

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146681.9 + phase: 2 /pseudo
(249 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
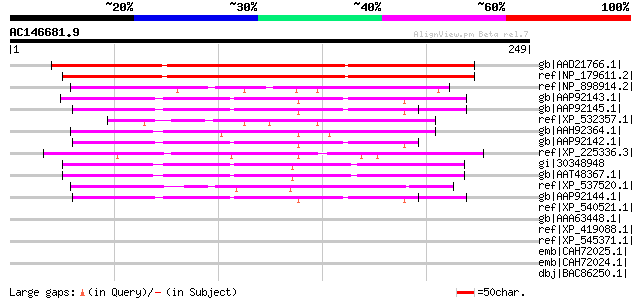
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD21766.1| hypothetical protein [Arabidopsis thaliana] gi|25... 177 2e-43
ref|NP_179611.2| leucine-rich repeat family protein [Arabidopsis... 176 4e-43
ref|NP_898914.2| I-kappa-B-related protein [Mus musculus] gi|425... 55 1e-06
gb|AAP92143.1| leucine-rich repeat protein N4C [synthetic constr... 54 5e-06
gb|AAP92145.1| leucine-rich repeat protein N6C [synthetic constr... 53 7e-06
ref|XP_532357.1| PREDICTED: similar to I-kappa-B-related protein... 52 1e-05
gb|AAH92364.1| Unknown (protein for IMAGE:7145273) [Danio rerio] 52 2e-05
gb|AAP92142.1| leucine-rich repeat protein N3C [synthetic constr... 50 8e-05
ref|XP_225336.3| PREDICTED: similar to CARMIL [Rattus norvegicus] 50 8e-05
gi|30348948 TPA: NOD3 [Homo sapiens] gi|30524928|ref|NP_849172.1... 48 3e-04
gb|AAT48367.1| caterpiller 16.2 [Homo sapiens] 48 3e-04
ref|XP_537520.1| PREDICTED: similar to chromosome 14 open readin... 47 5e-04
gb|AAP92144.1| leucine-rich repeat protein N5C [synthetic constr... 47 6e-04
ref|XP_540521.1| PREDICTED: similar to ribonuclease/angiogenin i... 46 0.001
gb|AAA63448.1| ribonuclease inhibitor 45 0.001
ref|XP_419088.1| PREDICTED: similar to leucine rich repeat conta... 45 0.001
ref|XP_545371.1| PREDICTED: similar to CARMIL [Canis familiaris] 45 0.002
emb|CAH72025.1| OTTHUMP00000016106 [Homo sapiens] gi|56202610|em... 45 0.002
emb|CAH72024.1| OTTHUMP00000039401 [Homo sapiens] gi|56202653|em... 45 0.002
dbj|BAC86250.1| unnamed protein product [Homo sapiens] 45 0.002
>gb|AAD21766.1| hypothetical protein [Arabidopsis thaliana] gi|25371295|pir||D84586
hypothetical protein At2g20210 [imported] - Arabidopsis
thaliana
Length = 271
Score = 177 bits (449), Expect = 2e-43
Identities = 104/203 (51%), Positives = 134/203 (65%), Gaps = 3/203 (1%)
Query: 21 FLI*FLSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMC 80
F+ +S RGN L + DAENL +A +MP LE LD+S NPIED G+R LI YF T
Sbjct: 22 FVSFLMSVRGNELDRYDAENLAHALLHMPGLESLDLSGNPIEDSGIRSLISYF--TKNPD 79
Query: 81 SRLACLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTPIEV 140
SRLA L LE C+LSC V LD+L +L LK LS+A+N LGS+VA A+ + IE
Sbjct: 80 SRLADLNLENCELSCCGVIEFLDTLSMLEKPLKFLSVADNALGSEVAEAVVNSFTISIES 139
Query: 141 LDASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVN 200
L+ GI L P GFL L L + L+ INISKNRGG++TARFLSKL+P AP+L+ ++
Sbjct: 140 LNIMGIGLGPLGFLALGRKLE-KVSKKLLSINISKNRGGLETARFLSKLIPLAPKLISID 198
Query: 201 ASSNCMPIESLSIISSALKFAKG 223
AS N MP E+L ++ +L+ AKG
Sbjct: 199 ASYNLMPPEALLMLCDSLRTAKG 221
>ref|NP_179611.2| leucine-rich repeat family protein [Arabidopsis thaliana]
Length = 604
Score = 176 bits (447), Expect = 4e-43
Identities = 103/198 (52%), Positives = 132/198 (66%), Gaps = 3/198 (1%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L+ RGN L + DAENL +A +MP LE LD+S NPIED G+R LI YF T SRLA
Sbjct: 360 LNLRGNELDRYDAENLAHALLHMPGLESLDLSGNPIEDSGIRSLISYF--TKNPDSRLAD 417
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTPIEVLDASG 145
L LE C+LSC V LD+L +L LK LS+A+N LGS+VA A+ + IE L+ G
Sbjct: 418 LNLENCELSCCGVIEFLDTLSMLEKPLKFLSVADNALGSEVAEAVVNSFTISIESLNIMG 477
Query: 146 IDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASSNC 205
I L P GFL L L + L+ INISKNRGG++TARFLSKL+P AP+L+ ++AS N
Sbjct: 478 IGLGPLGFLALGRKLE-KVSKKLLSINISKNRGGLETARFLSKLIPLAPKLISIDASYNL 536
Query: 206 MPIESLSIISSALKFAKG 223
MP E+L ++ +L+ AKG
Sbjct: 537 MPPEALLMLCDSLRTAKG 554
>ref|NP_898914.2| I-kappa-B-related protein [Mus musculus] gi|42557296|gb|AAH66068.1|
RIKEN cDNA 2810439M11 [Mus musculus]
Length = 1213
Score = 55.5 bits (132), Expect = 1e-06
Identities = 64/225 (28%), Positives = 91/225 (40%), Gaps = 49/225 (21%)
