

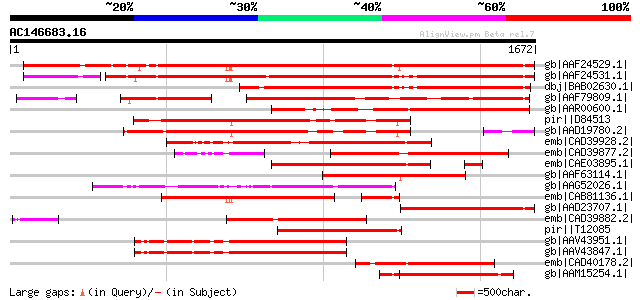
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.16 + phase: 0
(1672 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF24529.1| F7F22.15 [Arabidopsis thaliana] 1701 0.0
gb|AAF24531.1| F7F22.17 [Arabidopsis thaliana] 1516 0.0
dbj|BAB02630.1| retroelement pol polyprotein-like [Arabidopsis t... 1239 0.0
gb|AAF79809.1| T32E20.9 [Arabidopsis thaliana] 1125 0.0
gb|AAR00600.1| putative reverse transcriptase [Oryza sativa (jap... 870 0.0
pir||D84513 probable retroelement pol polyprotein [imported] - A... 849 0.0
gb|AAD19780.2| hypothetical protein [Arabidopsis thaliana] 809 0.0
emb|CAD39928.2| OSJNBa0091C12.6 [Oryza sativa (japonica cultivar... 736 0.0
emb|CAD39877.2| OSJNBb0058J09.16 [Oryza sativa (japonica cultiva... 723 0.0
emb|CAE03895.1| OSJNBb0026I12.3 [Oryza sativa (japonica cultivar... 689 0.0
gb|AAF63114.1| Hypothetical protein [Arabidopsis thaliana] gi|25... 651 0.0
gb|AAG52026.1| polyprotein, putative; 77260-80472 [Arabidopsis t... 645 0.0
emb|CAB81136.1| putative athila transposon protein [Arabidopsis ... 627 e-177
gb|AAD23707.1| putative retroelement pol polyprotein [Arabidopsi... 594 e-168
emb|CAD39882.2| OSJNBb0067G11.5 [Oryza sativa (japonica cultivar... 590 e-166
pir||T12085 reverse transcriptase homolog - fava bean (fragment)... 583 e-164
gb|AAV43951.1| putative polyprotein [Oryza sativa (japonica cult... 541 e-152
gb|AAV43847.1| putative polyprotein [Oryza sativa (japonica cult... 536 e-150
emb|CAD40178.2| OSJNBa0061A09.17 [Oryza sativa (japonica cultiva... 522 e-146
gb|AAM15254.1| putative retroelement pol polyprotein [Arabidopsi... 489 e-136
>gb|AAF24529.1| F7F22.15 [Arabidopsis thaliana]
Length = 1862
Score = 1701 bits (4406), Expect = 0.0
Identities = 899/1721 (52%), Positives = 1177/1721 (68%), Gaps = 127/1721 (7%)
Query: 45 NQFSGSPSEDPNLHISTFWRLSVTCKDD---QETVRLYLFPFSLKDKASNWFNSLKPGSI 101
N+F G P EDP H+ F RL K + ++ +L LFPFSL DKA W +L SI
Sbjct: 170 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSI 229
Query: 102 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQKNEESLFDVWEHFKEILRRCPHHGLEKW 161
T+WD ++ FL +FF ++TA+LR ++ F+QK ES + WE FK +C HHG K
Sbjct: 230 TTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQCSHHGFTKA 289
Query: 162 LIIHTFYNGLTSSTKLTVDTAAGGALMNKDFTTAYALIENMALNLFQWTEE-----KATI 216
++ T Y G+ ++ +DTA+ G NKD + L+EN+A + + E + T
Sbjct: 290 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSNGNYNENCDRTVRGTA 349
Query: 217 DPSPSKKEAGMDETSSIDYLSTKVNALSQRLDRMSTPTFSPPCETCDLSSHSNTDCNLSS 276
D ++ ++ AL+ +LDR+ D + D +
Sbjct: 350 DSDDKHRK--------------EIKALNDKLDRILLSQHKHVHFLVDDEQYEVQDGEGNQ 395
Query: 277 VEQLNFVQNGQRVVQKSHIELLMENYFLK-QSEQLQELKDQTRLLNNSLATLTTKIDSIS 335
+E+++++ N Q K + N L +S + +DQ ++
Sbjct: 396 LEEVSYINNNQGGY-KGYNNFKTNNPNLSYRSTNVANPQDQVYPPQQQ----QSQNKPFV 450
Query: 336 SHNKISETQLSQVVRK-VNHPNKM-NAVTLRSGKPL---EDPIRRTETDNSEKEICEPPS 390
+N+ ++ Q SQ+ K V +P + +A+TL SGK L E+P +T T++SE + E S
Sbjct: 451 PYNQATK-QTSQLPGKAVQNPKEYAHAITLHSGKALPTREEP--KTVTEDSEDQDGEDLS 507
Query: 391 -RETRAER------EKP---RVEENLPPPF--------KPKIPFPQRFAKSKLDEQFKKF 432
++ +A++ E+P ++++L PP +P IP A + + K+
Sbjct: 508 LKKDQADKPLDLSLEQPLDLSLQQSLDPPLDSFTRPTTRPVIPAASPTAPKPVAVKNKEK 567
Query: 433 IEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL-PP 491
+E+ +P + L +P KFLK+++ ++ + E V L+ ECSAIIQ K+ P
Sbjct: 568 VELR------IPLVDALALIPDSHKFLKDLIVERIQ-EVQGMVVLSHECSAIIQKKIIPK 620
Query: 492 KLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSV 551
KL DPGSF++PC +G + +CDLGASVSLMPLS+ +RLG + KS ++L LADRSV
Sbjct: 621 KLSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADRSV 680
Query: 552 KYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFN 611
+ P G++E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+IDVKKGK+ N
Sbjct: 681 RIPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLN 740
Query: 612 VGKE-TVEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGTK-L 669
+GK+ + F++ MK P+I+ I+ +D E D L L + L
Sbjct: 741 LGKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLEELAEEDHLNSALTKSGEDGFL 800
Query: 670 EGEAKAYEELLDRTLPMK---------GLSVEELVKEE-------PTLL----------- 702
E Y++LLD M+ G + E +V E P L
Sbjct: 801 HLETLGYQKLLDSHKAMEESEPFEELNGPATEVMVMSEEGSTRVQPALSRTYSSNHSTLS 860
Query: 703 ---PKE-------------APKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLL 746
P+E APKV+LK LP LRY FLGPNSTYPVI+NA L+ E LL
Sbjct: 861 TDEPREPIISTSDDWSELKAPKVDLKPLPKGLRYAFLGPNSTYPVIINAELNSDEVNLLL 920
Query: 747 YVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKL 806
L+KY +AIGY++ DIKGI+PSLC HRI LE + SIE RRLNPN+KEVVKKE+LKL
Sbjct: 921 SELRKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPHRRLNPNLKEVVKKEILKL 980
Query: 807 LDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATR 866
LDAGVIYPISDS WVSPV VPKK G+ V+KN+K+E I TRT+TG RMCIDYRKLN A+R
Sbjct: 981 LDAGVIYPISDSTWVSPVHCVPKKDGMIVVKNEKDELIPTRTITGHRMCIDYRKLNAASR 1040
Query: 867 KDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPF 926
KDHFPLPFIDQMLERLA H ++C+LDGYSGFFQIPIHPNDQEKTTFTCP+GTFAY+RMPF
Sbjct: 1041 KDHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEKTTFTCPYGTFAYKRMPF 1100
Query: 927 GLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVL 986
GLCNAPATFQRCM SIFSD +++++EVFMDDFSV+G +F CL NL +VL RCE+ NLVL
Sbjct: 1101 GLCNAPATFQRCMTSIFSDLIKEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNLVL 1160
Query: 987 NWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFI 1046
NWEKCHFMV+EGIVLGH + ++GIEVD+ K+E++ ++ PP +VK+IRSFLGHAGFYRRFI
Sbjct: 1161 NWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVKDIRSFLGHAGFYRRFI 1220
Query: 1047 KDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASD 1106
KDFS I +PLT LL K+ +F FDD CL++F +K+AL++AP+++ P+W+ PFEIMCDASD
Sbjct: 1221 KDFSKIARPLTRLLCKETEFKFDDDCLKSFQTIKDALVSAPVVRAPNWDYPFEIMCDASD 1280
Query: 1107 YAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIV 1166
YAVGAVLGQ+ DKK+H IYYAS+TLD AQ YATTEKELLAVV+A +KFR YLVGSK+ V
Sbjct: 1281 YAVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLAVVFAFEKFRSYLVGSKVTV 1340
Query: 1167 YTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELP 1226
YTDH+A+++L KKD KPRL+RWILLLQEFD+EI DKKG+EN ADHLSR+R ++ L
Sbjct: 1341 YTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGAADHLSRMR--IEEPLL 1398
Query: 1227 LDDSFPDDQLFLL-----------------AQTDAPWYADFVNFLAAGVLPPELNYQQKK 1269
+DDS P++QL ++ + ++PWYAD VN+LA GV PP L ++K
Sbjct: 1399 IDDSMPEEQLMVVEFFGKSYSGKEFHQLNAVEGESPWYADHVNYLACGVEPPNLTSYERK 1458
Query: 1270 KFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKIL 1329
KFF D+ HYYWDEPYL+ D I+RRC+ E+EV IL HCH S+YGGH +T KT KIL
Sbjct: 1459 KFFRDIHHYYWDEPYLYTLCKDKIYRRCVSEDEVEGILLHCHGSAYGGHFATFKTVSKIL 1518
Query: 1330 HSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSS 1389
+GFWWP++FKD F+SKCD CQR G+I++RNEMP N ILEVEIFDVWGIDFMGPFPSS
Sbjct: 1519 QAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEMPQNPILEVEIFDVWGIDFMGPFPSS 1578
Query: 1390 FGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRH 1449
+GN+YILVAVDYVSKWVEAIASPTNDA+VV+K+FK +IFPRFGVPRVVISDGG HFI++
Sbjct: 1579 YGNKYILVAVDYVSKWVEAIASPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKV 1638
Query: 1450 FEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYR 1509
FE LL+K GV+HK+ATPY+PQTSGQVE+SNR+IK ILEKTV +R DWS KLDDALWAYR
Sbjct: 1639 FENLLKKHGVKHKVATPYNPQTSGQVEISNREIKTILEKTVGITRKDWSAKLDDALWAYR 1698
Query: 1510 TAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRM 1569
T +KTPIG TPF L+YGKSCHLPVELE+KA WA++ LN D A +KR +QL++L+E+R+
Sbjct: 1699 TTFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLSDLDEIRL 1758
Query: 1570 DAYENARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTVY 1629
+AYE+++IYKERTK +HDKKII + F+ GD VLLFNSRLKLFPGKL+SRWSGPF + V
Sbjct: 1759 EAYESSKIYKERTKLFHDKKIITKDFQVGDQVLLFNSRLKLFPGKLKSRWSGPFCITEVR 1818
Query: 1630 PYGAIEIFSEETGSFTVNGQRLKIYNTGEVNEVVADFTLSD 1670
PYGA+ + + ++G FTVNGQRLK Y ++ V L +
Sbjct: 1819 PYGAVTL-TGKSGDFTVNGQRLKKYLADQILPEVTSVHLQE 1858
>gb|AAF24531.1| F7F22.17 [Arabidopsis thaliana]
Length = 1799
Score = 1516 bits (3924), Expect = 0.0
Identities = 802/1466 (54%), Positives = 1023/1466 (69%), Gaps = 144/1466 (9%)
Query: 305 KQSEQLQELKDQTRLLNNSLATLTTKIDSISSHNKISET--QLSQVVRK-VNHPNKM-NA 360
K SE +L LN + TL TK+ + H+ S Q SQ+ K V +P + +A
Sbjct: 374 KISELHNKLDCSYNDLNVKMETLDTKVRYLEGHSTSSSATKQTSQLPGKAVQNPKEYAHA 433
Query: 361 VTLRSGKPL---EDPIRRTETDNSEKEICEPPSRET-RAERE------------------ 398
+TLRSGK L E+P +T T++SE + E S E +A++
