

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.3 + phase: 0 /pseudo
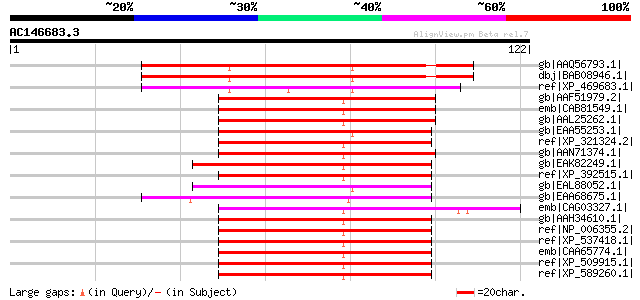
(122 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ56793.1| At5g43670 [Arabidopsis thaliana] gi|28058769|gb|A... 71 7e-12
dbj|BAB08946.1| protein transport protein SEC23 [Arabidopsis tha... 71 7e-12
ref|XP_469683.1| putative protein transport SEC23-related protei... 60 1e-08
gb|AAF51979.2| CG1250-PB, isoform B [Drosophila melanogaster] gi... 57 1e-07
emb|CAB81549.1| putative Sec23 protein [Drosophila melanogaster] 57 1e-07
gb|AAL25262.1| GH01163p [Drosophila melanogaster] 57 1e-07
gb|EAA55253.1| hypothetical protein MG06910.4 [Magnaporthe grise... 57 1e-07
ref|XP_321324.2| ENSANGP00000012825 [Anopheles gambiae str. PEST... 57 1e-07
gb|AAN71374.1| RE35250p [Drosophila melanogaster] 56 2e-07
gb|EAK82249.1| hypothetical protein UM01624.1 [Ustilago maydis 5... 55 3e-07
ref|XP_392515.1| PREDICTED: similar to ENSANGP00000012825 [Apis ... 55 4e-07
gb|EAL88052.1| protein transport protein Sec23, putative [Asperg... 54 9e-07
gb|EAA68675.1| conserved hypothetical protein [Gibberella zeae P... 53 1e-06
emb|CAG03327.1| unnamed protein product [Tetraodon nigroviridis] 53 1e-06
gb|AAH34610.1| Sec23a protein [Mus musculus] gi|34867969|ref|XP_... 52 3e-06
ref|NP_006355.2| SEC23-related protein A [Homo sapiens] 52 3e-06
ref|XP_537418.1| PREDICTED: similar to Sec23a protein [Canis fam... 52 3e-06
emb|CAA65774.1| Sec23 protein [Homo sapiens] gi|2498885|sp|Q1543... 52 3e-06
ref|XP_509915.1| PREDICTED: SEC23-related protein A [Pan troglod... 52 3e-06
ref|XP_589260.1| PREDICTED: similar to Protein transport protein... 52 3e-06
>gb|AAQ56793.1| At5g43670 [Arabidopsis thaliana] gi|28058769|gb|AAO29951.1| Unknown
protein [Arabidopsis thaliana]
gi|18422356|ref|NP_568626.1| transport protein, putative
[Arabidopsis thaliana]
Length = 794
Score = 70.9 bits (172), Expect = 7e-12
Identities = 43/86 (50%), Positives = 53/86 (61%), Gaps = 10/86 (11%)
Query: 32 ITVAIEDIHSTYNFKSRNR---STGVAFSAALGLLECCFVNTGSRIMVFAS-----GPGL 83
+T A E+I + K +R STG A S ALGLLE C V TGSRIMVF S GPG+
Sbjct: 277 LTSAFEEIIPLVDVKPGHRPHRSTGAAISTALGLLEGCSVTTGSRIMVFTSGPATRGPGI 336
Query: 84 VLDSDFWQSMRTHNDIYICNGHLAVY 109
++DSD S+RTH D I GH++ Y
Sbjct: 337 IVDSDLSNSIRTHRD--IITGHVSYY 360
>dbj|BAB08946.1| protein transport protein SEC23 [Arabidopsis thaliana]
Length = 736
Score = 70.9 bits (172), Expect = 7e-12
Identities = 43/86 (50%), Positives = 53/86 (61%), Gaps = 10/86 (11%)
Query: 32 ITVAIEDIHSTYNFKSRNR---STGVAFSAALGLLECCFVNTGSRIMVFAS-----GPGL 83
+T A E+I + K +R STG A S ALGLLE C V TGSRIMVF S GPG+
Sbjct: 219 LTSAFEEIIPLVDVKPGHRPHRSTGAAISTALGLLEGCSVTTGSRIMVFTSGPATRGPGI 278
Query: 84 VLDSDFWQSMRTHNDIYICNGHLAVY 109
++DSD S+RTH D I GH++ Y
