSQEFLLPDVIRIWDSLFAD 344
Score = 94.0 bits (232), Expect = 8e-18
Identities = 49/139 (35%), Positives = 76/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 16 LKEPSIVLEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRGLYSQFLR 75
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M + R +T +HPL+ S WN +F+D +
Sbjct: 76 EMIIQPGIAKANM-----------------GVFREDVTFEDHPLNPNPDSRWNTYFKDNE 118
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 119 VLLQIDKDVRRLCPDISFF 137
>ref|XP_520299.1| PREDICTED: similar to TBC1 domain family, member 13 [Pan
troglodytes]
Length = 450
Score = 146 bits (369), Expect = 1e-33
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 255 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 314
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 315 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQ-EQNIKPQFFAFRWLTL 373
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 374 LLSQEFLLPDVIRIWDSLFAD 394
Score = 96.3 bits (238), Expect = 2e-18
Identities = 50/139 (35%), Positives = 77/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 66 LKEPSIALEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRELYAQFLR 125
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M +SR +T +HPL+ S WN +F+D +
Sbjct: 126 EMIIQPGIAKANM-----------------GVSREDVTFEDHPLNPNPDSRWNTYFKDNE 168
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 169 VLLQIDKDVRRLCPDISFF 187
>dbj|BAD92802.1| TBC1 domain family, member 13 variant [Homo sapiens]
Length = 355
Score = 146 bits (369), Expect = 1e-33
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 160 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 219
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 220 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQ-EQNIKPQFFAFRWLTL 278
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 279 LLSQEFLLPDVIRIWDSLFAD 299
Score = 76.3 bits (186), Expect = 2e-12
Identities = 39/109 (35%), Positives = 61/109 (55%), Gaps = 17/109 (15%)
Query: 147 KLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILINPSEITRRMFNSASYDADDVKCETRG 206
K+LL YLP +R+ W+S LAK+R Y +F ++++I P M
Sbjct: 1 KILLNYLPLERASWTSILAKQRELYAQFLREMIIQPGIAKANMG---------------- 44
Query: 207 MLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFF 255
+SR +T +HPL+ S WN +F+D +++ QID+DV+R PD+ FF
Sbjct: 45 -VSREDVTFEDHPLNPNPDSRWNTYFKDNEVLLQIDKDVRRLCPDISFF 92
>ref|NP_060671.2| TBC1 domain family, member 13 [Homo sapiens]
Length = 400
Score = 146 bits (369), Expect = 1e-33
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 205 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 264
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 265 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQ-EQNIKPQFFAFRWLTL 323
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 324 LLSQEFLLPDVIRIWDSLFAD 344
Score = 96.3 bits (238), Expect = 2e-18
Identities = 50/139 (35%), Positives = 77/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 16 LKEPSIALEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRELYAQFLR 75
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M +SR +T +HPL+ S WN +F+D +
Sbjct: 76 EMIIQPGIAKANM-----------------GVSREDVTFEDHPLNPNPDSRWNTYFKDNE 118
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 119 VLLQIDKDVRRLCPDISFF 137
>ref|NP_648245.2| CG5978-PA [Drosophila melanogaster] gi|7295061|gb|AAF50388.1|
CG5978-PA [Drosophila melanogaster]
Length = 403
Score = 139 bits (351), Expect = 1e-31
Identities = 67/141 (47%), Positives = 99/141 (69%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPG YVQGMNE++ P++YV +DPD A +EAD FFCF
Sbjct: 211 EAHWEVVQRILFIYAKLNPGQGYVQGMNEIVGPIYYVMASDPDLTYRAHAEADCFFCFTA 270
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L+S RD F + LD++ GI+ + RLS +LK D ++ L + +++PQ+Y+FRW+TL
Sbjct: 271 LMSEIRDFFIKTLDDAEGGIKFMMARLSNMLKSKDLSIYELLR-SQELHPQYYSFRWLTL 329
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D LRIWD++ +D
Sbjct: 330 LLSQEFPLPDVLRIWDSVFAD 350
Score = 109 bits (273), Expect = 1e-22
Identities = 58/136 (42%), Positives = 85/136 (61%), Gaps = 11/136 (8%)
Query: 121 VIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILI 180
+ID+++LRK+A G+PD R+ WKLLLGYL P RS W++ LA+KR+ YK+F +++++
Sbjct: 21 LIDLKQLRKLAFNGVPDVQSFRALSWKLLLGYLGPRRSSWTTTLAQKRALYKQFIEELVL 80
Query: 181 NPSEITRRMFNSASYD-ADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIE 239
P + N AS D D K ++ G + +HPLS G S WN FF D + +
Sbjct: 81 PPGHSS----NRASVDGGDGDKVDSGG------VGLQDHPLSEGPESAWNTFFNDNEFLL 130
Query: 240 QIDRDVKRTHPDMHFF 255
QID+DV+R PD+ FF
Sbjct: 131 QIDKDVRRLCPDISFF 146
>gb|AAL13771.1| LD24460p [Drosophila melanogaster]
Length = 403
Score = 139 bits (351), Expect = 1e-31
Identities = 67/141 (47%), Positives = 99/141 (69%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPG YVQGMNE++ P++YV +DPD A +EAD FFCF
Sbjct: 211 EAHWEVVQRILFIYAKLNPGQGYVQGMNEIVGPIYYVMASDPDLTYRAHAEADCFFCFTA 270
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L+S RD F + LD++ GI+ + RLS +LK D ++ L + +++PQ+Y+FRW+TL
Sbjct: 271 LMSEIRDFFIKTLDDAEGGIKFMMARLSNMLKSKDLSIYELLR-SQELHPQYYSFRWLTL 329
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D LRIWD++ +D
Sbjct: 330 LLSQEFPLPDVLRIWDSVFAD 350
Score = 109 bits (273), Expect = 1e-22
Identities = 58/136 (42%), Positives = 85/136 (61%), Gaps = 11/136 (8%)
Query: 121 VIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILI 180
+ID+++LRK+A G+PD R+ WKLLLGYL P RS W++ LA+KR+ YK+F +++++
Sbjct: 21 LIDLKQLRKLAFNGVPDVQSFRALSWKLLLGYLGPRRSSWTTTLAQKRALYKQFIEELVL 80
Query: 181 NPSEITRRMFNSASYD-ADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIE 239
P + N AS D D K ++ G + +HPLS G S WN FF D + +
Sbjct: 81 PPGHSS----NRASVDGGDGDKVDSGG------VGLQDHPLSEGPESAWNTFFNDNEFLL 130
Query: 240 QIDRDVKRTHPDMHFF 255
QID+DV+R PD+ FF
Sbjct: 131 QIDKDVRRLCPDISFF 146
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.327 0.139 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 713,211,721
Number of Sequences: 2540612
Number of extensions: 28744955
Number of successful extensions: 243732
Number of sequences better than 10.0: 1210
Number of HSP's better than 10.0 without gapping: 409
Number of HSP's successfully gapped in prelim test: 818
Number of HSP's that attempted gapping in prelim test: 237045
Number of HSP's gapped (non-prelim): 4149
length of query: 424
length of database: 863,360,394
effective HSP length: 131
effective length of query: 293
effective length of database: 530,540,222
effective search space: 155448285046
effective search space used: 155448285046
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146683.4