Sbjct: 434 ITLRSGKALPTREEP--KTVTEDSEDQDGEDLSLEKDQADKPLDLSLEQPLDLSLQQSLD 491
Query: 399 -----------------------KPRVEEN-----LPPPFKPKIPFPQRFAKSKLDEQFK 430
KP +N +PPP+K ++PFP R K+ D+
Sbjct: 492 PPLDSFTRPTTRPVIPAASPTAPKPVAVKNKEKVFVPPPYKSQLPFPGRHKKALADKYRA 551
Query: 431 KFIEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL- 489
F + + ++ + +P + L +P KFLK+++ ++ + E V L+ ECSAIIQ K+
Sbjct: 552 MFAKNIKEVELRIPLVDALALIPDSHKFLKDLIVERIQ-EVQGMVVLSHECSAIIQKKII 610
Query: 490 PPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADR 549
P KL DPGSF++PC +G + +CDLGASVSLMPLS+ +RLG + KS ++L LADR
Sbjct: 611 PKKLSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADR 670
Query: 550 SVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLA 609
SV+ P G++E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+IDVKKGK+
Sbjct: 671 SVRIPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKID 730
Query: 610 FNVGKE-TVEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGTK 668
N+GK+ + F++ MK P+I+ I+ +D E D L L +
Sbjct: 731 LNLGKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLEELAEEDHLNSALTKSGEDG 790
Query: 669 -LEGEAKAYEELLDRTLPMK---------GLSVEELVKEE-------PTLL--------- 702
L E Y++LLD M+ G + E +V E P L
Sbjct: 791 FLHLETLGYQKLLDSHKAMEESEPFEELNGPATEVMVMSEEGSTRVQPALSRTYSSNHST 850
Query: 703 -----PKE-------------APKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEK 744
P+E APKV+LK LP LRY FLGPNSTYPVI+NA L+ E
Sbjct: 851 LSTDEPREPIIPTSDDWSELKAPKVDLKPLPKGLRYAFLGPNSTYPVIINAELNSDEVNL 910
Query: 745 LLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVL 804
LL LKKY +AIGY++ DIKGI+PSLC HRI LE + SIE QRRLNPN+KEVVKKE+L
Sbjct: 911 LLSELKKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPQRRLNPNLKEVVKKEIL 970
Query: 805 KLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKA 864
KLLDAGVIYPISD + VPKK G+TV+KN+K+E I TRT+TG RMCIDYRKLN A
Sbjct: 971 KLLDAGVIYPISD------MHCVPKKDGMTVVKNEKDELIPTRTITGHRMCIDYRKLNAA 1024
Query: 865 TRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRM 924
+RKDHFPLPFIDQMLERLA H ++C+LDGY+GFFQIPIHPNDQEKTTFTCP+GTFAY+RM
Sbjct: 1025 SRKDHFPLPFIDQMLERLANHPYYCFLDGYNGFFQIPIHPNDQEKTTFTCPYGTFAYKRM 1084
Query: 925 PFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNL 984
PFGLCNAPATFQRCM SIFSD +E+++EVFMDDFSV+G +F CL NL +VL RCE+ NL
Sbjct: 1085 PFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNL 1144
Query: 985 VLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRR 1044
VLNWEKCHFMV+EGIVLGH + ++GIEVD+ K+E++ ++ PP +VK+IRSFLGHAGFYRR
Sbjct: 1145 VLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVKDIRSFLGHAGFYRR 1204
Query: 1045 FIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDA 1104
FIKDFS I +PLT LL K+ +F FDD CL++F +K+AL++AP+++ P+W+ PFEIMCDA
Sbjct: 1205 FIKDFSKIARPLTRLLCKETEFKFDDDCLKSFQTIKDALVSAPVVRAPNWDYPFEIMCDA 1264
Query: 1105 SDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKI 1164
SDYAVGAVLGQ+ DKK+H IYYAS+TLD AQ YATTEKELL VV+A +KFR YLVGSK+
Sbjct: 1265 SDYAVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLVVVFAFEKFRSYLVGSKV 1324
Query: 1165 IVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDE 1224
VYTDH+A+++L KKD KPRL+RWILLLQEFD+EI DKKG+EN ADHLSR+R ++
Sbjct: 1325 TVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGAADHLSRMR--IEEP 1382
Query: 1225 LPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPY 1284
L +DDS P++QL ++ +F +G K F+ L + P+
Sbjct: 1383 LLIDDSMPEEQLMVV---------EFFGKSYSG------------KEFHQLNAVEGESPW 1421
Query: 1285 LFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHL 1344
RC+ E+EV IL HCH S+YGGH +T KT KIL +GFWWP++FKD
Sbjct: 1422 -----------RCVSEDEVEGILLHCHGSAYGGHFATFKTVSKILQAGFWWPTMFKDAQE 1470
Query: 1345 FISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSK 1404
F+SKCD CQR G+I++RNEMP N ILEVEIFDVWGIDFMGPFPSS+GN+YILVAVDYVSK
Sbjct: 1471 FVSKCDSCQRKGNISRRNEMPQNPILEVEIFDVWGIDFMGPFPSSYGNKYILVAVDYVSK 1530
Query: 1405 WVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIA 1464
WVEAIASPTNDA+VV+K+FK +IFPRFGVPRVVISDGG HFI++ FE LL+K GV+HK+A
Sbjct: 1531 WVEAIASPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVFENLLKKHGVKHKVA 1590
Query: 1465 TPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLV 1524
TPY+PQTSGQVE+SNR+IK ILEKTV +R DWS KLDDALWAYRTA+KTPIG TPF L+
Sbjct: 1591 TPYNPQTSGQVEISNREIKTILEKTVGITRKDWSAKLDDALWAYRTAFKTPIGTTPFNLL 1650
Query: 1525 YGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKT 1584
YGKSCHLPVELE+KA WA++ LN D A +KR +QL++L+E+R++AYE+++IYKERTK
Sbjct: 1651 YGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLSDLDEIRLEAYESSKIYKERTKL 1710
Query: 1585 WHDKKIIKRHFKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSF 1644
+HDKKII + F+ G VLLFNSRLKLFPGKL+SRWSGPF + V PYGA+ + + ++G F
Sbjct: 1711 FHDKKIITKDFQFGVQVLLFNSRLKLFPGKLKSRWSGPFCITEVRPYGAVTL-AGKSGDF 1769
Query: 1645 TVNGQRLKIYNTGEVNEVVADFTLSD 1670
TVNGQRLK Y ++ V L +
Sbjct: 1770 TVNGQRLKKYLADQILPEVTSVHLQE 1795
Score = 135 bits (341), Expect = 9e-30
Identities = 81/251 (32%), Positives = 127/251 (50%), Gaps = 22/251 (8%)
Query: 45 NQFSGSPSEDPNLHISTFWRLSVTCKDD---QETVRLYLFPFSLKDKASNWFNSLKPGSI 101
N+F G P EDP H+ F RL K + ++ +L LFPFSL DKA W +L SI
Sbjct: 35 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSI 94
Query: 102 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQKNEESLFDVWEHFKEILRRCPHHGLEKW 161
T+WD ++ FL +FF ++TA+LR ++ F+QK ES + WE FK +CPHHG K
Sbjct: 95 TTWDDCKKVFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQCPHHGFTKA 154
Query: 162 LIIHTFYNGLTSSTKLTVDTAAGGALMNKDFTTAYALIENMALNLFQWTEE-----KATI 216
++ T Y G+ ++ +DTA+ G NKD + L+EN+A + + E+ + T
Sbjct: 155 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSDGNYNEDCDRTVRGTA 214
Query: 217 DPSPSKKEAGMDETSSIDYLSTKVNALSQRLDRMSTPTFSPPCETCDLSSHSNTDCNLSS 276
D S D ++ AL+ +LDR+ D + D +
Sbjct: 215 D--------------SDDKHRKEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQDGEGNQ 260
Query: 277 VEQLNFVQNGQ 287
+E+++++ N Q
Sbjct: 261 LEEVSYINNNQ 271
>dbj|BAB02630.1| retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 897
Score = 1239 bits (3206), Expect = 0.0
Identities = 599/929 (64%), Positives = 736/929 (78%), Gaps = 47/929 (5%)
Query: 731 VIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRR 790
+I+NA L+ E L L+KY +AIGY++ DIKGI+PSLC HRI LE + SIE QRR
Sbjct: 1 MIINAELNSDEVNLQLSELRKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPQRR 60
Query: 791 LNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVT 850
LNPN DAGVIYPISDS WVS V VPKKGG+TV+KN+K+E I TRT+T
Sbjct: 61 LNPNF------------DAGVIYPISDSTWVSLVYCVPKKGGMTVVKNEKDELIPTRTIT 108
Query: 851 GWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKT 910
G RMCIDYRKLN A+RKDHFPLPFIDQMLERLA H ++C+LDGYSGFFQIPIHPNDQEKT
Sbjct: 109 GHRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEKT 168
Query: 911 TFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLT 970
TFTCP+GTFAY+RMPFGLCNAPATFQRCM SIFSD +E+++EVFMDDFS +G +F CL
Sbjct: 169 TFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSGYGPSFSSCLL 228
Query: 971 NLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVK 1030
NL +VL RCE+ NLVLNWEKCHFMV+EGIVLGH + ++GIEVD+ K+E++ ++ PP +VK
Sbjct: 229 NLGRVLTRCEETNLVLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVK 288
Query: 1031 EIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQ 1090
EIRSFLGHAGFYRRFIKDFS I +PLT LL K+ +F FD+ CL++F +K+AL++AP+++
Sbjct: 289 EIRSFLGHAGFYRRFIKDFSKIVRPLTRLLCKETEFEFDEDCLKSFQTIKDALVSAPVVR 348
Query: 1091 PPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVY 1150
P+W+ PFEIMCDASDY VGAVLGQ+ DKK+H IYYAS+TLD AQ YATTEKELLAVV+
Sbjct: 349 APNWDYPFEIMCDASDYVVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLAVVF 408
Query: 1151 AIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVV 1210
A +KFR YLVGSK+ VYTDH+A+++L KKD KPRL+RWILLLQEFD+EI DKKG+EN
Sbjct: 409 AFEKFRSYLVGSKVTVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGA 468
Query: 1211 ADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKK 1270
ADHL R+R ++ LP+DDS P++QL ++ +F +G K
Sbjct: 469 ADHLLRMR--IEEPLPIDDSMPEEQLMVV---------EFFGKSYSG------------K 505
Query: 1271 FFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILH 1330
F+ L + P+ RC+ E+EV IL HCH S+YGGH +T KT KIL
Sbjct: 506 EFHQLNDVEGESPW-----------RCVSEDEVEGILLHCHCSAYGGHFATFKTVSKILQ 554
Query: 1331 SGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSF 1390
+GFWWP++FKD F+SKCD CQR +I++ NEMP N I+EVEIFDVWGIDFMG FPSS+
Sbjct: 555 AGFWWPTMFKDAQEFVSKCDSCQRKDNISRINEMPQNPIVEVEIFDVWGIDFMGLFPSSY 614