Sbjct: 279 IVDSDLSNSIRTHRD--IITGHVSYY 302
>ref|XP_469683.1| putative protein transport SEC23-related protein [Oryza sativa
(japonica cultivar-group)] gi|51964252|ref|XP_506911.1|
PREDICTED OJ1365_D05.6 gene product [Oryza sativa
(japonica cultivar-group)] gi|40539042|gb|AAR87299.1|
putative protein transport SEC23-related protein [Oryza
sativa (japonica cultivar-group)]
Length = 757
Score = 60.1 bits (144), Expect = 1e-08
Identities = 38/84 (45%), Positives = 50/84 (59%), Gaps = 9/84 (10%)
Query: 32 ITVAIEDIHSTYNFKSRNR---STGVAFSAALGLLE-CCFVNTGSRIMVFAS-----GPG 82
IT AIED+ S +R +TG A S A+ LLE CC N G RIMVF S GPG
Sbjct: 241 ITSAIEDLRSMSACPRGHRPLRATGAAISTAVALLEGCCSPNAGGRIMVFTSGPTTVGPG 300
Query: 83 LVLDSDFWQSMRTHNDIYICNGHL 106
LV+++D +++R+H DI+ N L
Sbjct: 301 LVVETDLGKAIRSHRDIFNGNAPL 324
>gb|AAF51979.2| CG1250-PB, isoform B [Drosophila melanogaster]
gi|23170394|gb|AAF51978.2| CG1250-PA, isoform A
[Drosophila melanogaster] gi|24644351|ref|NP_730979.1|
CG1250-PB, isoform B [Drosophila melanogaster]
gi|24644349|ref|NP_730978.1| CG1250-PA, isoform A
[Drosophila melanogaster]
Length = 773
Score = 57.0 bits (136), Expect = 1e-07
Identities = 28/56 (50%), Positives = 37/56 (66%), Gaps = 5/56 (8%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDIY 100
RSTG A S A+GLLEC + NTG RIM F + GPG V+D + +R+H+DI+
Sbjct: 270 RSTGAALSIAVGLLECTYPNTGGRIMTFVGGPCSQGPGQVVDDELKHPIRSHHDIH 325
>emb|CAB81549.1| putative Sec23 protein [Drosophila melanogaster]
Length = 769
Score = 57.0 bits (136), Expect = 1e-07
Identities = 28/56 (50%), Positives = 37/56 (66%), Gaps = 5/56 (8%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDIY 100
RSTG A S A+GLLEC + NTG RIM F + GPG V+D + +R+H+DI+
Sbjct: 266 RSTGAALSIAVGLLECTYPNTGGRIMTFVGGPCSQGPGQVVDDELKHPIRSHHDIH 321
>gb|AAL25262.1| GH01163p [Drosophila melanogaster]
Length = 312
Score = 57.0 bits (136), Expect = 1e-07
Identities = 28/56 (50%), Positives = 37/56 (66%), Gaps = 5/56 (8%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDIY 100
RSTG A S A+GLLEC + NTG RIM F + GPG V+D + +R+H+DI+
Sbjct: 68 RSTGAALSIAVGLLECTYPNTGGRIMTFVGGPCSQGPGQVVDDELKHPIRSHHDIH 123
>gb|EAA55253.1| hypothetical protein MG06910.4 [Magnaporthe grisea 70-15]
gi|39978051|ref|XP_370413.1| hypothetical protein
MG06910.4 [Magnaporthe grisea 70-15]
Length = 763
Score = 57.0 bits (136), Expect = 1e-07
Identities = 29/55 (52%), Positives = 39/55 (70%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVFAS-----GPGLVLDSDFWQSMRTHNDI 99
R TGVA S A+GLLE F N+G RIM+FA+ GPG+V+ S+ + MR+H+DI
Sbjct: 265 RCTGVALSVAVGLLESSFQNSGGRIMLFAAGPATEGPGMVVSSELREPMRSHHDI 319
>ref|XP_321324.2| ENSANGP00000012825 [Anopheles gambiae str. PEST]
gi|55233595|gb|EAA01238.2| ENSANGP00000012825 [Anopheles
gambiae str. PEST]
Length = 772
Score = 56.6 bits (135), Expect = 1e-07