Query: 30 GNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEM---------- 79
GN L A L PNL LD+S N + EGLR L+ +G + +
Sbjct: 918 GNRLGDACATELLATLGTTPNLVLLDLSSNHLGQEGLRQLVEGSSGQAALQNLEELDLSM 977
Query: 80 -----------------CSRLACLKLEACDLSCDVVNHLLDSLPVLNGT------LKSLS 116
C L+ L+L+AC S + L L G LK+LS
Sbjct: 978 NPLGDGCGQALASLLRACPMLSTLRLQACGFSS---SFFLSHQAALGGAFQDAVHLKTLS 1034
Query: 117 IAENCLGSKVAGALGRFLST----PIEVLDASGI--DLLPSGFLELQNMLTIEEELSLVI 170
++ N LG A AL R L T ++ LD S + SG +E +E +L
Sbjct: 1035 LSYNLLG---APALARVLQTLPACTLKRLDLSSVAASKSNSGIIEPVIKYLTKEGCALAH 1091
Query: 171 INISKNRGGIQTARFLSKLLPQAPRLVDVNASSN----CMPIESL 211
+ +S N G + R LS+ LP P L ++ S+N C +E L
Sbjct: 1092 LTLSANCLGDKAVRELSRCLPCCPSLTSLDLSANPEVSCASLEEL 1136
>gb|AAP92143.1| leucine-rich repeat protein N4C [synthetic construct]
Length = 305
Score = 53.5 bits (127), Expect = 5e-06
Identities = 55/199 (27%), Positives = 94/199 (46%), Gaps = 10/199 (5%)
Query: 25 FLSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLA 84
+L N+L + ++L P+L +LD+S+N + D G+R L+ G + +RL
Sbjct: 43 YLDLNWNDLTEAGMKDLASVLRSNPSLRELDLSNNKLGDAGVRLLL---QGLLDPGTRLE 99
Query: 85 CLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVL 141
L L DL+ + L L N +L+ L+++ N LG L + L P +E L
Sbjct: 100 NLDLNESDLTEAGLKDLASVLR-SNPSLRELTLSNNKLGDAGVRLLLQGLLDPGTRLEKL 158
Query: 142 DASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRLVDVN 200
D DL +G +L ++L SL +++S N+ G R L + LL RL ++
Sbjct: 159 DLDQTDLTEAGMKDLASVL--RSNPSLRELSLSSNKLGDAGVRLLLQGLLDPGTRLEKLD 216
Query: 201 ASSNCMPIESLSIISSALK 219
+ N + L ++S L+
Sbjct: 217 LNQNDLTEADLKDLASVLR 235
>gb|AAP92145.1| leucine-rich repeat protein N6C [synthetic construct]
Length = 415
Score = 53.1 bits (126), Expect = 7e-06
Identities = 54/193 (27%), Positives = 93/193 (47%), Gaps = 10/193 (5%)
Query: 31 NNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
N+L + ++L P+L +L +S+N + D G+R L+ G + +RL LKL++
Sbjct: 49 NDLTEAGLKDLASVLRSNPSLRELSLSNNKLGDAGVRLLL---QGLLDPGTRLESLKLQS 105
Query: 91 CDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLDASGID 147
DL+ + L L N +L+ L+++ N LG L + L P +E LD + D
Sbjct: 106 TDLTEAGLKDLASVLR-SNPSLRELNLSTNKLGDAGVRLLLQGLLDPGTRLEKLDLNDTD 164
Query: 148 LLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRLVDVNASSNCM 206
L +G +L ++L SL +++S N+ G R L + LL RL + N +
Sbjct: 165 LTEAGVKDLASVL--RSNPSLRELSLSTNKLGDAGVRLLLQGLLDPGTRLEKLYLEDNDL 222
Query: 207 PIESLSIISSALK 219
L ++S L+
Sbjct: 223 TEAGLKDLASVLR 235
Score = 46.6 bits (109), Expect = 6e-04
Identities = 51/170 (30%), Positives = 81/170 (47%), Gaps = 10/170 (5%)
Query: 31 NNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
N+L + ++L P+L +L++SDN + D G+R L+ G + +RL L+L
Sbjct: 220 NDLTEAGLKDLASVLRSNPSLRELNLSDNKLGDAGVRLLL---QGLLDPGTRLEELQLRN 276
Query: 91 CDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLDASGID 147
DL+ V L L N +L+ LS++ N LG L + L P +E L D
Sbjct: 277 TDLTEAGVEDLASVLR-SNPSLRELSLSNNKLGDAGVRLLLQGLLDPGTRLEKLYLRNTD 335
Query: 148 LLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRL 196
L +G +L ++L SL +++S N+ G R L + LL RL
Sbjct: 336 LTEAGMKDLASVL--RSNPSLRELSLSTNKLGDAGVRLLLQGLLDPGTRL 383
Score = 44.7 bits (104), Expect = 0.002
Identities = 53/198 (26%), Positives = 90/198 (44%), Gaps = 10/198 (5%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L + +L + ++L P+L +L++S N + D G+R L+ G + +RL
Sbjct: 101 LKLQSTDLTEAGLKDLASVLRSNPSLRELNLSTNKLGDAGVRLLL---QGLLDPGTRLEK 157
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLD 142
L L DL+ V L L N +L+ LS++ N LG L + L P +E L
Sbjct: 158 LDLNDTDLTEAGVKDLASVLR-SNPSLRELSLSTNKLGDAGVRLLLQGLLDPGTRLEKLY 216
Query: 143 ASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRLVDVNA 201
DL +G +L ++L SL +N+S N+ G R L + LL RL ++
Sbjct: 217 LEDNDLTEAGLKDLASVL--RSNPSLRELNLSDNKLGDAGVRLLLQGLLDPGTRLEELQL 274