Query: 1391 GNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHF 1450
GN+YILVA+DYVSKWVEAIA PTNDA+VV+K+FK +IFPRFGVPRVVISDGG HFI++ F
Sbjct: 615 GNKYILVAIDYVSKWVEAIAIPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVF 674
Query: 1451 EKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRT 1510
E LL+K GV+HK+ATPYHPQTSGQVE+S+R+IK ILEKTV +R DWS KLDDALWAYRT
Sbjct: 675 ENLLKKHGVKHKVATPYHPQTSGQVEISDREIKTILEKTVGITRKDWSAKLDDALWAYRT 734
Query: 1511 AYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMD 1570
A+KTPIG TPF ++YGKSCHLPVELE+KA WA++ LN D A +KR +QL++L+E+R++
Sbjct: 735 AFKTPIGTTPFNILYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLSDLDEIRLE 794
Query: 1571 AYENARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTVYP 1630
AYE+++IYKERTK +HDKKII + F+ GD VLLFNSRLKLFPGKL+SRWSGPF + V P
Sbjct: 795 AYESSKIYKERTKLFHDKKIITKDFQVGDQVLLFNSRLKLFPGKLKSRWSGPFCITKVRP 854
Query: 1631 YGAIEIFSEETGSFTVNGQRLKIYNTGEV 1659
YGA+ + + ++G FTVNGQRLK Y ++
Sbjct: 855 YGAVTL-AGKSGDFTVNGQRLKKYLADQI 882
>gb|AAF79809.1| T32E20.9 [Arabidopsis thaliana]
Length = 1586
Score = 1125 bits (2910), Expect = 0.0
Identities = 554/901 (61%), Positives = 668/901 (73%), Gaps = 113/901 (12%)
Query: 754 KAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIY 813
KAIGY++DDIKGI+P+LC HRI LE + SIE QRRLNPN+KEVVKKE+LKLLDAGVIY
Sbjct: 779 KAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIY 838
Query: 814 PISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLP 873
PISDS WVSPV VPKKGG+TV+KN K+E I TRT+TG RMCI+YRKLN A+RK+HFPLP
Sbjct: 839 PISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLP 898
Query: 874 FIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPA 933
FID MLERLA H ++C+LD YSGFFQIPIHPNDQ KTTFTCP+GTFAY+RMPFGLCNAPA
Sbjct: 899 FIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPA 958
Query: 934 TFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHF 993
TFQRCM SIFSD +E+++EVFMDDFSV+GS+F CL NL +VL+RCE+ NLVLNWEKCHF
Sbjct: 959 TFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHF 1018
Query: 994 MVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSIT 1053
MVREGIVLG + + GIEVD+AKI+++ ++ PP +VK+IRSFLGHAGFYR FIKDFS +
Sbjct: 1019 MVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLA 1078
Query: 1054 KPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVL 1113
+PLT LL K+ +F FDD CL AF +KEALITAPI+Q P+W+ PFEI+
Sbjct: 1079 RPLTRLLCKETEFAFDDECLTAFKLIKEALITAPIVQAPNWDFPFEII------------ 1126
Query: 1114 GQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAI 1173
T+D AQV YATTEKELLAVV+A +KFR YLVGSK+ +YTDH+A+
Sbjct: 1127 ----------------TMDDAQVRYATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAAL 1170
Query: 1174 KYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPD 1233
+++ KKD KPRL+RWILLLQEFD+EI DKKG+EN
Sbjct: 1171 RHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIEN------------------------- 1205
Query: 1234 DQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGI 1293
PWYAD VN+L +G PP L+ +KKKFF D+ H+YWDEPYL+ D I
Sbjct: 1206 --------EKLPWYADHVNYLVSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKI 1257
Query: 1294 FRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQ 1353
+R C+ E+E+ IL HCH +YGGH +T KT KIL +
Sbjct: 1258 YRTCVSEDEIEGILLHCHGFAYGGHFATFKTMSKILQA---------------------- 1295
Query: 1354 RTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPT 1413
EVE FDVWGIDFMGPFPSS+GN+YILVA+DYVSKWVEAIAS T
Sbjct: 1296 -----------------EVENFDVWGIDFMGPFPSSYGNKYILVAIDYVSKWVEAIASHT 1338
Query: 1414 NDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSG 1473
NDA+VV+K+FK +IFPRFGVPR+VISDGG HFI++ FE LL+K GV+HK
Sbjct: 1339 NDARVVLKLFKTIIFPRFGVPRIVISDGGKHFINKGFENLLKKHGVKHK----------- 1387
Query: 1474 QVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPV 1533
VE+SNR+IKAILEKTV ++R DWS KL+D LWAYRTA+KTPIG TPF L+YGKSCHLPV
Sbjct: 1388 -VEISNREIKAILEKTVGSTRKDWSAKLNDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPV 1446
Query: 1534 ELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKR 1593
ELE+KA WA++ LN D A +KR +QLN+L ++R++AYE+++IYKERTK++HDKKI+ R
Sbjct: 1447 ELEYKAMWAVKLLNFDIKTAEEKRLIQLNDLNKIRLEAYESSKIYKERTKSFHDKKIVSR 1506
Query: 1594 HFKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKI 1653
FK GD VLLFNSRL+LFPGKL+SRWSGPF V V PYGAI + + + G FTVNGQRLK
Sbjct: 1507 DFKVGDQVLLFNSRLRLFPGKLKSRWSGPFSVTAVRPYGAITL-AGKNGDFTVNGQRLKK 1565
Query: 1654 Y 1654
Y
Sbjct: 1566 Y 1566
Score = 216 bits (551), Expect = 4e-54
Identities = 131/321 (40%), Positives = 198/321 (60%), Gaps = 34/321 (10%)
Query: 353 NHPNKMNAVTLRSGKPL---EDPIRRTET-------------DNSEKEICEP----PSRE 392
N +A+TLRSGK L E P + TE +++EK I EP P+R
Sbjct: 459 NSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEKAIEEPILHQPTRP 518
Query: 393 TRAER----EKP---RVEEN--LPPPFKPKIPFPQRFAKSKLDEQFKKFIE-MMNKIYID 442
EKP + +EN +PPP+KP +PFP RF K + +++K +E + + +
Sbjct: 519 LAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMI-QKYKALLEKQLKNLEVT 577
Query: 443 VPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNK-LPPKLKDPGSFSI 501
+P + L +P K++K++++++ K E V L+ ECSAIIQ K +P KL DPGSF++
Sbjct: 578 MPLVDCLALIPDSNKYVKDMITERIK-EVQGMVVLSHECSAIIQQKIIPKKLGDPGSFTL 636
Query: 502 PCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDV 561
PC +G K +CDLGASVSLMPL + ++LG + K ++L LADRSV+ G++ED+
Sbjct: 637 PCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILADRSVRISHGLLEDL 696
Query: 562 PVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKE-TVEFE 620
PV +G V +P DFVV+EM+E+ + P++LGRPFLA A AIIDVKKGK+ N+G++ + F+
Sbjct: 697 PVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKIDLNLGRDLKMTFD 756
Query: 621 LAKLMKGPSIKDSYCMIDIID 641
+ MK P+I+ + I+ +D
Sbjct: 757 ITNTMKKPTIEGNIFWIEEMD 777
Score = 141 bits (355), Expect = 2e-31
Identities = 76/194 (39%), Positives = 109/194 (56%), Gaps = 22/194 (11%)
Query: 22 IVYPTVQGSNFKIKPALLILVQQNQFSGSPSEDPNLHISTFWRLSVTCKDD---QETVRL 78
IV P VQ +NF+IK L+ +VQ N+F G P EDP H+ F RL K + ++ +L
Sbjct: 61 IVPPPVQNNNFEIKSGLIAMVQSNKFHGLPMEDPLDHLDEFDRLCSLTKINRVSEDGFKL 120
Query: 79 YLFPFSLKDKASNWFNSLKPGSITSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQKNEES 138
LFPFSL DKA W SL GSITSW+ ++ FL +FF S+TA+LR + FTQ N E+
Sbjct: 121 RLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNET 180
Query: 139 LFDVWEHFKEILRRCPHHGLEKWLIIHTFYNGLTSSTKLTVDTAAGGALMNKDFTTAYAL 198
++ WE FK +CPHH ++ +DTA+ G +NKD + +
Sbjct: 181 FYEAWERFKGYQTQCPHH-------------------EMLLDTASNGNFLNKDVEDGWEV 221
Query: 199 IENMALNLFQWTEE 212
+EN+A + + E+
Sbjct: 222 VENLAQSDGNYNED 235
>gb|AAR00600.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|50901488|ref|XP_463177.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 743
Score = 870 bits (2247), Expect = 0.0
Identities = 448/823 (54%), Positives = 552/823 (66%), Gaps = 103/823 (12%)
Query: 833 LTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 892
+TV+ N +NE I RTVTGWRMCIDYRKLNKAT++DHFPLPFID+MLER+A HS FC+LD
Sbjct: 1 MTVVANAQNELIPQRTVTGWRMCIDYRKLNKATKEDHFPLPFIDEMLERMANHSFFCFLD 60
Query: 893 GYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIME 952
GYSG+ QIPIHP DQ KTTFTCP+GT+AYRRM FGLCNAPA+FQRCMM
Sbjct: 61 GYSGYHQIPIHPEDQSKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMM------------ 108
Query: 953 VFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEV 1012
S F D + ++ +V + + V G FD+
Sbjct: 109 ----------SIFSDMIEDIMEVF------------------MDDFSVYGK-TFDQ---- 135
Query: 1013 DRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSC 1072
+ PP ++K IRSFLGHAGFYRRFIKDFS+I +PLT+LL KDA F DD+C
Sbjct: 136 ----------IRPPVNIKGIRSFLGHAGFYRRFIKDFSTIARPLTNLLAKDAPFESDDAC 185
Query: 1073 LQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLD 1132
L++F LK+AL++APIIQPPDW L FEIMCDASD+AVGA
Sbjct: 186 LKSFEVLKKALVSAPIIQPPDWTLSFEIMCDASDFAVGA--------------------- 224
Query: 1133 GAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILL 1192
FR YLV +K+I+YTDH+A+KY L KKDAKPRL+RWILL
Sbjct: 225 ----------------------FRSYLVRAKVIIYTDHAALKYPLTKKDAKPRLLRWILL 262
Query: 1193 LQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVN 1252
LQEFDLEIKDKKGVEN VADHLSRL+ TN E P++D DD L + ++ PWYA+ VN
Sbjct: 263 LQEFDLEIKDKKGVENSVADHLSRLQITNMPEQPINDFLRDDMLMTVRDSN-PWYANIVN 321
Query: 1253 FLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHS 1312
++ +PP N + K + + + WDEPYL+R SDG+ RRC+P E I+ CH+
Sbjct: 322 YMVFKYIPPGKNQRNLKY---ESRRHIWDEPYLYRVCSDGLLRRCVPTEEGLKIIERCHA 378
Query: 1313 SSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEV 1372