Identities = 28/55 (50%), Positives = 37/55 (66%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RSTG A S A+GLLEC + NTG RIM+F + GPG V+D + +R+H+DI
Sbjct: 269 RSTGAALSIAVGLLECTYPNTGGRIMLFVGGPCSQGPGQVVDDELKHPIRSHHDI 323
>gb|AAN71374.1| RE35250p [Drosophila melanogaster]
Length = 773
Score = 55.8 bits (133), Expect = 2e-07
Identities = 28/56 (50%), Positives = 36/56 (64%), Gaps = 5/56 (8%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDIY 100
RSTG A S A+GLLEC + NTG RIM F + GPG V+D + +R+H DI+
Sbjct: 270 RSTGAALSIAVGLLECTYPNTGGRIMTFVGGPCSQGPGQVVDDELKHPIRSHYDIH 325
>gb|EAK82249.1| hypothetical protein UM01624.1 [Ustilago maydis 521]
gi|49069900|ref|XP_399239.1| hypothetical protein
UM01624.1 [Ustilago maydis 521]
Length = 773
Score = 55.5 bits (132), Expect = 3e-07
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 5/61 (8%)
Query: 44 NFKSRNRSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHND 98
N K R TGVA S A+G+LE F NTG+R+M+F GPG+V+ ++ + +R+H+D
Sbjct: 263 NDKRSQRCTGVALSVAVGMLETTFPNTGARVMLFCGGPATEGPGMVVSTELRERIRSHHD 322
Query: 99 I 99
I
Sbjct: 323 I 323
>ref|XP_392515.1| PREDICTED: similar to ENSANGP00000012825 [Apis mellifera]
Length = 768
Score = 55.1 bits (131), Expect = 4e-07
Identities = 28/55 (50%), Positives = 37/55 (66%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RSTGVA + A GLLE + NTG+RIM+F + GPG V+ D Q +R+H+DI
Sbjct: 265 RSTGVALAVATGLLEASYANTGARIMLFVGGPCSQGPGQVVTDDLRQPIRSHHDI 319
>gb|EAL88052.1| protein transport protein Sec23, putative [Aspergillus fumigatus
Af293]
Length = 767
Score = 53.9 bits (128), Expect = 9e-07
Identities = 31/61 (50%), Positives = 37/61 (59%), Gaps = 5/61 (8%)
Query: 44 NFKSRNRSTGVAFSAALGLLECCFVNTGSRIMVFAS-----GPGLVLDSDFWQSMRTHND 98
N K R TGVA S A+GLLE F N G RIMVF S GPG V+ + + MR+H+D
Sbjct: 245 NDKRPLRCTGVALSVAVGLLETSFQNAGGRIMVFTSGPATEGPGHVVGPELKEPMRSHHD 304
Query: 99 I 99
I
Sbjct: 305 I 305
>gb|EAA68675.1| conserved hypothetical protein [Gibberella zeae PH-1]
gi|46110070|ref|XP_382093.1| conserved hypothetical
protein [Gibberella zeae PH-1]
Length = 771
Score = 53.1 bits (126), Expect = 1e-06
Identities = 31/77 (40%), Positives = 44/77 (56%), Gaps = 9/77 (11%)
Query: 32 ITVAIEDIHS----TYNFKSRNRSTGVAFSAALGLLECCFVNTGSRIMVFA-----SGPG 82
+T A+E + N + R TGVA S A+GLLE F N G RIM+FA GPG
Sbjct: 243 LTKALESLQKDPWPVANDRRNLRCTGVALSVAVGLLESSFQNAGGRIMLFAGGPATEGPG 302
Query: 83 LVLDSDFWQSMRTHNDI 99
+V+ + + +R+H+DI
Sbjct: 303 MVVGPELREPIRSHHDI 319
>emb|CAG03327.1| unnamed protein product [Tetraodon nigroviridis]
Length = 674
Score = 53.1 bits (126), Expect = 1e-06
Identities = 34/79 (43%), Positives = 47/79 (59%), Gaps = 8/79 (10%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDIYICNG 104
RSTGVA S A+GLLE F NTG+R+M+F GPG+V+ + +R+ +DI N