Query: 202 SSNCMPIESLSIISSALK 219
+ + + ++S L+
Sbjct: 275 RNTDLTEAGVEDLASVLR 292
Score = 36.2 bits (82), Expect = 0.86
Identities = 30/98 (30%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L R +L + E+L P+L +L +S+N + D G+R L+ G + +RL
Sbjct: 272 LQLRNTDLTEAGVEDLASVLRSNPSLRELSLSNNKLGDAGVRLLL---QGLLDPGTRLEK 328
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLG 123
L L DL+ + L L N +L+ LS++ N LG
Sbjct: 329 LYLRNTDLTEAGMKDLASVLR-SNPSLRELSLSTNKLG 365
>ref|XP_532357.1| PREDICTED: similar to I-kappa-B-related protein [Canis familiaris]
Length = 1551
Score = 52.4 bits (124), Expect = 1e-05
Identities = 48/169 (28%), Positives = 82/169 (48%), Gaps = 22/169 (13%)
Query: 48 MPNLEDLDISDNPIED---EGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDS 104
+ NL++LD+S NP+ D + L +++ + C L+ L L+AC L LL
Sbjct: 1039 LQNLQELDLSMNPLGDGCGQALAFVL-------QACPSLSTLHLQACGLG---PGFLLSH 1088
Query: 105 LPVLNGT------LKSLSIAENCLG-SKVAGALGRFLSTPIEVLDASGI--DLLPSGFLE 155
L LK+LS++ N LG + +A AL + ++ L+ S + SG +E
Sbjct: 1089 QAALGSAFQDATHLKTLSLSYNILGTTALARALQSLPARTLQRLELSSVAASKSDSGLVE 1148
Query: 156 LQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASSN 204
EE +L +++S N G + + LS+ LP P L+ ++ S+N
Sbjct: 1149 PVVRYLTEEGCALSHLSLSANHLGDKAVKDLSRCLPCCPSLISLDLSAN 1197
>gb|AAH92364.1| Unknown (protein for IMAGE:7145273) [Danio rerio]
Length = 485
Score = 52.0 bits (123), Expect = 2e-05
Identities = 50/181 (27%), Positives = 82/181 (44%), Gaps = 10/181 (5%)
Query: 30 GNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLE 89
G LRK + A P LE+L++S NP+ D + L A C L+ L L+
Sbjct: 214 GEGLRKASDAFETRSQAAFPCLEELNLSMNPLGDGWTQAL----ASLLSSCPLLSSLSLQ 269
Query: 90 ACDLSCDVVNH---LLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP-IEVLDASG 145
AC LS + LL + G ++S+ ++ N LGS + + L + L+ S
Sbjct: 270 ACGLSARFLQQHRLLLANAMASTGNMRSVCLSHNALGSTGFELVLKTLPMHCLTHLELSA 329
Query: 146 IDLLPSG--FLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASS 203
+ PS +E+ L + + L +N+S N + L++ LP P LV ++ S+
Sbjct: 330 VCRGPSDQPSMEILTKLLAQGDCPLTHLNLSGNELTDHSVLLLARCLPVCPSLVSLDLSA 389
Query: 204 N 204
N
Sbjct: 390 N 390
>gb|AAP92142.1| leucine-rich repeat protein N3C [synthetic construct]
Length = 244
Score = 49.7 bits (117), Expect = 8e-05
Identities = 50/170 (29%), Positives = 84/170 (49%), Gaps = 10/170 (5%)
Query: 31 NNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
N+L + ++L P+L +L +S+N + D G+R L+ G + +RL LKL++
Sbjct: 49 NDLTEAGLKDLASVLRSNPSLRELSLSNNKLGDAGVRLLL---QGLLDPGTRLESLKLQS 105
Query: 91 CDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLDASGID 147
DL+ + L L N +L+ L+++ N LG L + L P +E LD + D
Sbjct: 106 TDLTEAGLKDLASVLR-SNPSLRELNLSTNKLGDAGVRLLLQGLLDPGTRLEKLDLNDTD 164
Query: 148 LLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRL 196
L +G +L ++L SL +++S N+ G R L + LL RL
Sbjct: 165 LTEAGVKDLASVL--RSNPSLRELSLSTNKLGDAGVRLLLQGLLDPGTRL 212
Score = 33.9 bits (76), Expect = 4.3
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L + +L + ++L P+L +L++S N + D G+R L+ G + +RL
Sbjct: 101 LKLQSTDLTEAGLKDLASVLRSNPSLRELNLSTNKLGDAGVRLLL---QGLLDPGTRLEK 157
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLG 123
L L DL+ V L L N +L+ LS++ N LG
Sbjct: 158 LDLNDTDLTEAGVKDLASVLR-SNPSLRELSLSTNKLG 194
>ref|XP_225336.3| PREDICTED: similar to CARMIL [Rattus norvegicus]
Length = 1118
Score = 49.7 bits (117), Expect = 8e-05
Identities = 61/223 (27%), Positives = 94/223 (41%), Gaps = 19/223 (8%)
Query: 17 ISREFLI*FLSHRGNNLRKDDAENLGYAFAYMPN--LEDLDISDNPIEDEGLRYLIPYFA 74
+SR L+ L LR D A+ L A A+ PN L ++++ NP+ED G+ L FA
Sbjct: 439 VSRSNLLEELVLENAGLRTDFAQKLASALAHNPNSGLHTINLAGNPLEDRGVSSLSIQFA 498
Query: 75 GTSEMCSRLACLKLEACDLSCDVVNHLLDSL---PVLNGTLKSLSIAENCLGSKVAGALG 131
++ L L L LS VN L SL P+ TL L ++ N L +
Sbjct: 499 ---KLPKGLKHLNLSKTSLSPKGVNSLCQSLSANPLTASTLTHLDLSGNVLRGDDLSHMY 555