S YGGH +T KI SGF+WP++++D F+ +C CQR G IT R+ MPL L+V
Sbjct: 379 SPYGGHYGVFRTQAKIWQSGFFWPTMYEDSKEFVRRCTSCQRQGGITARDAMPLTYNLQV 438
Query: 1373 EIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFG 1432
EIFDVWGIDFMGPFP S +YILVAVDYVSKWV+A+ DA+ KMF ++IFP+F
Sbjct: 439 EIFDVWGIDFMGPFPKSRNCEYILVAVDYVSKWVKAMPCSATDARHAKKMFIEIIFPKFD 498
Query: 1433 VPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVST 1492
PR+VISDGGSHFI + F LL+++G +H +ATPYHPQTSGQ E SN+QIK IL+KTV+
Sbjct: 499 TPRMVISDGGSHFIDKTFRDLLREMGAKHNVATPYHPQTSGQAETSNKQIKNILQKTVNK 558
Query: 1493 SRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNL 1552
T W ++L DAL AY AYKTPI M+P+++VY K C LPVELEH+AYWAIRN N+D
Sbjct: 559 MGTGWKDRLPDALCAYWIAYKTPIRMSPYQIVYRKPCRLPVELEHRAYWAIRNWNMDFEG 618
Query: 1553 AGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLKLF- 1611
AG+ RK+Q+ ELEE R AY NA+IYKERTK WHDK+ + FK GD VL+FNSR+KLF
Sbjct: 619 AGEWRKMQIAELEEWREKAYHNAKIYKERTKRWHDKRKKIKKFKPGDKVLMFNSRVKLFG 678
Query: 1612 PGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKIY 1654
GKLRS+W GPF V +GAI + E F VNGQRLKI+
Sbjct: 679 HGKLRSKWEGPFDVIDTSSHGAITLRDELGNIFKVNGQRLKIF 721
>pir||D84513 probable retroelement pol polyprotein [imported] - Arabidopsis
thaliana
Length = 841
Score = 849 bits (2193), Expect = 0.0
Identities = 459/919 (49%), Positives = 589/919 (63%), Gaps = 128/919 (13%)
Query: 394 RAEREKPRVEENLPPPFKPKIPFPQRFAKSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMP 453
R + + + ++ + PP+ PK+PFP R K + ++++ F E+M ++ P
Sbjct: 4 RKQTLEEKAKKTVLPPYVPKLPFPGRQRKIQREKEYSLFDEIMRQLQRYSP--------- 54
Query: 454 TYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVIGSEVVKKA 513
ECSAI+QN +P K +DPGSF +P IG +
Sbjct: 55 --------------------------ECSAILQNVIPVKREDPGSFVLPSRIGEYTFDRC 88
Query: 514 MCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPAD 573
+CDLGA VSLMP S+ +RLG T+M+L L DRS+ +P G+ EDV V+VG YIP D
Sbjct: 89 LCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYIPTD 148
Query: 574 FVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDS 633
FV++E++E+ + ++LGRPFL A+IDV+K K+ +G EF + ++M P+ +
Sbjct: 149 FVIIELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIGDIVQEFNMERIMSKPTTECQ 208
Query: 634 YCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGT-KLEGEAKA-YEELLDRTLPMKG--- 688
+DI+D V E D L+ L GEA + +LD + PM
Sbjct: 209 TFWVDIMDELVNELLAELNTEDPLQTVLTKEESEFGYLGEATTRFARILDSSSPMTKVVA 268
Query: 689 ---LSVEELVKEEPTLLPKE--------APKVELKTLPSNLRYEFLGPNSTYPVIVNASL 737
L E+ K PK+ APK+ELK LP+ LR FLGPNSTY VI+N L
Sbjct: 269 FAELGDNEVEKALVVSSPKDCDDWSELNAPKMELKPLPARLRVAFLGPNSTYLVIINPEL 328
Query: 738 DEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKE 797
+ VE+ LL L+KY KA+GY++DDI GI+P+LCMH I LE + S+EHQRRLN N+++
Sbjct: 329 NNVESALLLCELRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRD 388
Query: 798 VVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCID 857
VVKKE++KLLDAG+IYPISDS WVSPV VVPKKGG+ VIKN+KNE I TRTVTG RMCID
Sbjct: 389 VVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCID 448
Query: 858 YRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFG 917
YRKLN ATRKD+FPL FIDQMLERL+ ++C+LDGY GFFQI IHP+DQEKTTFTCP+G
Sbjct: 449 YRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYG 508
Query: 918 TFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLE 977
TFAYRRMPFGLCNAPATFQ CM IFSD +E MEVF+DDF VL+
Sbjct: 509 TFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDF---------------LVLQ 553
Query: 978 RCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLG 1037
RCE +LVLNWEK HFMVR+GIVLGH + ++G+EVDRAKIEI++ FLG
Sbjct: 554 RCEDKHLVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKIEIMR-------------FLG 600
Query: 1038 HAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLP 1097
HAGFYRRFIKDFS I +P T LL K+ FDD+CLQAF +KE+L++A I+QPP+W LP
Sbjct: 601 HAGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPEWELP 660
Query: 1098 FEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQ 1157
FE++CDASDY VGAVLGQR DKK+HAIYYAS+TLDGAQV
Sbjct: 661 FEVICDASDYVVGAVLGQRKDKKLHAIYYASRTLDGAQV--------------------- 699
Query: 1158 YLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRL 1217
IV+T H+A++YLL+KKDAKPRL+RWILLLQ+FDLEIKDKKG+EN VAD+LSRL
Sbjct: 700 -------IVHTVHAALRYLLSKKDAKPRLLRWILLLQDFDLEIKDKKGIENEVADNLSRL 752
Query: 1218 RETN----KDELP------LDDSFPDD---------QLFLL--AQTDAPWYADFVNFLAA 1256
R +D LP ++D + D+ +L L +++ PWYADF N+L+
Sbjct: 753 RVQEEVLMRDILPGENLASIEDCYMDEVGRLRVSTLELMTLHTGESNLPWYADFANYLSC 812
Query: 1257 GVLPPELNYQQKKKFFNDL 1275
V PP+ KKK ++
Sbjct: 813 EVPPPDFTGYFKKKLLKEV 831
>gb|AAD19780.2| hypothetical protein [Arabidopsis thaliana]
Length = 1048
Score = 809 bits (2089), Expect = 0.0
Identities = 449/953 (47%), Positives = 590/953 (61%), Gaps = 135/953 (14%)
Query: 364 RSGKPLEDPIRRTETDNSEKEICEPPSRETRAEREKPRVEENLP----PPFKPKIPFPQR 419
+SG+ L +PI + E + + E +E +EE PP+ PK+PFP R
Sbjct: 45 KSGREL-NPILKKEKAKEKSKAASDDLEEDNTGKESDTLEEKAKKTVLPPYVPKLPFPGR 103
Query: 420 FAKSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTE 479
K + ++++ F E+M ++ + +PF +++ + Y K LK+IL+ KR +EE + ++
Sbjct: 104 QRKIQREKEYSLFDEIMRQLQVKLPFLDLVQNVSIYRKHLKDILTNKRTLEEGHVL-ISH 162
Query: 480 ECSAIIQNKLPPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKS 539
ECSAI+QN FS +IG + +CDLGA VSLMP S+ +RLG
Sbjct: 163 ECSAILQNV--------ALFSR--MIGEYTFDRCLCDLGAGVSLMPFSVAKRLGDTNFTP 212
Query: 540 TRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGA 599
T+M+L L DRS+ +P G+ EDV V+VG YIP DFV++E++E+ + ++LGRPFL A
Sbjct: 213 TKMSLVLGDRSISFPVGVAEDVQVRVGNFYIPTDFVIIELDEEPRHRLILGRPFLNIVAA 272
Query: 600 IIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEV 659
+IDV+K K+ +G EF + ++M P+ + +DI+D V E D L+
Sbjct: 273 LIDVRKSKINLRIGDIVQEFNMERIMSKPTTECQTFWVDIMDELVNELLAELNTEDPLQT 332
Query: 660 CLVNNAGT-KLEGEAKA-YEELLDRTLPMKG------LSVEELVKEEPTLLPKE------ 705
L GEA + +LD + PM L E+ K PK+
Sbjct: 333 VLTKEESEFGYLGEATTRFARILDSSSPMTKVVAFAELGDNEVEKALVVSSPKDCDDWSE 392
Query: 706 --APKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKAIGYTIDDI 763
APK+ELK LP+ LR FLGPNSTY VI+N L+ VE+ LL L+KY KA+GY++DDI
Sbjct: 393 LNAPKMELKPLPARLRVAFLGPNSTYLVIINPELNNVESALLLCELRKYRKALGYSLDDI 452
Query: 764 KGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSP 823
GI+P+LCMH I LE + S+EHQRRLN N+++VVKKE++KLLDAG+IYPISDS WVSP
Sbjct: 453 TGISPTLCMHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLDAGIIYPISDSTWVSP 512
Query: 824 VQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLA 883
V VVPKKGG+ VIKN+KNE I TRTVTG RMCIDYRKLN ATRKD+FPL FIDQMLERL+
Sbjct: 513 VHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKDNFPLSFIDQMLERLS 572
Query: 884 KHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIF 943
++C+LDGY GFFQI IHP+DQEKTTFTCP+GTFAYRRMPFGLCNAPATFQ CM IF
Sbjct: 573 NQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGLCNAPATFQHCMKYIF 632
Query: 944 SDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGH 1003
SD +E M ERCE +LVLNWEK HFMVR+GIVLGH
Sbjct: 633 SDMIEDFM-------------------------ERCEDKHLVLNWEKSHFMVRDGIVLGH 667
Query: 1004 LVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKD 1063
+ ++G+EVDRAKIE++ M PP SVK+ ++
Sbjct: 668 KISEKGVEVDRAKIEVMMSMQPPNSVKD-----------------------------HEE 698
Query: 1064 ADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHA 1123
FDD+CLQAF +KE+L++A I+QPP+W LPFE++CDASDY VGAVLGQR DKK+HA
Sbjct: 699 QKSEFDDTCLQAFNIVKESLVSALIVQPPEWELPFEVICDASDYVVGAVLGQRKDKKLHA 758
Query: 1124 IYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAK 1183
IYYAS+TLDGAQV IV+T H+A++YLL+KKDAK
Sbjct: 759 IYYASRTLDGAQV----------------------------IVHTVHAALRYLLSKKDAK 790
Query: 1184 PRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETN----KDELP------LDDSFPD 1233
PRL+RWILLLQ+FDLEIKDKKG+EN VAD+LSRLR +D LP ++D + D
Sbjct: 791 PRLLRWILLLQDFDLEIKDKKGIENEVADNLSRLRVQEEVLMRDILPGENLASIEDCYMD 850
Query: 1234 D---------QLFLL--AQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDL 1275
+ +L L +++ PWYADF N+L+ V PP+ KKK ++
Sbjct: 851 EVGRLRVSTLELMTLHTGESNLPWYADFANYLSCEVPPPDFTGYFKKKLLKEV 903
Score = 136 bits (343), Expect = 5e-30
Identities = 74/163 (45%), Positives = 95/163 (57%), Gaps = 30/163 (18%)
Query: 1510 TAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRK-LQLNELEELR 1568
T YKTP+G TPF L+YGKSCHLPVELE+KA WA + +N D AG++R+ LQLNEL+E++
Sbjct: 912 TTYKTPVGTTPFNLIYGKSCHLPVELEYKALWATKLMNYDIKPAGERRRLLQLNELDEIQ 971
Query: 1569 MDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTV 1628