Sbjct: 236 RSTGVALSVAVGLLEGAFPNTGARVMLFIGGPPTQGPGMVVGDELKTPIRSWHDIQKDNA 295
Query: 105 -HL--AVYTSKLLRGCRCV 120
HL A TS+ L+ C +
Sbjct: 296 RHLKKATKTSRELKVCGAI 314
>gb|AAH34610.1| Sec23a protein [Mus musculus] gi|34867969|ref|XP_347237.1|
PREDICTED: SEC23A (S. cerevisiae) (predicted) [Rattus
norvegicus] gi|22450209|gb|AAL92480.1| Sec23-like A
protein [Mus musculus] gi|67906177|ref|NP_033173.2|
SEC23A [Mus musculus] gi|27735257|sp|Q01405|SC23A_MOUSE
Protein transport protein Sec23A (SEC23-related protein
A)
Length = 765
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RS+GVA S A+GLLEC F NTG+RIM+F GPG+V+ + +R+ +DI
Sbjct: 262 RSSGVALSIAVGLLECTFPNTGARIMMFIGGPATQGPGMVVGDELKTPIRSWHDI 316
>ref|NP_006355.2| SEC23-related protein A [Homo sapiens]
Length = 765
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RS+GVA S A+GLLEC F NTG+RIM+F GPG+V+ + +R+ +DI
Sbjct: 262 RSSGVALSIAVGLLECTFPNTGARIMMFIGGPATQGPGMVVGDELKTPIRSWHDI 316
>ref|XP_537418.1| PREDICTED: similar to Sec23a protein [Canis familiaris]
Length = 765
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RS+GVA S A+GLLEC F NTG+RIM+F GPG+V+ + +R+ +DI
Sbjct: 262 RSSGVALSIAVGLLECTFPNTGARIMMFIGGPATQGPGMVVGDELKTPIRSWHDI 316
>emb|CAA65774.1| Sec23 protein [Homo sapiens] gi|2498885|sp|Q15436|SC23A_HUMAN
Protein transport protein Sec23A (SEC23-related protein
A)
Length = 765
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RS+GVA S A+GLLEC F NTG+RIM+F GPG+V+ + +R+ +DI
Sbjct: 262 RSSGVALSIAVGLLECTFPNTGARIMMFIGGPATQGPGMVVGDELKTPIRSWHDI 316
>ref|XP_509915.1| PREDICTED: SEC23-related protein A [Pan troglodytes]
Length = 649
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RS+GVA S A+GLLEC F NTG+RIM+F GPG+V+ + +R+ +DI
Sbjct: 94 RSSGVALSIAVGLLECTFPNTGARIMMFIGGPATQGPGMVVGDELKTPIRSWHDI 148
>ref|XP_589260.1| PREDICTED: similar to Protein transport protein Sec23A
(SEC23-related protein A) [Bos taurus]
Length = 196
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 5/55 (9%)
Query: 50 RSTGVAFSAALGLLECCFVNTGSRIMVF-----ASGPGLVLDSDFWQSMRTHNDI 99
RS+GVA S A+GLLEC F NTG+RIM+F GPG+V+ + +R+ +DI
Sbjct: 44 RSSGVALSIAVGLLECTFPNTGARIMMFIGGPATQGPGMVVGDELKTPIRSWHDI 98
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.135 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 203,476,471
Number of Sequences: 2540612
Number of extensions: 7274918
Number of successful extensions: 17550
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 17415
Number of HSP's gapped (non-prelim): 99
length of query: 122
length of database: 863,360,394
effective HSP length: 98
effective length of query: 24
effective length of database: 614,380,418
effective search space: 14745130032
effective search space used: 14745130032
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146683.3