Query: 132 RFLSTP--IEVLDASGIDLLPSGFLELQNMLTIEEELS-LVIINISK----NRGGIQTAR 184
FL+ P I LD S + LE+ + L L ++N+S+ +R G +
Sbjct: 556 NFLAQPNTIVHLDLSNTEC----SLEMVCSALLRGCLQCLAVLNLSRSVFSHRKGKEVPP 611
Query: 185 FLSKLLPQAPRLVDVNASSNCMPIESLSIISSALKFAKGIYGV 227
+ + L+ +N S + E L + L + GV
Sbjct: 612 SFKQFFSSSLALIQINLSGTKLSPEPLKALLLGLACNHSLKGV 654
Score = 37.0 bits (84), Expect = 0.50
Identities = 44/148 (29%), Positives = 75/148 (49%), Gaps = 12/148 (8%)
Query: 30 GNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLK-- 87
G+ LR A+ L A + N+ LDISDN +E + L LI + + + LA K
Sbjct: 664 GHCLRSGGAQVLEGCIAEIHNITSLDISDNGLESD-LSTLIVWLS-RNRSIQHLALGKNF 721
Query: 88 --LEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVA---GALGRFLSTPIEVLD 142
+++ +L+ V+++L+ + + L+SLS+A++ L ++V ALG +T + +D
Sbjct: 722 NNMKSKNLT-PVLDNLVQMIQDEDSPLQSLSLADSKLKTEVTIIINALGS--NTSLTKVD 778
Query: 143 ASGIDLLPSGFLELQNMLTIEEELSLVI 170
SG + G L L I +L VI
Sbjct: 779 ISGNSMGDMGAKMLAKALQINTKLRTVI 806
>gi|30348948 TPA: NOD3 [Homo sapiens] gi|30524928|ref|NP_849172.1| NOD3 protein
[Homo sapiens]
Length = 1112
Score = 47.8 bits (112), Expect = 3e-04
Identities = 50/195 (25%), Positives = 87/195 (43%), Gaps = 9/195 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
LS R N++ + A+ + +A L++LD++ N + D+G R + A L
Sbjct: 886 LSLRENSISPEGAQAIAHALCANSTLKNLDLTANLLHDQGARAI----AVAVRENRTLTS 941
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFL--STPIEVLDA 143
L L+ + L +L LN +L SL + EN +G A A+ R L +T + L
Sbjct: 942 LHLQWNFIQAGAAQALGQALQ-LNRSLTSLDLQENAIGDDGACAVARALKVNTALTALYL 1000
Query: 144 SGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASS 203
+ SG L L + +L I+++ N G+ A+ L+ L L +N
Sbjct: 1001 QVASIGASGAQVLGEALAVNR--TLEILDLRGNAIGVAGAKALANALKVNSSLRRLNLQE 1058
Query: 204 NCMPIESLSIISSAL 218
N + ++ I++AL
Sbjct: 1059 NSLGMDGAICIATAL 1073
Score = 39.3 bits (90), Expect = 0.10
Identities = 43/196 (21%), Positives = 83/196 (41%), Gaps = 9/196 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L RGN++ A+ L A L L + N + D+G R + A L+
Sbjct: 746 LDLRGNSIGPQGAKALADALKINRTLTSLSLQGNTVRDDGARSMAEALASN----RTLSM 801
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFL--STPIEVLDA 143
L L+ + + D+L N +LK L + N +G A AL L + +E LD
Sbjct: 802 LHLQKNSIGPMGAQRMADALK-QNRSLKELMFSSNSIGDGGAKALAEALKVNQGLESLDL 860
Query: 144 SGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASS 203
+ +G L L + +L+ +++ +N + A+ ++ L L +++ ++
Sbjct: 861 QSNSISDAGVAALMGALCTNQ--TLLSLSLRENSISPEGAQAIAHALCANSTLKNLDLTA 918
Query: 204 NCMPIESLSIISSALK 219
N + + I+ A++
Sbjct: 919 NLLHDQGARAIAVAVR 934
Score = 35.4 bits (80), Expect = 1.5
Identities = 54/213 (25%), Positives = 87/213 (40%), Gaps = 39/213 (18%)
Query: 44 AFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA-CDLSCDVVNHLL 102
A AY+ L +SD ++ L + S + L C KL + D V LL
Sbjct: 650 ALAYL-----LQVSDACAQEANLSLSLSQGVLQSLLPQLLYCRKLRLDTNQFQDPVMELL 704
Query: 103 DSLPVLNGT---LKSLSIAENCLGSKVAGALGRFL--STPIEVLDASGIDLLPSGFLELQ 157
S VL+G ++ +S+AEN + +K A AL R L + + LD G + P G L
Sbjct: 705 GS--VLSGKDCRIQKISLAENQISNKGAKALARSLLVNRSLTSLDLRGNSIGPQGAKALA 762
Query: 158 NMLTIEEELS--------------------------LVIINISKNRGGIQTARFLSKLLP 191
+ L I L+ L ++++ KN G A+ ++ L
Sbjct: 763 DALKINRTLTSLSLQGNTVRDDGARSMAEALASNRTLSMLHLQKNSIGPMGAQRMADALK 822
Query: 192 QAPRLVDVNASSNCMPIESLSIISSALKFAKGI 224
Q L ++ SSN + ++ ALK +G+
Sbjct: 823 QNRSLKELMFSSNSIGDGGAKALAEALKVNQGL 855
>gb|AAT48367.1| caterpiller 16.2 [Homo sapiens]
Length = 1065
Score = 47.8 bits (112), Expect = 3e-04
Identities = 50/195 (25%), Positives = 87/195 (43%), Gaps = 9/195 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
LS R N++ + A+ + +A L++LD++ N + D+G R + A L
Sbjct: 839 LSLRENSISPEGAQAIAHALCANSTLKNLDLTANLLHDQGARAI----AVAVRENRTLTS 894
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFL--STPIEVLDA 143
L L+ + L +L LN +L SL + EN +G A A+ R L +T + L