GD VLLFNS+LK+FPGKLRS+WSGPF + +
Sbjct: 972 -----------------------------GDKVLLFNSKLKIFPGKLRSKWSGPFVIMEM 1002
Query: 1629 YPYGAIEIFSEETGSFTVNGQRLKIYNTGEVNEVVADFTLSDP 1671
P G+ ++ F+ NGQRLK Y E L+DP
Sbjct: 1003 RPNGSAILWDNNGKPFSTNGQRLKPYLAHSTLEEGTAVPLTDP 1045
>emb|CAD39928.2| OSJNBa0091C12.6 [Oryza sativa (japonica cultivar-group)]
gi|50921841|ref|XP_471281.1| OSJNBa0091C12.6 [Oryza
sativa (japonica cultivar-group)]
Length = 781
Score = 736 bits (1900), Expect = 0.0
Identities = 410/846 (48%), Positives = 541/846 (63%), Gaps = 90/846 (10%)
Query: 498 SFSIPCVIGSEVVKKAMCDLGASVSLMPLSL-YERLGIGELKSTRMTLQLADRSVKYPAG 556
S +I C IG V+ A+ + ++M + ++ LG L+ T T +++ S G
Sbjct: 22 SSTIMCHIGGNSVR-ALYNPSVGANIMSATFAFDCLGDRSLEPTVRTFRISTNSTTEGLG 80
Query: 557 IIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKK-GKLAFNVGKE 615
II VP + V + DF V E+ + + IL+G P ++DV + G L +G+
Sbjct: 81 IISGVPTRHNSVEVILDFHVFEIYDFD---ILIGHPI----EKLLDVPETGVLNLKIGR- 132
Query: 616 TVEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGTKLEGEAKA 675
+ + L S+ +S + + I E +A + + LE + EGE +
Sbjct: 133 -IATSVPMLQSTNSLMESLPVPEPI-----EGVMAISPFETLESIFDESIEEFNEGEDET 186
Query: 676 YEELLDRTLPMKGLSVEELVKEEPTLLPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNA 735
E + +L K + LP AP +ELK LPS+L Y FL ++ VI++
Sbjct: 187 GETM-------------DLPKTD---LPSRAP-IELKPLPSSLSYAFLNSDAESLVIISD 229
Query: 736 SLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNM 795
L E E +L+ +L+K+ A GY++ D+KGI+P+LC HRILLE PS E QRRLN M
Sbjct: 230 KLSEREMARLIAILEKHSAAFGYSLQDLKGISPTLCTHRILLEPSSTPSREPQRRLNNAM 289
Query: 796 KEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMC 855
+EV+KKEVLKLL G+IYP+ S+WVSPVQVVPKKGG+TV++N NE I RTVTGWRMC
Sbjct: 290 REVIKKEVLKLLHTGIIYPVPYSEWVSPVQVVPKKGGMTVVENSNNELIPQRTVTGWRMC 349
Query: 856 IDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCP 915
IDYRKLNKAT+KDHF LPFID+MLERLA HS FC+LDGYS
Sbjct: 350 IDYRKLNKATKKDHFSLPFIDEMLERLANHSFFCFLDGYS-------------------- 389
Query: 916 FGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKV 975
+Y ++P ++ DD ++G F+ CL NL+KV
Sbjct: 390 ----SYHQIP---------------------------IYPDD-QIYGKTFNHCLENLDKV 417
Query: 976 LERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSF 1035
L+RC++ +LVLNWEK HFMVREGIVLGH + +RG+EVDRAKIE+I+++ PP + +RSF
Sbjct: 418 LQRCQEKDLVLNWEKYHFMVREGIVLGHQISERGVEVDRAKIEVIRQLPPPVNDMGVRSF 477
Query: 1036 LGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWN 1095
LGHAGFYRRFIKDFS I +PLT+LL D FDD C+++F LKE+LI+APIIQP DW+
Sbjct: 478 LGHAGFYRRFIKDFSKIARPLTALLANDVPVDFDDECMKSFKILKESLISAPIIQPSDWS 537
Query: 1096 LPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKF 1155
L FEIMCDASD+AVGAVLGQ D+K HAI YASKTL GAQ+NYATTEKELLAVV+AI+KF
Sbjct: 538 LLFEIMCDASDFAVGAVLGQTKDRKHHAITYASKTLTGAQLNYATTEKELLAVVFAIEKF 597
Query: 1156 RQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLS 1215
R YLVG+K++VYTDH+A+KYLL KKDAKPR+IRWILLLQEFD+EI DKKGVEN VADHLS
Sbjct: 598 RSYLVGAKVVVYTDHAALKYLLTKKDAKPRVIRWILLLQEFDIEIMDKKGVENYVADHLS 657
Query: 1216 RLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDL 1275
R++ TN +L ++D DD + +D PWYA VNF+A G +PP + KK+ +
Sbjct: 658 RMQITNMQKLLINDYLRDDMRLKVIDSD-PWYATIVNFMATGHVPP---VENKKRLIYES 713
Query: 1276 KHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWW 1335
+ + WD PYLFR SDG+ R C+ +E I+ CH++ YGGH +T KI SGF+W
Sbjct: 714 RRHLWDAPYLFRVCSDGLLRICVSTDEGIMIIEKCHAAPYGGHYGAFRTHAKIWQSGFFW 773
Query: 1336 PSLFKD 1341
PS++ D
Sbjct: 774 PSMYDD 779
>emb|CAD39877.2| OSJNBb0058J09.16 [Oryza sativa (japonica cultivar-group)]
gi|50922333|ref|XP_471527.1| OSJNBb0058J09.16 [Oryza
sativa (japonica cultivar-group)]
Length = 1045
Score = 723 bits (1867), Expect = 0.0
Identities = 356/566 (62%), Positives = 429/566 (74%), Gaps = 22/566 (3%)
Query: 1022 KMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKE 1081
++ PP ++K I SFLGHAGFYRRFIKDFS+I +PLT+LL KDA F F+D+CL++F LK+
Sbjct: 500 RLPPPVNIKGICSFLGHAGFYRRFIKDFSTIARPLTNLLAKDAPFEFNDACLRSFKILKK 559
Query: 1082 ALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATT 1141
AL++APIIQPPDW LPFEIMCDASD+AVGAVLGQ DKK HAI YASKTL GAQ+NYATT
Sbjct: 560 ALVSAPIIQPPDWTLPFEIMCDASDFAVGAVLGQTKDKKHHAICYASKTLTGAQLNYATT 619
Query: 1142 EKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIK 1201
EKELL VV+AIDKFR YLVG+K+I+YTDH+A+KYLL KKDAKPRL+RWILLLQEFD+EIK
Sbjct: 620 EKELLTVVFAIDKFRSYLVGAKVIIYTDHAALKYLLTKKDAKPRLLRWILLLQEFDIEIK 679
Query: 1202 DKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPP 1261
DKKGVEN VADHLSRL TN E P++D DD L +PP
Sbjct: 680 DKKGVENYVADHLSRLHITNMQEQPINDLLRDDMLMTC-------------------IPP 720
Query: 1262 ELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHAST 1321
N Q+K K+ + + WDEPYL+R SDG+ RRC+P +E I+ CH+S YGGH
Sbjct: 721 GEN-QRKLKY--ESHRHIWDEPYLYRVCSDGLLRRCVPTDEGLKIIERCHASPYGGHYGA 777
Query: 1322 QKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGID 1381
+T KI SGF+WP++++D FI +C CQR G IT R+ MPL L+VEIFDVWGID
Sbjct: 778 FRTQAKIWQSGFFWPTMYEDSKEFIRRCTSCQRQGGITARDAMPLTYNLQVEIFDVWGID 837
Query: 1382 FMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDG 1441
FMGPFP S +YILVAVDYVSKWVEA+ DA+ KMF + IFPRFG R+VISD
Sbjct: 838 FMGPFPKSRNCEYILVAVDYVSKWVEAMPCSAADARHAKKMFTETIFPRFGTRRMVISDR 897
Query: 1442 GSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKL 1501
G HFI + F LL+++ +H +ATPYHPQTSGQ E SN+QIK IL+KTV+ T+W ++L
Sbjct: 898 GFHFIDKTFRDLLREMEAKHNVATPYHPQTSGQAETSNKQIKNILQKTVNKMGTEWKDRL 957
Query: 1502 DDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQL 1561
DALWAYRTAYKTPIGM+P+++VYGKSC LPVELEH+AYWAIRN N+D AG+ RK+Q+
Sbjct: 958 QDALWAYRTAYKTPIGMSPYQIVYGKSCRLPVELEHRAYWAIRNSNMDFEGAGEWRKMQI 1017
Query: 1562 NELEELRMDAYENARIYKERTKTWHD 1587
ELEE R AY NA+IYKERTK WHD
Sbjct: 1018 AELEEWREMAYHNAKIYKERTKRWHD 1043
Score = 112 bits (279), Expect = 1e-22
Identities = 91/296 (30%), Positives = 147/296 (48%), Gaps = 46/296 (15%)
Query: 524 MPLSLYERLGIGELKST--RMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEE 581
+PLS + +G L+ T +T ++D ++ I++DV V E DF V E+++
Sbjct: 210 VPLSPPNPMELGFLRETVRELTSIMSDEWLRE---IVQDVLVYFEEREAILDFHVFEIQD 266
Query: 582 DN---QVPI---LLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDSYC 635
+ ++PI L+ P L + K+ + ++ F A++ ++ D
Sbjct: 267 FDILIRLPIEQLLINTPHLGSL---------KITLGENEFSIPFSRARI----TLTDPLP 313
Query: 636 MIDIIDHCVKECSLASTAHDGLEVCLVNNAGTKLEGEAKAYEELLDRTLPMKGLSVEELV 695
I++++ A H+ E L + ++ EA E L
Sbjct: 314 EIELVEEVT-----AVPPHESPEALLEDEVPDFIKEEADPGETL---------------- 352
Query: 696 KEEPTLLPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKA 755
+ P + P P +ELK LP LRY FL + PVI++ L + ET++L VL+K+
Sbjct: 353 -DLPIMEPPPQPPLELKPLPLGLRYAFLHNDREAPVIISDKLSKDETQRLPTVLEKHRSV 411
Query: 756 IGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGV 811
+GY++ D++GIN +LC HRI ++ PS E QRRLN M+EVVKKEVLKLL AG+
Sbjct: 412 LGYSLQDLRGINLALCTHRIPIDPKSTPSREPQRRLNNEMQEVVKKEVLKLLHAGI 467
Score = 43.1 bits (100), Expect = 0.080
Identities = 26/132 (19%), Positives = 61/132 (45%), Gaps = 6/132 (4%)
Query: 119 SKTAQLRGKLYQFTQKNEESLFDVWEHFKEILRRCPHHGLEKWLIIHTFYNGLTSSTKLT 178
++ R ++ F Q ES+ W F +++ P L +++++ F+ GL +
Sbjct: 39 TRITAFRVEILSFKQIKNESIGATWSRFTNLVQSGPTLSLPEYVLLQYFHTGLDKESAFY 98
Query: 179 VDTAAGGALMNKDFTTAYALIENMALNLFQWTEEKATIDPSPSKKEAGMDETSSIDYL-- 236
+D GG+ M+K + +++ + N T+ +P P + + E +I+ L
Sbjct: 99 LDITVGGSFMHKTPSEGRTILDRILENTSFMTQPN---EPQPKASLSKIVEPLTIEPLTE 155
Query: 237 -STKVNALSQRL 247
ST +++ +++
Sbjct: 156 PSTSASSIDEKV 167
>emb|CAE03895.1| OSJNBb0026I12.3 [Oryza sativa (japonica cultivar-group)]
gi|50921891|ref|XP_471306.1| OSJNBb0026I12.3 [Oryza
sativa (japonica cultivar-group)]
Length = 589
Score = 689 bits (1777), Expect = 0.0
Identities = 336/507 (66%), Positives = 409/507 (80%), Gaps = 5/507 (0%)
Query: 833 LTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 892
+TV+ N NE I RTVTGWRMCIDY+KLNKAT+KD+FPLP ID+MLERLA HS FC+LD
Sbjct: 1 MTVVANVNNELIPQRTVTGWRMCIDYQKLNKATKKDNFPLPVIDEMLERLANHSLFCFLD 60
Query: 893 GYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIME 952
GYSG+ QIPIHP DQ KTTFTCP+GT+AYRRM FGLCNAPA+FQRCMMSIFSD +E IME
Sbjct: 61 GYSGYHQIPIHPKDQCKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMMSIFSDMIEAIME 120
Query: 953 VFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEV 1012
VFMDDFS++G FD CL NL+KV +RC++ +LVLNWEKCHFMV EGIVLGH V +RGIEV
Sbjct: 121 VFMDDFSLYGKTFDHCLQNLDKVFQRCQEKDLVLNWEKCHFMVHEGIVLGHRVSERGIEV 180
Query: 1013 DRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSC 1072
DRAKIE+I ++ PP ++K IR+FLGHAGF+RRFIK+FS+I +PLT+LL KDA F FDD C
Sbjct: 181 DRAKIEVIDQLPPPVNIKGIRNFLGHAGFHRRFIKEFSTIARPLTNLLAKDAPFEFDDVC 240
Query: 1073 LQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLD 1132
L++F LK+AL++A IIQPPD LPFEIMCDASD+ VGAVLGQ DKK+HAI YAS+TL
Sbjct: 241 LKSFKTLKKALVSALIIQPPDGMLPFEIMCDASDFIVGAVLGQTKDKKLHAICYASETLA 300
Query: 1133 GAQVNYATTEKELLAVVYAIDKFRQY-LVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWIL 1191
GAQ NYATTEKELLAVV+AIDKFR+ LVG+K+IVYT+H+A+KYLL KKDAKPRL+RWI
Sbjct: 301 GAQSNYATTEKELLAVVFAIDKFRRSDLVGAKVIVYTEHAALKYLLTKKDAKPRLLRWIP 360
Query: 1192 LLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFV 1251
LLQEFDLEIKDKKGVEN VADHLSRL+ TN E P++ DD L + +D PWYA+ V
Sbjct: 361 LLQEFDLEIKDKKGVENSVADHLSRLQLTNMQEPPINYFLQDDMLMAVRNSD-PWYANIV 419
Query: 1252 NFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCH 1311
N++ + +P N Q+K K+ + + WDEPYL+R S+G+ RRC+P+ E I+ CH
Sbjct: 420 NYMVSKYVPQGEN-QRKLKY--ESHCHIWDEPYLYRVCSNGLLRRCVPKEEGFKIIERCH 476
Query: 1312 SSSYGGHASTQKTSFKILHSGFWWPSL 1338
++ Y GH +T KI S F+WP++
Sbjct: 477 AAPYRGHYGAFRTQAKIWQSRFFWPTM 503
Score = 79.7 bits (195), Expect = 8e-13
Identities = 35/57 (61%), Positives = 44/57 (76%)
Query: 1450 FEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALW 1506
F +LL+K+ +H IATPYHPQT+GQ E+SN+QIK IL+KTV T W +KL DALW
Sbjct: 505 FRELLRKMNAKHNIATPYHPQTNGQAEMSNKQIKNILQKTVHEMGTGWKDKLPDALW 561
>gb|AAF63114.1| Hypothetical protein [Arabidopsis thaliana] gi|25405147|pir||B96502
hypothetical protein F28H19.8 [imported] - Arabidopsis
thaliana
Length = 640
Score = 651 bits (1679), Expect = 0.0
Identities = 307/472 (65%), Positives = 379/472 (80%), Gaps = 18/472 (3%)
Query: 997 EGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPL 1056
EGIVLGH + +GIEVD+AKI+++ ++ P +VK+IRSFLGHAGFYRRFIKDFS I +PL
Sbjct: 170 EGIVLGHKISGKGIEVDKAKIDVMVRLQPLKTVKDIRSFLGHAGFYRRFIKDFSKIARPL 229
Query: 1057 TSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQR 1116
T LL K+A+F FD+ CL+AF +KEAL+ API+Q P+WN PFEIMCDASDYAVGAVLGQR
Sbjct: 230 TRLLCKEAEFDFDEDCLKAFHSIKEALVLAPIVQAPNWNHPFEIMCDASDYAVGAVLGQR 289
Query: 1117 NDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYL 1176
DKK+H IYYAS+TLD Q YATTEKELL VV+A +KFR YLVGSK+ VYTDH+A+K++
Sbjct: 290 IDKKLHVIYYASRTLDDTQSRYATTEKELLVVVFAFEKFRSYLVGSKVTVYTDHAALKHI 349
Query: 1177 LNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQL 1236
KKD KPRL+RWIL LQEFD+EI DK+G EN VADHLSR+R T + +P+DDS P++QL
Sbjct: 350 YAKKDTKPRLLRWILFLQEFDMEIVDKRGSENGVADHLSRMRMT--EAVPIDDSMPEEQL 407
Query: 1237 FLLAQT---------------DAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWD 1281
+ + PWY D VN+L AG++P EL+ QKKKFF D+ HYYWD
Sbjct: 408 LAVGTSFDKFKIETMCATRSGKLPWYVDLVNYLTAGIVPSELSSYQKKKFFRDINHYYWD 467
Query: 1282 EPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKD 1341
EP L+++G DG+FRRCI E EV +L CH+S YGGH +T KT K+L +G WWPS+FKD
Sbjct: 468 EPVLYKKGVDGLFRRCIAEEEVQGVLELCHNSPYGGHFATFKTVQKVLQAGLWWPSMFKD 527
Query: 1342 VHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMGPF-PSSFGNQYILVAVD 1400
H I++CD+CQR G+I++RNEMP N ILEVE+FDVWGIDFMGPF P+S+GN+YILVAVD
Sbjct: 528 AHEHITRCDRCQRVGNISRRNEMPQNPILEVEVFDVWGIDFMGPFEPASYGNKYILVAVD 587
Query: 1401 YVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEK 1452
YVSKWVEAIASPTND++VV+K+FK +IFPRFG+PRVVISDGGSHFI++ FEK
Sbjct: 588 YVSKWVEAIASPTNDSKVVLKLFKTIIFPRFGIPRVVISDGGSHFINKVFEK 639
>gb|AAG52026.1| polyprotein, putative; 77260-80472 [Arabidopsis thaliana]
gi|25405064|pir||B96492 probable polyprotein, 77260-80472
[imported] - Arabidopsis thaliana
Length = 884
Score = 645 bits (1665), Expect = 0.0
Identities = 397/975 (40%), Positives = 556/975 (56%), Gaps = 141/975 (14%)
Query: 265 SSHSNTDCNLSSVEQLNFVQNGQRVVQKSHIELLMENYFLKQSEQLQELKDQTRLLNNSL 324
S+ S +++ Q+ +V + + ++ F +QS + ++ T + N
Sbjct: 28 SNISTKSVDIARKTQVQYVAHVDSLATSEKMDQQRYMSFNQQSTHQKPVQQTTSFIQNHA 87
Query: 325 ATLTTKIDSISSHNKISETQLSQVVR-----KVNHPNKMNAVTLRSGKPLEDPIRRTETD 379
++ T+ K E+ L Q+++ + +K + IRR E +
Sbjct: 88 SS--TQAHPPPDTTKTIESMLEQILKGQEKQTTDFDHKTGCI-----------IRRCEWE 134
Query: 380 NSEKEICEPPSRETRAEREKPRVEENLPPPFKPKIPFPQRFAKSKLDEQFKKFIEMMNKI 439
+S E + + + + R +V KPK+PFP+ KSK + + MM+K+
Sbjct: 135 DSNIEQLDQ-AIDYKQRRSAAKVT-------KPKVPFPKSPRKSKQELDDARCKVMMDKL 186
Query: 440 YIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSF 499
+++P + + P +F+K I++K + E+ + ++ + S IIQNK+P KL D
Sbjct: 187 IVEMPLIDAVKSSPMIRQFVKRIVTKDM-LTETVVMTMSTQVSDIIQNKIPQKLPDR--- 242
Query: 500 SIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIE 559
+L+ T++TL LADRSV+ GII
Sbjct: 243 ------------------------------------DLEPTQITLVLADRSVRRSDGIIC 266
Query: 560 DVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEF 619
DVPV+VG YIP D VV+ E++ + P++LGRPFL T GAIIDV+KG + NVG T++F
Sbjct: 267 DVPVQVGTSYIPTDLVVLSYEKEPKDPLILGRPFLVTTGAIIDVRKGIIGLNVGDLTMQF 326
Query: 620 ELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGTKLEGEAKAYEEL 679
++ K++K P+I +D I E + T L+ L+ + ++ K Y +L
Sbjct: 327 DINKVVKKPTIDGKTFYLDTISFLADEFLMKMTLAVPLKHALIPS----IDKVPKGYGKL 382
Query: 680 LDRTLPMKGLSVEELVKEEPTLLPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDE 739
LDR V +LV +E E +G +S I + SL+
Sbjct: 383 LDRIE-----HVMQLVAQE----------------------ELIGTSSQAATIGDWSLE- 414
Query: 740 VETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVV 799
K PK D+K + P L L E+ + LN ++
Sbjct: 415 -----------KAPKV------DLKPLPPGL--RYAFLGENSTYHVIVNASLNKVELTLL 455
Query: 800 KKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYR 859
++ K A + Y + D +SP + + +K++ S+ R + +
Sbjct: 456 LSKLRKYRKA-LGYSLDDITGISPDLCMHR----IHLKDESKPSVEHRRRLNPNLKDAVK 510
Query: 860 K-----LNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTC 914
K LN ATRK+HFPL FIDQ+LERL+ H ++C LDGYSGFFQIPIHP+DQEKT FTC
Sbjct: 511 KEIMKKLNAATRKNHFPLQFIDQLLERLSNHKYYCVLDGYSGFFQIPIHPDDQEKTMFTC 570
Query: 915 PFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEK 974
P+GTFAY RMPFGLCNAPA F+RCMMSIF+D +E +EVFMDDFSV+GS+F+ CL NL K
Sbjct: 571 PYGTFAYSRMPFGLCNAPAIFERCMMSIFTDMIENFIEVFMDDFSVYGSSFEACLENLRK 630
Query: 975 VLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRS 1034
VL RCE+ NLVLNWEKCHFMV+EGIVLGH V GIEV++AKIE++ + SV
Sbjct: 631 VLARCEEKNLVLNWEKCHFMVQEGIVLGHKVSGAGIEVNKAKIEVMTSLQALDSVN---- 686
Query: 1035 FLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDW 1094
RF+KDFS I +PLT+LL KD F F+ C AF ++K AL++API+QP DW
Sbjct: 687 --------LRFVKDFSKIARPLTALLCKDVKFDFNSECHDAFNQIKNALVSAPIVQPLDW 738
Query: 1095 NLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDK 1154
NLPFEIMCDA DYAVGAVLGQR K+HAI+YAS+TLD AQ N ATTEKELLAVV+A DK
Sbjct: 739 NLPFEIMCDAGDYAVGAVLGQRKGNKLHAIHYASRTLDDAQRNNATTEKELLAVVFAFDK 798
Query: 1155 FRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHL 1214
FR YLVGSK+IV+TDHSA+KYL+ KKDAKPRL+RWILLLQEFD+E++D+KG+EN VADHL
Sbjct: 799 FRSYLVGSKVIVHTDHSALKYLMQKKDAKPRLLRWILLLQEFDIEVRDRKGIENGVADHL 858
Query: 1215 SRLRETNKDELPLDD 1229
SR++ +D++P++D
Sbjct: 859 SRIKV--EDDIPIND 871
>emb|CAB81136.1| putative athila transposon protein [Arabidopsis thaliana]
gi|25407395|pir||D85075 probable athila transposon
protein [imported] - Arabidopsis thaliana
Length = 724
Score = 627 bits (1616), Expect = e-177
Identities = 328/596 (55%), Positives = 413/596 (69%), Gaps = 46/596 (7%)
Query: 483 AIIQNKL-PPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTR 541
AI Q K+ P KL DPGSF++PC +G K+ +CDLGA VSLMPLS+ +RLG + KS
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 542 MTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAII 601
++L LADRSV+ P + E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 602 DVKKGKLAFNVGKET-VEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVC 660
DVKKGK+ N+GK + F++ MK P+IK I+ ID E D L
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLEERAEEDHLYSA 182
Query: 661 LVNNAGTK-LEGEAKAYEELLDRTLPMK---------GLSVEELVKEEP----------- 699
L L E Y++LLD M+ G E +V E
Sbjct: 183 LTKRGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPETEVMVMSEEGSTQVQPAHSR 242
Query: 700 ----------------TLLPK-------EAPKVELKTLPSNLRYEFLGPNSTYPVIVNAS 736
++P +APKV+LK+LP LRY F GPNSTYPVI+N
Sbjct: 243 TYSTNNSTSTISNSGELIIPTSDDWSELKAPKVDLKSLPKGLRYVFFGPNSTYPVIINVE 302
Query: 737 LDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMK 796
L++ E LL L+KY +AIGY++ DIK I+PSLC HRI LE + SIE QRRLN N+K
Sbjct: 303 LNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNLK 362
Query: 797 EVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCI 856
EVVKKE+LKLLDAGVIYPISDS WV PV VPKKGG+TV+KN+K+E I TRT+TG R+CI