Sbjct: 895 LHLQWNFIQAGAAQALGQALQ-LNRSLTSLDLQENAIGDDGACAVARALKVNTALTALYL 953
Query: 144 SGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASS 203
+ SG L L + +L I+++ N G+ A+ L+ L L +N
Sbjct: 954 QVASIGASGAQVLGEALAVNR--TLEILDLRGNAIGVAGAKALANALKVNSSLRRLNLQE 1011
Query: 204 NCMPIESLSIISSAL 218
N + ++ I++AL
Sbjct: 1012 NSLGMDGAICIATAL 1026
Score = 39.3 bits (90), Expect = 0.10
Identities = 43/196 (21%), Positives = 83/196 (41%), Gaps = 9/196 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L RGN++ A+ L A L L + N + D+G R + A L+
Sbjct: 699 LDLRGNSIGPQGAKALADALKINRTLTSLSLQGNTVRDDGARSMAEALASN----RTLSM 754
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFL--STPIEVLDA 143
L L+ + + D+L N +LK L + N +G A AL L + +E LD
Sbjct: 755 LHLQKNSIGPMGAQRMADALK-QNRSLKELMFSSNSIGDGGAKALAEALKVNQGLESLDL 813
Query: 144 SGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASS 203
+ +G L L + +L+ +++ +N + A+ ++ L L +++ ++
Sbjct: 814 QSNSISDAGVAALMGALCTNQ--TLLSLSLRENSISPEGAQAIAHALCANSTLKNLDLTA 871
Query: 204 NCMPIESLSIISSALK 219
N + + I+ A++
Sbjct: 872 NLLHDQGARAIAVAVR 887
Score = 35.4 bits (80), Expect = 1.5
Identities = 54/213 (25%), Positives = 87/213 (40%), Gaps = 39/213 (18%)
Query: 44 AFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA-CDLSCDVVNHLL 102
A AY+ L +SD ++ L + S + L C KL + D V LL
Sbjct: 603 ALAYL-----LQVSDACAQEANLSLSLSQGVLQSLLPQLLYCRKLRLDTNQFQDPVMELL 657
Query: 103 DSLPVLNGT---LKSLSIAENCLGSKVAGALGRFL--STPIEVLDASGIDLLPSGFLELQ 157
S VL+G ++ +S+AEN + +K A AL R L + + LD G + P G L
Sbjct: 658 GS--VLSGKDCRIQKISLAENQISNKGAKALARSLLVNRSLTSLDLRGNSIGPQGAKALA 715
Query: 158 NMLTIEEELS--------------------------LVIINISKNRGGIQTARFLSKLLP 191
+ L I L+ L ++++ KN G A+ ++ L
Sbjct: 716 DALKINRTLTSLSLQGNTVRDDGARSMAEALASNRTLSMLHLQKNSIGPMGAQRMADALK 775
Query: 192 QAPRLVDVNASSNCMPIESLSIISSALKFAKGI 224
Q L ++ SSN + ++ ALK +G+
Sbjct: 776 QNRSLKELMFSSNSIGDGGAKALAEALKVNQGL 808
>ref|XP_537520.1| PREDICTED: similar to chromosome 14 open reading frame 166B [Canis
familiaris]
Length = 643
Score = 47.0 bits (110), Expect = 5e-04
Identities = 56/193 (29%), Positives = 84/193 (43%), Gaps = 22/193 (11%)
Query: 30 GNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLE 89
GNN R + AE L A A ++ LD+S N D+G Y+ A + L+
Sbjct: 239 GNNFRDESAELLCQALAANYQIKTLDLSHNQFSDKGGEYMGQMLA---------LNVGLQ 289
Query: 90 ACDLSCDVVNHLLDSLPV-------LNGTLKSLSIAENCLGSKVAGALGRF--LSTPIEV 140
+ DLS NH V NGTL+ L ++ N G++ A ALG L++ +
Sbjct: 290 SLDLSW---NHFYIRGAVALCNGLRANGTLQKLDLSMNGFGNEGATALGEVLRLNSSLVY 346
Query: 141 LDASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVN 200
LD S D+ G ++ L E L ++ + AR S+ L QA R +V+
Sbjct: 347 LDISINDISNEGISKISKGLEFNESLKVLKHSPPVWAPLAPPARPESRAL-QAMRGREVS 405
Query: 201 ASSNCMPIESLSI 213
S+C +S I
Sbjct: 406 LQSSCSDSKSFKI 418
>gb|AAP92144.1| leucine-rich repeat protein N5C [synthetic construct]
Length = 358
Score = 46.6 bits (109), Expect = 6e-04
Identities = 51/170 (30%), Positives = 81/170 (47%), Gaps = 10/170 (5%)
Query: 31 NNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
N+L + ++L P+L +L++SDN + D G+R L+ G + +RL L+L
Sbjct: 163 NDLTEAGLKDLASVLRSNPSLRELNLSDNKLGDAGVRLLL---QGLLDPGTRLEELQLRN 219
Query: 91 CDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLDASGID 147
DL+ V L L N +L+ LS++ N LG L + L P +E L D
Sbjct: 220 TDLTEAGVEDLASVLR-SNPSLRELSLSNNKLGDAGVRLLLQGLLDPGTRLEKLYLRNTD 278
Query: 148 LLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRL 196
L +G +L ++L SL +++S N+ G R L + LL RL
Sbjct: 279 LTEAGMKDLASVL--RSNPSLRELSLSTNKLGDAGVRLLLQGLLDPGTRL 326
Score = 46.2 bits (108), Expect = 8e-04
Identities = 53/193 (27%), Positives = 89/193 (45%), Gaps = 10/193 (5%)
Query: 31 NNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
N+L + ++L P+L +L++S N + D G+R L+ G + +RL L L
Sbjct: 49 NDLTEAGLKDLASVLRSNPSLRELNLSHNKLGDAGVRLLL---QGLLDPGTRLEKLDLND 105