Sbjct: 363 EVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCI 422
Query: 857 DYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPF 916
DYRKLN A+RKDHFPLPF +QMLE LA H + C+LDGYSGFFQIPIHPNDQEKTTFTCP+
Sbjct: 423 DYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPY 482
Query: 917 GTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVL 976
GTFAY+RMPFGLCNAP TFQRCM SIFSD +EK++EVFMDDFSV+G +F CL NL +VL
Sbjct: 483 GTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVL 542
Query: 977 ERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEI 1032
+ E+ NLVLNWEKC+FMV+EGIVLGH + ++GIEVD+ KI+++ ++ PP +VK+I
Sbjct: 543 TKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 146 bits (368), Expect = 7e-33
Identities = 73/121 (60%), Positives = 95/121 (78%), Gaps = 2/121 (1%)
Query: 1119 KKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLN 1178
K + IYYAS+TLD AQ YATTEKELL VV+A KFR YLV SK+ VYTDH+A++++
Sbjct: 593 KTVKDIYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYA 652
Query: 1179 KKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFL 1238
KKD KPRL+R ILLLQEFD+EI +KKG+EN ADHLSR+R ++ + +DDS P++QL +
Sbjct: 653 KKDTKPRLLRGILLLQEFDMEIVEKKGIENGAADHLSRMR--IEEPILIDDSMPEEQLMV 710
Query: 1239 L 1239
+
Sbjct: 711 V 711
>gb|AAD23707.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25411232|pir||G84476 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 466
Score = 594 bits (1532), Expect = e-168
Identities = 278/427 (65%), Positives = 342/427 (79%), Gaps = 1/427 (0%)
Query: 1244 APWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEV 1303
+PWYAD VN+LA G+ PP L ++K FF D+ HYYWDEPYL+ D I+R + E+EV
Sbjct: 37 SPWYADHVNYLACGIEPPNLTSYERKNFFRDIHHYYWDEPYLYTLCKDKIYRSYVSEDEV 96
Query: 1304 SSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNE 1363
IL HCH S+YGGH +T KT KIL +GFWWP++FKD F+SKCD CQR G+I++RNE
Sbjct: 97 EGILLHCHDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNE 156
Query: 1364 MPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMF 1423
MP N ILEV+IFDVWGIDFMGPFPSS+GN+YILV VDYVSKWVEAIASPTNDA+VV+K+F
Sbjct: 157 MPQNPILEVDIFDVWGIDFMGPFPSSYGNKYILVVVDYVSKWVEAIASPTNDAKVVLKLF 216
Query: 1424 KKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIK 1483
K +IFPRFGV VVISDGG HFI++ FE LL+K GV+HK+ATPYHPQTSGQVE+SNR+IK
Sbjct: 217 KTIIFPRFGVSWVVISDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSGQVEISNREIK 276
Query: 1484 AILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAI 1543
ILEKTV +R DWS KLDDALWAY+TA+KTPIG TPF L+ KSCHL VELE+KA WA+
Sbjct: 277 TILEKTVGITRKDWSTKLDDALWAYKTAFKTPIGTTPFNLLCVKSCHLHVELEYKAMWAV 336
Query: 1544 RNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLL 1603
+ LN D A +KR +QL+EL+E+R++AYE+++IYKERTK +HDKKII + F+ GD VLL
Sbjct: 337 KLLNFDIKTAEEKRLIQLSELDEIRLEAYESSKIYKERTKLFHDKKIITKDFQVGDQVLL 396
Query: 1604 FNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKIYNTGEVNEVV 1663
FNSRLK+FPGKL+SRWSGPF + V PYGA+ + + ++G FTVNGQRLK Y ++ V
Sbjct: 397 FNSRLKIFPGKLKSRWSGPFCITEVRPYGAVTL-AGKSGVFTVNGQRLKKYLANQILPEV 455
Query: 1664 ADFTLSD 1670
L +
Sbjct: 456 TSIHLQE 462
>emb|CAD39882.2| OSJNBb0067G11.5 [Oryza sativa (japonica cultivar-group)]
gi|50922247|ref|XP_471484.1| OSJNBb0067G11.5 [Oryza
sativa (japonica cultivar-group)]
Length = 791
Score = 590 bits (1521), Expect = e-166
Identities = 292/450 (64%), Positives = 346/450 (76%), Gaps = 18/450 (4%)
Query: 690 SVEELVKEEP-----TLLPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEK 744
S EE+ P LL E P + LRY+FL + PVI++ L E ET+
Sbjct: 355 SAEEVTAVPPHESPEALLEDEVPDFIKEEADPGLRYDFLHNDREAPVIISDKLSEDETQH 414
Query: 745 LLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVL 804
LL VL+K+ +GY++ D++GINP+LC HRIL++ + PS E QR LN M+EVVKKEVL
Sbjct: 415 LLTVLEKHRSILGYSLQDLRGINPALCTHRILIDPESTPSREPQRWLNNAMREVVKKEVL 474
Query: 805 KLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKA 864
KLL AG+IYPI S+WVSPVQVVPKKGG+TV+ WRMCIDYRKLNKA
Sbjct: 475 KLLHAGIIYPIPYSEWVSPVQVVPKKGGMTVVYR-------------WRMCIDYRKLNKA 521
Query: 865 TRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRM 924
T+KDHFPLPFID+MLERLA HS FC+LDGYSG+ QIPIHP DQ KTTFTCP+GT+AYRRM
Sbjct: 522 TKKDHFPLPFIDEMLERLANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPYGTYAYRRM 581
Query: 925 PFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNL 984
FGLCN PA+FQRCMMSIFSD +E IMEVFMDDFSV+G CL NL+KVL+RC++ +L
Sbjct: 582 SFGLCNTPASFQRCMMSIFSDMIEDIMEVFMDDFSVYGKTLGHCLQNLDKVLQRCQEKDL 641
Query: 985 VLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRR 1044
VLN EKCHFMVREGIVLGH V +RG+EVDRAKI++I ++ PP ++K I SF GHAGFYRR
Sbjct: 642 VLNREKCHFMVREGIVLGHRVSERGVEVDRAKIDVIDQLPPPMNIKGIHSFFGHAGFYRR 701
Query: 1045 FIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDA 1104
FIKDFS+I +PLT+LL KDA FDD CL++F LK+AL++APIIQPPDW LPFEIMCDA
Sbjct: 702 FIKDFSTIARPLTNLLAKDAPIEFDDVCLKSFEILKKALVSAPIIQPPDWTLPFEIMCDA 761
Query: 1105 SDYAVGAVLGQRNDKKMHAIYYASKTLDGA 1134
SD+AVGAVLGQ DKK HAI YASKTL GA
Sbjct: 762 SDFAVGAVLGQTKDKKHHAICYASKTLTGA 791
Score = 80.5 bits (197), Expect = 5e-13
Identities = 49/148 (33%), Positives = 75/148 (50%), Gaps = 11/148 (7%)
Query: 10 YSIPSSDEPCTIIVYPTVQGSNFKIKPALLILVQQNQFSGSPSEDPNLHISTFWRLSVTC 69
Y P S EP +Y T ++I+P L+ + ++N FSG E+P H+ F ++
Sbjct: 16 YEPPPSSEP----IYTT----GYEIRPELISMFRENPFSGFDLENPYHHLRDFEQVCSCL 67
Query: 70 KDD---QETVRLYLFPFSLKDKASNWFNSLKPGSITSWDQLRREFLCRFFPPSKTAQLRG 126
K QETV LFPFSL+++A W+ S SW++LR F FFP ++ LR
Sbjct: 68 KIRGMRQETVWWKLFPFSLQERAKQWYTSTVGCVNGSWEKLRDRFCLTFFPVTRITALRV 127
Query: 127 KLYQFTQKNEESLFDVWEHFKEILRRCP 154
++ F Q ES+ W F +++ P
Sbjct: 128 EILNFKQIENESIGAAWSRFTNLVQSGP 155
>pir||T12085 reverse transcriptase homolog - fava bean (fragment)
gi|2522228|dbj|BAA22787.1| reverse transcriptase-like
protein [Vicia faba]
Length = 407
Score = 583 bits (1502), Expect = e-164
Identities = 275/396 (69%), Positives = 329/396 (82%), Gaps = 4/396 (1%)
Query: 854 MCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFT 913
+CIDYR+LN ATRKDHFPLPFIDQMLERLA H ++C+LDGYSG+ QI + P DQEKTTFT
Sbjct: 1 VCIDYRRLNLATRKDHFPLPFIDQMLERLADHEYYCFLDGYSGYNQIAVVPEDQEKTTFT 60
Query: 914 CPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLE 973
CPFG F+YRR+PFGLCNAPATFQRCM SIF+D +EK MEVFMDDFSV G +FD+CL+NL
Sbjct: 61 CPFGIFSYRRIPFGLCNAPATFQRCMQSIFADMLEKYMEVFMDDFSVFGKSFDNCLSNLA 120
Query: 974 KVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIR 1033
VLERC++ NL+LNWEKCHFMVREGIVLGH + +GIEVD+AKIE+I K+ PPT+ K IR
Sbjct: 121 LVLERCQESNLILNWEKCHFMVREGIVLGHKISYKGIEVDQAKIEVISKLHPPTNEKGIR 180
Query: 1034 SFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPD 1093
SFLGH GFYRRFI+DFS I KPLT+LL+KD DF FD C+ AF LK L++API+ PD
Sbjct: 181 SFLGHVGFYRRFIRDFSKIEKPLTTLLVKDKDFVFDKECVVAFETLKGKLVSAPIVVAPD 240
Query: 1094 WNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAID 1153
W LPFEIMCDASD AVGAVLGQR +K++H IYYAS L+ AQ+NYATTEKELL +VYA D
Sbjct: 241 WYLPFEIMCDASDIAVGAVLGQRREKRLHVIYYASHVLNPAQMNYATTEKELLVMVYAFD 300
Query: 1154 KFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADH 1213
KFRQY++GSK+IVYTDH+A+KYL +K+D+KPRL+RWILLLQEFD+EI+DK+G EN VADH
Sbjct: 301 KFRQYMLGSKVIVYTDHAALKYLFSKQDSKPRLLRWILLLQEFDVEIRDKRGCENTVADH 360
Query: 1214 LSRLR--ETNKDELPLDDSFPDDQLFLLAQTDAPWY 1247
LSR+ E K + P+ D F D+ +L PW+
Sbjct: 361 LSRMSHIEETKVKQPIKDEFADEH--ILVVIGVPWF 394
>gb|AAV43951.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 963
Score = 541 bits (1394), Expect = e-152
Identities = 311/678 (45%), Positives = 414/678 (60%), Gaps = 47/678 (6%)
Query: 398 EKPRVE-ENLPPPFKPKIPFPQRFAKSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMPTYA 456
E+P VE E + P I F F DE + + +K P + P
Sbjct: 327 EQPSVENEEIQTPDCATILFKDGF-----DEDYGNTLNYFSK---RKPLVPLPPPDPMEL 378
Query: 457 KFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVI-GSEVVKKAMC 515
+FL+E + + I E + E S +I+ P++ IPC + G++V
Sbjct: 379 RFLRETIRELTTIMSDEWLREAELSSEVIRINTAPRI-------IPCHLEGNDVSILYSP 431
Query: 516 DLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFV 575
+GA++ + + L + T + ++ GI++DVPV + DF
Sbjct: 432 TVGANLVSGSFA-FAYLSDKAVTPTNKFFKHPSGNIIKRFGIMQDVPVCFEDKEAILDFH 490
Query: 576 VMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDSYC 635
V E+++ IL+G P + K+ + ++ F A+++ + +
Sbjct: 491 VFEIQD---FDILIGLPIEQLLINTPRLDSLKITLGETEFSIPFSRARIVLTDPLPEIES 547
Query: 636 MIDIIDHCVKECSLASTAHDGLEVCLVNNAGTKLEGEAKAYEELLDRTLPMKGLSVEELV 695
++I A H+ E L + ++ EA E L
Sbjct: 548 TEEVI---------AVPPHESPEALLEDEVPDFIKEEADPGETL---------------- 582
Query: 696 KEEPTLLPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKA 755
+ P + P P +ELK LP +L Y FL + PVI++ L E ET++LL VL+K+
Sbjct: 583 -DLPIMEPPPWPPLELKPLPPSLHYAFLHHDREAPVIISDKLFEDETQRLLAVLEKHCSV 641
Query: 756 IGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPI 815
+GY++ D+KGINP LC+HRI ++ + PS E QRRLN M+EVVKKEVLKLL AG+IYP+
Sbjct: 642 LGYSLQDLKGINPVLCIHRIPIDPESTPSREPQRRLNNAMREVVKKEVLKLLHAGIIYPV 701
Query: 816 SDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFI 875
S+WVSPVQVVPKKGG+ V+ N +NE I RTVT WRMCIDYRKLNKAT+KDHFPLPFI
Sbjct: 702 PYSEWVSPVQVVPKKGGIMVVANAQNELIPQRTVTEWRMCIDYRKLNKATKKDHFPLPFI 761
Query: 876 DQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATF 935
D+MLERLA HS FC+LDGYSG+ QIPIHP DQ KTTFTCP+GT+AYRRM FGLCNAPA+F
Sbjct: 762 DEMLERLANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPYGTYAYRRMSFGLCNAPASF 821
Query: 936 QRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMV 995
QRCMMSIFSD +E IMEVFMDDF V+G F L NL+KVL+RC++ +LVLNWEKCHFMV
Sbjct: 822 QRCMMSIFSDMIEYIMEVFMDDFLVYGKTFGHYLQNLDKVLQRCQEKDLVLNWEKCHFMV 881
Query: 996 REGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKP 1055
REGIVLGH V +RGIEVDRAKI++I ++ PP ++K IRSFLGHA FYRRFI+D+S+I +P
Sbjct: 882 REGIVLGHRVSERGIEVDRAKIDVIDQLPPPVNIKGIRSFLGHASFYRRFIEDYSTIVRP 941
Query: 1056 LTSLLLKDADFTFDDSCL 1073
LT+LL KDA F FDD+CL
Sbjct: 942 LTNLLAKDAPFEFDDACL 959
>gb|AAV43847.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1005
Score = 536 bits (1381), Expect = e-150
Identities = 309/676 (45%), Positives = 412/676 (60%), Gaps = 47/676 (6%)
Query: 398 EKPRVE-ENLPPPFKPKIPFPQRFAKSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMPTYA 456
E+P VE E + P I F F DE + + +K P + P
Sbjct: 332 EQPSVENEEIQTPDCATILFKDGF-----DEDYGNTLNYFSK---RKPLVPLPPPDPMEL 383
Query: 457 KFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVI-GSEVVKKAMC 515
+FL+E + + I E + E S +I+ P++ IPC + G++V
Sbjct: 384 RFLRETIRELTTIMSDEWLREAELSSEVIRINTAPRI-------IPCHLEGNDVSILYSP 436
Query: 516 DLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFV 575
+GA++ + + L + T + ++ GI++DVPV + DF
Sbjct: 437 TVGANLVSGSFA-FAYLSDKAVTPTNKFFKHPSGNIIKRFGIMQDVPVCFEDKEAILDFH 495
Query: 576 VMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDSYC 635
V E+++ IL+G P + K+ + ++ F A+++ + +
Sbjct: 496 VFEIQD---FDILIGLPIEQLLINTPRLDSLKITLGETEFSIPFSRARIVLTDPLPEIES 552
Query: 636 MIDIIDHCVKECSLASTAHDGLEVCLVNNAGTKLEGEAKAYEELLDRTLPMKGLSVEELV 695
++I A H+ E L + ++ EA E L
Sbjct: 553 TEEVI---------AVPPHESPEALLEDEVPDFIKEEADPGETL---------------- 587
Query: 696 KEEPTLLPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKA 755
+ P + P P +ELK LP +L Y FL + PVI++ L E ET++LL VL+K+
Sbjct: 588 -DLPIMEPPPWPPLELKPLPPSLHYAFLHHDREAPVIISDKLFEDETQRLLAVLEKHCSV 646
Query: 756 IGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPI 815
+GY++ D+KGINP LC+HRI ++ + PS E QRRLN M+EVVKKEVLKLL AG+IYP+
Sbjct: 647 LGYSLQDLKGINPVLCIHRIPIDPESTPSREPQRRLNNAMREVVKKEVLKLLHAGIIYPV 706
Query: 816 SDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFI 875
S+WVSPVQVVPKKGG+ V+ N +NE I RTVT WRMCIDYRKLNKAT+KDHFPLPFI
Sbjct: 707 PYSEWVSPVQVVPKKGGIMVVANAQNELIPQRTVTEWRMCIDYRKLNKATKKDHFPLPFI 766
Query: 876 DQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATF 935
D+MLERLA HS FC+LDGYSG+ QIPIHP DQ KTTFTCP+GT+AYRRM FGLCNAPA+F
Sbjct: 767 DEMLERLANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPYGTYAYRRMSFGLCNAPASF 826
Query: 936 QRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMV 995
QRCMMSIFSD +E IMEVFMDDF V+G F L NL+KVL+RC++ +LVLNWEKCHFMV
Sbjct: 827 QRCMMSIFSDMIEYIMEVFMDDFLVYGKTFGHYLQNLDKVLQRCQEKDLVLNWEKCHFMV 886
Query: 996 REGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKP 1055
REGIVLGH V +RGIEVDRAKI++I ++ PP ++K IRSFLGHA FYRRFI+D+S+I +P
Sbjct: 887 REGIVLGHRVSERGIEVDRAKIDVIDQLPPPVNIKGIRSFLGHASFYRRFIEDYSTIVRP 946
Query: 1056 LTSLLLKDADFTFDDS 1071
LT+LL KDA F FDD+
Sbjct: 947 LTNLLAKDAPFEFDDA 962
>emb|CAD40178.2| OSJNBa0061A09.17 [Oryza sativa (japonica cultivar-group)]
gi|50921885|ref|XP_471303.1| OSJNBa0061A09.17 [Oryza
sativa (japonica cultivar-group)]
Length = 430
Score = 522 bits (1345), Expect = e-146
Identities = 254/444 (57%), Positives = 318/444 (71%), Gaps = 15/444 (3%)
Query: 1101 MCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLV 1160
MCDAS++AVGAVLGQ D+K HAI YASKTL GAQ+NY+TTEKELLAVV
Sbjct: 1 MCDASNFAVGAVLGQTKDRKQHAIAYASKTLTGAQLNYSTTEKELLAVV----------- 49
Query: 1161 GSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRET 1220
G+K++VYTDH+A+KYLL KKDAKPRLIRWIL+LQEFD+EI +KKG+EN VADHLSR++ T
Sbjct: 50 GAKVVVYTDHAALKYLLTKKDAKPRLIRWILILQEFDIEINNKKGIENSVADHLSRMQIT 109
Query: 1221 NKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYW 1280
N EL ++D DD L + +D PWYA VNF+ A +PP N KK+ + + W
Sbjct: 110 NMQELSINDYLRDDMLLKVTDSD-PWYAPIVNFMVAWHVPPGEN---KKRLSYESRKRLW 165
Query: 1281 DEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFK 1340
D PYL+R SD + RRC+P +E I+ C + YG H +T KI SGF+WP+++
Sbjct: 166 DAPYLYRVCSDSLLRRCVPADEGMEIIEKCRVAPYGSHYGAFRTHAKIWQSGFFWPTMYG 225
Query: 1341 DVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVD 1400
D FI +C CQ+ G IT ++ MPL L+VE+FDVWGIDFMGPFP + +YILVAVD
Sbjct: 226 DTKEFIRRCTSCQKHGGITAQDAMPLTYNLQVELFDVWGIDFMGPFPKFYDCEYILVAVD 285
Query: 1401 YVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVR 1460
YVSKWVEA+ + + +MF ++IFP FG PR VISDGGSHFI + F+ LQ+LG +
Sbjct: 286 YVSKWVEAMPCRAANTKHAHRMFDEIIFPCFGTPRTVISDGGSHFIDKTFQNFLQELGAQ 345
Query: 1461 HKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTP 1520
H I TPYHP+T GQ E N+QI IL+KTV+ W NKL +ALWAYRT Y PIGM+P
Sbjct: 346 HNIVTPYHPKTGGQAETRNKQIMNILQKTVNEMGKAWKNKLPNALWAYRTTYNMPIGMSP 405
Query: 1521 FKLVYGKSCHLPVELEHKAYWAIR 1544
++LVYGK+CHLPVELEHKA+WAIR
Sbjct: 406 YQLVYGKTCHLPVELEHKAHWAIR 429
>gb|AAM15254.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25411471|pir||B84505 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 930
Score = 489 bits (1259), Expect = e-136
Identities = 228/364 (62%), Positives = 282/364 (76%), Gaps = 6/364 (1%)
Query: 1241 QTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPE 1300
+ D PWYAD VN+LAA V P KK+F +++ Y WDEPYL++ DGI+RRCI
Sbjct: 133 EKDYPWYADIVNYLAADVEPDNFTDYNKKRFLREIRRYQWDEPYLYKHSYDGIYRRCIAA 192
Query: 1301 NEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITK 1360
EV SIL+HCHSSSYGGH +T KT K+L + FWWP++F+D FIS+CD CQR G I+K
Sbjct: 193 TEVPSILSHCHSSSYGGHFATFKTVSKVLQADFWWPTMFRDTQKFISQCDPCQRRGKISK 252
Query: 1361 RNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVI 1420
RNEMP N LEVE+FD WGIDFMGPFP S N YILV VDYVSKWVEAIAS ND+ VV+
Sbjct: 253 RNEMPPNFRLEVEVFDRWGIDFMGPFPPSNKNLYILVDVDYVSKWVEAIASLKNDSAVVM 312
Query: 1421 KMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNR 1480
K+FK +IFPRFGVPR+VISDG HFI++ EKLL + GV+H++ATPYHPQTSGQVEVSNR
Sbjct: 313 KLFKSIIFPRFGVPRIVISDGDKHFINKILEKLLLQYGVQHRVATPYHPQTSGQVEVSNR 372
Query: 1481 QIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAY 1540
QIK ILEKTV ++ +WS KL DALWAY+TA+KTP+G TPF L+YGK+CHLPVELEHKA
Sbjct: 373 QIKEILEKTVGKAKKEWSYKLYDALWAYKTAFKTPLGTTPFHLLYGKACHLPVELEHKAA 432
Query: 1541 WAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDL 1600
WA++ +N D AG+ +EL+E+R+ AY+N+++YKERTK +HDKKI+ R F+ D
Sbjct: 433 WAVKMMNFDIKSAGE------SELDEIRIHAYDNSKLYKERTKAYHDKKILTRTFEPNDQ 486
Query: 1601 VLLF 1604
+L F
Sbjct: 487 ILRF 490
Score = 83.6 bits (205), Expect = 5e-14
Identities = 37/63 (58%), Positives = 53/63 (83%), Gaps = 2/63 (3%)
Query: 1177 LNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQL 1236
+ KKDAKPRL+ WILLLQEFD+E++DKKGVEN VADHLSR+R D++P++D P++ +
Sbjct: 1 MQKKDAKPRLLIWILLLQEFDIEVRDKKGVENGVADHLSRIR--IDDDVPINDFLPEENI 58
Query: 1237 FLL 1239
+++
Sbjct: 59 YMI 61
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,880,293,716
Number of Sequences: 2540612
Number of extensions: 126537968
Number of successful extensions: 407204
Number of sequences better than 10.0: 37337
Number of HSP's better than 10.0 without gapping: 4886
Number of HSP's successfully gapped in prelim test: 32464
Number of HSP's that attempted gapping in prelim test: 352675
Number of HSP's gapped (non-prelim): 45683
length of query: 1672
length of database: 863,360,394
effective HSP length: 142
effective length of query: 1530
effective length of database: 502,593,490
effective search space: 768968039700
effective search space used: 768968039700
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 82 (36.2 bits)
Medicago: description of AC146683.16