Query: 91 CDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLDASGID 147
DL+ V L L N +L+ LS++ N LG L + L P +E L D
Sbjct: 106 TDLTEAGVKDLASVLR-SNPSLRELSLSTNKLGDAGVRLLLQGLLDPGTRLEKLYLEDND 164
Query: 148 LLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSK-LLPQAPRLVDVNASSNCM 206
L +G +L ++L SL +N+S N+ G R L + LL RL ++ + +
Sbjct: 165 LTEAGLKDLASVL--RSNPSLRELNLSDNKLGDAGVRLLLQGLLDPGTRLEELQLRNTDL 222
Query: 207 PIESLSIISSALK 219
+ ++S L+
Sbjct: 223 TEAGVEDLASVLR 235
Score = 36.2 bits (82), Expect = 0.86
Identities = 30/98 (30%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query: 26 LSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLAC 85
L R +L + E+L P+L +L +S+N + D G+R L+ G + +RL
Sbjct: 215 LQLRNTDLTEAGVEDLASVLRSNPSLRELSLSNNKLGDAGVRLLL---QGLLDPGTRLEK 271
Query: 86 LKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLG 123
L L DL+ + L L N +L+ LS++ N LG
Sbjct: 272 LYLRNTDLTEAGMKDLASVLR-SNPSLRELSLSTNKLG 308
>ref|XP_540521.1| PREDICTED: similar to ribonuclease/angiogenin inhibitor 1 [Canis
familiaris]
Length = 296
Score = 45.8 bits (107), Expect = 0.001
Identities = 43/148 (29%), Positives = 68/148 (45%), Gaps = 9/148 (6%)
Query: 48 MPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDSLPV 107
MP L +L +SDNP+ED GL+ L G + L L+LE C+L+ L +L
Sbjct: 146 MPTLRELHLSDNPLEDAGLQLLC---EGLLDPQCHLEKLQLEYCNLTAASCESLASALR- 201
Query: 108 LNGTLKSLSIAENCLGSKVAGALGRFL---STPIEVLDASGIDLLPSGFLELQNMLTIEE 164
K L+++ N +G L + L + +E L+ D+ G L ++L
Sbjct: 202 NKQHFKELAVSNNEIGEAGVRVLCQGLVESACQLETLNLGDCDVANDGCASLASLLLANR 261
Query: 165 ELSLVIINISKNRGGIQTARFLSKLLPQ 192
SL +++S NR Q R L + + Q
Sbjct: 262 --SLRELDLSNNRMNDQGIRRLMESVEQ 287
Score = 35.4 bits (80), Expect = 1.5
Identities = 27/89 (30%), Positives = 43/89 (47%), Gaps = 4/89 (4%)
Query: 32 NLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEAC 91
NL E+L A + ++L +S+N I + G+R L G E +L L L C
Sbjct: 187 NLTAASCESLASALRNKQHFKELAVSNNEIGEAGVRVLC---QGLVESACQLETLNLGDC 243
Query: 92 DLSCDVVNHLLDSLPVLNGTLKSLSIAEN 120
D++ D L SL + N +L+ L ++ N
Sbjct: 244 DVANDGCASLA-SLLLANRSLRELDLSNN 271
>gb|AAA63448.1| ribonuclease inhibitor
Length = 375
Score = 45.4 bits (106), Expect = 0.001
Identities = 45/155 (29%), Positives = 69/155 (44%), Gaps = 14/155 (9%)
Query: 48 MPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDL---SCDVVNHLLDS 104
+P L +L +SDNP+ D GLR L G + L L+LE C L SC+ + +L +
Sbjct: 26 LPTLRELHLSDNPLGDAGLRLLC---EGLLDPQCHLEKLQLEYCRLTAASCEPLASVLRA 82
Query: 105 LPVLNGTLKSLSIAENCLGSKVAGALGRFL---STPIEVLDASGIDLLPSGFLELQNMLT 161
LK L+++ N +G A LG+ L + +E L L P+ +L ++
Sbjct: 83 ----TRALKELTVSNNDIGEAGARVLGQGLADSACQLETLRLENCGLTPANCKDLCGIVA 138
Query: 162 IEEELSLVIINISKNRGGIQTARFLSKLLPQAPRL 196
+ L + + S G A LL A RL
Sbjct: 139 SQASLRELDLG-SNGLGDAGIAELCPGLLSPASRL 172
Score = 43.1 bits (100), Expect = 0.007
Identities = 51/184 (27%), Positives = 77/184 (41%), Gaps = 16/184 (8%)
Query: 46 AYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDSL 105
A +L +LD+ N + D G+ L P G SRL L L CD++ L L
Sbjct: 138 ASQASLRELDLGSNGLGDAGIAELCP---GLLSPASRLKTLWLWECDITASGCRDLCRVL 194
Query: 106 PVLNGTLKSLSIAENCLGSKVAGALGRFLSTP---IEVLDASGIDLLPSGFLELQNMLTI 162
TLK LS+A N LG + A L L P +E L L + + MLT
Sbjct: 195 QA-KETLKELSLAGNKLGDEGARLLCESLLQPGCQLESLWVKSCSLTAACCQHVSLMLT- 252
Query: 163 EEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDV-------NASSNCMPIESLSIIS 215
+ L+ + +S N+ G + L + L Q + V +S C + SL + +
Sbjct: 253 -QNKHLLELQLSSNKLGDSGIQELCQALSQPGTTLRVLCLGDCEVTNSGCSSLASLLLAN 311
Query: 216 SALK 219
+L+
Sbjct: 312 RSLR 315
>ref|XP_419088.1| PREDICTED: similar to leucine rich repeat containing 16 [Gallus
gallus]
Length = 1391
Score = 45.4 bits (106), Expect = 0.001
Identities = 52/195 (26%), Positives = 85/195 (42%), Gaps = 21/195 (10%)
Query: 33 LRKDDAENLGYAFAYMPN--LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
LR D A+ L A ++ PN L ++++ NP+ED G+ L FA ++ L L L
Sbjct: 224 LRTDFAQKLASALSHNPNSGLHTINLASNPLEDRGVSSLSIQFA---KLPKGLKHLNLSK 280
Query: 91 CDLSCDVVNHLLDSL---PVLNGTLKSLSIAENCLGSKVAGALGRFLSTP--IEVLDAS- 144
LS VN L SL ++ TL L ++ N L +L FL+ P + LD S
Sbjct: 281 TSLSPKGVNSLSQSLSANSLIANTLVYLDLSGNALRGDDLSSLYNFLAQPNALVHLDLSN 340
Query: 145 ---GIDLLPSGFLE--LQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDV 199
+D++ L LQ++ + +L +R G + + + L+ +
Sbjct: 341 TECALDMVCGALLRGCLQHLAVLSLSRTLF-----SHRKGKEVPPSFKQFFSSSLALMQI 395
Query: 200 NASSNCMPIESLSII 214
N S +P E L +
Sbjct: 396 NLSGTKLPPEPLKAL 410
Score = 33.9 bits (76), Expect = 4.3
Identities = 42/145 (28%), Positives = 73/145 (49%), Gaps = 12/145 (8%)
Query: 33 LRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLK----L 88
LR A+ L A + N+ LDISDN +E + L L+ + + + LA K +
Sbjct: 432 LRSGGAQVLEGCIAEIRNITSLDISDNGLESD-LSTLVVWLS-KNRSIRHLALGKNFNNM 489
Query: 89 EACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVA---GALGRFLSTPIEVLDASG 145
++ +L+ V+++L+ + + L+SLS+A++ L ++V ALG +T + +D SG
Sbjct: 490 KSKNLT-PVLDNLVQMIQDEDSPLQSLSLADSKLKTEVTIIINALGS--NTSLTKVDISG 546
Query: 146 IDLLPSGFLELQNMLTIEEELSLVI 170
+ G L L I +L VI
Sbjct: 547 NAMGDMGAKMLAKALQINTKLRTVI 571
>ref|XP_545371.1| PREDICTED: similar to CARMIL [Canis familiaris]
Length = 2423
Score = 45.1 bits (105), Expect = 0.002
Identities = 58/212 (27%), Positives = 90/212 (42%), Gaps = 29/212 (13%)
Query: 33 LRKDDAENLGYAFAYMPN--LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
LR D A+ L A A+ PN L ++++ NP+ED G+ L FA ++ L L L
Sbjct: 1119 LRTDFAQKLASALAHNPNSGLHTINLAGNPLEDRGVSSLSIQFA---KLPKGLKHLNLSK 1175
Query: 91 CDLSCDVVNHLLDSL---PVLNGTLKSLSIAENCLGSKVAGALGRFLSTP--IEVLDAS- 144
LS VN L SL P+ TL L ++ N L + FL+ P I LD S
Sbjct: 1176 TSLSPKGVNSLSQSLSANPLTATTLTHLDLSGNVLRGDDLSYMYSFLAQPNAIAHLDLSN 1235
Query: 145 ---GIDLLPSGFLE--LQNMLTIEEELSLVIINISK----NRGGIQTARFLSKLLPQAPR 195
+D++ + L LQ L ++N+S+ +R G + + +
Sbjct: 1236 TECSLDMVCAALLRGCLQ---------YLAVLNLSRAVFSHRKGKEVPPSFKQFFSSSLA 1286
Query: 196 LVDVNASSNCMPIESLSIISSALKFAKGIYGV 227
L+ +N S + E L + L + GV
Sbjct: 1287 LMQINLSGTKLSPEPLKALLLGLACNHNLKGV 1318
Score = 33.9 bits (76), Expect = 4.3
Identities = 43/145 (29%), Positives = 71/145 (48%), Gaps = 12/145 (8%)
Query: 33 LRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLK----L 88
LR A+ L A + N+ LDISDN +E + L LI + + + LA K +
Sbjct: 1327 LRSGGAQVLEGCIAEIHNITSLDISDNGLESD-LSTLIVWLS-KNRSIQHLALGKNFNNM 1384
Query: 89 EACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVA---GALGRFLSTPIEVLDASG 145
++ +L+ V++ L+ + L+SLS+A++ L ++V ALG +T + +D SG
Sbjct: 1385 KSKNLT-PVLDSLVQMIQDEESPLQSLSLADSKLKTEVTIIINALGS--NTSLTKVDISG 1441
Query: 146 IDLLPSGFLELQNMLTIEEELSLVI 170
+ G L L I +L VI
Sbjct: 1442 NGMGDMGAKMLAKALQINTKLRTVI 1466
>emb|CAH72025.1| OTTHUMP00000016106 [Homo sapiens] gi|56202610|emb|CAI21584.1|
OTTHUMP00000016106 [Homo sapiens]
Length = 1214
Score = 44.7 bits (104), Expect = 0.002
Identities = 58/212 (27%), Positives = 89/212 (41%), Gaps = 29/212 (13%)
Query: 33 LRKDDAENLGYAFAYMPN--LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
LR D A+ L A A+ PN L ++++ NP+ED G+ L FA ++ L L L
Sbjct: 101 LRTDFAQKLASALAHNPNSGLHTINLAGNPLEDRGVSSLSIQFA---KLPKGLKHLNLSK 157
Query: 91 CDLSCDVVNHLLDSL---PVLNGTLKSLSIAENCLGSKVAGALGRFLSTP--IEVLDAS- 144
LS VN L SL P+ TL L ++ N L + FL+ P I LD S
Sbjct: 158 TSLSPKGVNSLSQSLSANPLTASTLVHLDLSGNVLRGDDLSHMYNFLAQPNAIVHLDLSN 217
Query: 145 ---GIDLLPSGFLE--LQNMLTIEEELSLVIINISK----NRGGIQTARFLSKLLPQAPR 195
+D++ L LQ L ++N+S+ +R G + + +
Sbjct: 218 TECSLDMVCGALLRGCLQ---------YLAVLNLSRTVFSHRKGKEVPPSFKQFFSSSLA 268
Query: 196 LVDVNASSNCMPIESLSIISSALKFAKGIYGV 227
L+ +N S + E L + L + GV
Sbjct: 269 LMHINLSGTKLSPEPLKALLLGLACNHNLKGV 300
Score = 34.7 bits (78), Expect = 2.5
Identities = 43/145 (29%), Positives = 72/145 (49%), Gaps = 12/145 (8%)
Query: 33 LRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLK----L 88
LR A+ L A + N+ LDISDN +E + L LI + + + LA K +
Sbjct: 309 LRSGGAQVLEGCIAEIHNITSLDISDNGLESD-LSTLIVWLS-KNRSIQHLALGKNFNNM 366
Query: 89 EACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVA---GALGRFLSTPIEVLDASG 145
++ +L+ V+++L+ + L+SLS+A++ L ++V ALG +T + +D SG
Sbjct: 367 KSKNLT-PVLDNLVQMIQDEESPLQSLSLADSKLKTEVTIIINALGS--NTSLTKVDISG 423
Query: 146 IDLLPSGFLELQNMLTIEEELSLVI 170
+ G L L I +L VI
Sbjct: 424 NGMGDMGAKMLAKALQINTKLRTVI 448
>emb|CAH72024.1| OTTHUMP00000039401 [Homo sapiens] gi|56202653|emb|CAI21665.1|
OTTHUMP00000039401 [Homo sapiens]
gi|56202609|emb|CAI21583.1| OTTHUMP00000039401 [Homo
sapiens]
Length = 1371
Score = 44.7 bits (104), Expect = 0.002
Identities = 58/212 (27%), Positives = 89/212 (41%), Gaps = 29/212 (13%)
Query: 33 LRKDDAENLGYAFAYMPN--LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
LR D A+ L A A+ PN L ++++ NP+ED G+ L FA ++ L L L
Sbjct: 258 LRTDFAQKLASALAHNPNSGLHTINLAGNPLEDRGVSSLSIQFA---KLPKGLKHLNLSK 314
Query: 91 CDLSCDVVNHLLDSL---PVLNGTLKSLSIAENCLGSKVAGALGRFLSTP--IEVLDAS- 144
LS VN L SL P+ TL L ++ N L + FL+ P I LD S
Sbjct: 315 TSLSPKGVNSLSQSLSANPLTASTLVHLDLSGNVLRGDDLSHMYNFLAQPNAIVHLDLSN 374
Query: 145 ---GIDLLPSGFLE--LQNMLTIEEELSLVIINISK----NRGGIQTARFLSKLLPQAPR 195
+D++ L LQ L ++N+S+ +R G + + +
Sbjct: 375 TECSLDMVCGALLRGCLQ---------YLAVLNLSRTVFSHRKGKEVPPSFKQFFSSSLA 425
Query: 196 LVDVNASSNCMPIESLSIISSALKFAKGIYGV 227
L+ +N S + E L + L + GV
Sbjct: 426 LMHINLSGTKLSPEPLKALLLGLACNHNLKGV 457
Score = 34.7 bits (78), Expect = 2.5
Identities = 43/145 (29%), Positives = 72/145 (49%), Gaps = 12/145 (8%)
Query: 33 LRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLK----L 88
LR A+ L A + N+ LDISDN +E + L LI + + + LA K +
Sbjct: 466 LRSGGAQVLEGCIAEIHNITSLDISDNGLESD-LSTLIVWLS-KNRSIQHLALGKNFNNM 523
Query: 89 EACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVA---GALGRFLSTPIEVLDASG 145
++ +L+ V+++L+ + L+SLS+A++ L ++V ALG +T + +D SG
Sbjct: 524 KSKNLT-PVLDNLVQMIQDEESPLQSLSLADSKLKTEVTIIINALGS--NTSLTKVDISG 580
Query: 146 IDLLPSGFLELQNMLTIEEELSLVI 170
+ G L L I +L VI
Sbjct: 581 NGMGDMGAKMLAKALQINTKLRTVI 605
>dbj|BAC86250.1| unnamed protein product [Homo sapiens]
Length = 499
Score = 44.7 bits (104), Expect = 0.002
Identities = 58/212 (27%), Positives = 89/212 (41%), Gaps = 29/212 (13%)
Query: 33 LRKDDAENLGYAFAYMPN--LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEA 90
LR D A+ L A A+ PN L ++++ NP+ED G+ L FA ++ L L L
Sbjct: 258 LRTDFAQKLASALAHNPNSGLHTINLAGNPLEDRGVSSLSIQFA---KLPKGLKHLNLSK 314
Query: 91 CDLSCDVVNHLLDSL---PVLNGTLKSLSIAENCLGSKVAGALGRFLSTP--IEVLDAS- 144
LS VN L SL P+ TL L ++ N L + FL+ P I LD S
Sbjct: 315 TSLSPKGVNSLSQSLSANPLTASTLVHLDLSGNVLRGDDLSHMYNFLAQPNAIVHLDLSN 374
Query: 145 ---GIDLLPSGFLE--LQNMLTIEEELSLVIINISK----NRGGIQTARFLSKLLPQAPR 195
+D++ L LQ L ++N+S+ +R G + + +
Sbjct: 375 TECSLDMVCGALLRGCLQ---------YLAVLNLSRTVFSHRKGKEVPPSFKQFFSSSLA 425
Query: 196 LVDVNASSNCMPIESLSIISSALKFAKGIYGV 227
L+ +N S + E L + L + GV
Sbjct: 426 LMHINLSGTKLSPEPLKALLLGLACNHNLKGV 457
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.330 0.146 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 403,358,945
Number of Sequences: 2540612
Number of extensions: 16117352
Number of successful extensions: 47958
Number of sequences better than 10.0: 273
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 268
Number of HSP's that attempted gapping in prelim test: 47482
Number of HSP's gapped (non-prelim): 669
length of query: 249
length of database: 863,360,394
effective HSP length: 125
effective length of query: 124
effective length of database: 545,783,894
effective search space: 67677202856
effective search space used: 67677202856
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146681.9


