

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.11 + phase: 0
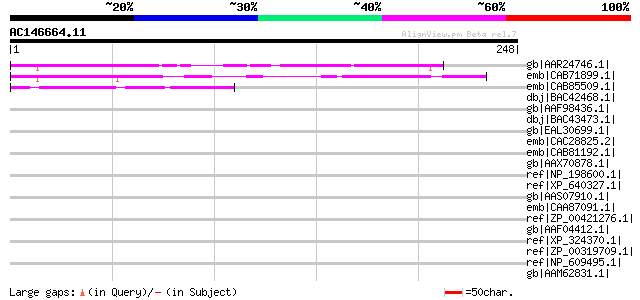
(248 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR24746.1| At1g64700 [Arabidopsis thaliana] gi|38454064|gb|A... 134 2e-30
emb|CAB71899.1| putative protein [Arabidopsis thaliana] gi|30102... 111 2e-23
emb|CAB85509.1| putative protein [Arabidopsis thaliana] gi|97580... 58 2e-07
dbj|BAC42468.1| unknown protein [Arabidopsis thaliana] gi|283728... 39 0.13
gb|AAF98436.1| Hypothetical protein [Arabidopsis thaliana] gi|15... 36 0.85
dbj|BAC43473.1| unknown protein [Arabidopsis thaliana] 36 0.85
gb|EAL30699.1| GA18776-PA [Drosophila pseudoobscura] 36 0.85
emb|CAC28825.2| related to a-agglutinin core protein AGA1 [Neuro... 35 1.5
emb|CAB81192.1| hypothetical protein [Arabidopsis thaliana] gi|4... 35 1.5
gb|AAX70878.1| hypothetical protein, conserved [Trypanosoma brucei] 35 1.9
ref|NP_198600.1| expressed protein [Arabidopsis thaliana] gi|483... 35 1.9
ref|XP_640327.1| hypothetical protein DDB0218364 [Dictyostelium ... 35 1.9
gb|AAS07910.1| oxidoreductase, short-chain dehydrogenase/reducta... 35 1.9
emb|CAA87091.1| secreted acid phosphatase 2 (SAP2) [Leishmania m... 34 3.2
ref|ZP_00421276.1| Haemagluttinin motif:Hep_Hag [Burkholderia vi... 34 3.2
gb|AAF04412.1| unknown protein [Arabidopsis thaliana] 34 3.2
ref|XP_324370.1| predicted protein [Neurospora crassa] gi|289173... 34 3.2
ref|ZP_00319709.1| COG1705: Muramidase (flagellum-specific) [Oen... 34 4.2
ref|NP_609495.1| CG4751-PA [Drosophila melanogaster] gi|7297828|... 33 5.5
gb|AAM62831.1| unknown [Arabidopsis thaliana] gi|26451664|dbj|BA... 33 5.5
>gb|AAR24746.1| At1g64700 [Arabidopsis thaliana] gi|38454064|gb|AAR20726.1|
At1g64700 [Arabidopsis thaliana]
gi|5042407|gb|AAD38246.1| Hypothetical Protein
[Arabidopsis thaliana] gi|15217713|ref|NP_176651.1|
expressed protein [Arabidopsis thaliana]
gi|25404486|pir||B96670 hypothetical protein F13O11.2
[imported] - Arabidopsis thaliana
gi|6633824|gb|AAF19683.1| F1N19.27 [Arabidopsis
thaliana]
Length = 203
Score = 134 bits (337), Expect = 2e-30
Identities = 92/217 (42%), Positives = 121/217 (55%), Gaps = 28/217 (12%)
Query: 1 MGNCLLGANSDP--DTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLC 58
MGNCL G D D LIKVI DG ++EFY P+T F+++ F GHA+F D KPL
Sbjct: 1 MGNCLFGGLGDEEEDLLIKVIKSDGGVLEFYSPVTAGFVSHGFSGHALFSAVDLLWKPLA 60
Query: 59 QFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPA 118
LV GQSYYL P ++S K + +C HVRS S + +
Sbjct: 61 HDHLLVPGQSYYLFP-NIVSDELK-TFVGSC---------------HVRSNSESLSAI-T 102
Query: 119 PYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCIT 178
PYRMSLDY + +LKR+ T+ FS ++ + K + RR+S K IWKV L I
Sbjct: 103 PYRMSLDYNH----RVLKRSYTDVFSRNSHIRTRQKEKKTRRRRTSSKG-AIWKVNLIIN 157
Query: 179 PEKLLEILSQEVSTKELIESVRIVAK---CGVTAASS 212
E+LL+ILS++ T ELIESVR VAK +T++SS
Sbjct: 158 TEELLQILSEDGRTNELIESVRAVAKGETSSITSSSS 194
>emb|CAB71899.1| putative protein [Arabidopsis thaliana] gi|30102914|gb|AAP21375.1|
At3g61920 [Arabidopsis thaliana]
gi|17065044|gb|AAL32676.1| putative protein [Arabidopsis
thaliana] gi|15228644|ref|NP_191751.1| expressed protein
[Arabidopsis thaliana] gi|11281203|pir||T47984
hypothetical protein F21F14.90 - Arabidopsis thaliana
Length = 187
Score = 111 bits (277), Expect = 2e-23
Identities = 85/242 (35%), Positives = 111/242 (45%), Gaps = 70/242 (28%)
Query: 1 MGNCLLGANSDP-------DTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTND-- 51
MGNC+ N D+LIKV+T +G +ME +PPI FITNEF GH I +
Sbjct: 1 MGNCVFKGNGGSRKLYDKDDSLIKVVTPNGGVMELHPPIFAEFITNEFPGHVIHDSLSLR 60
Query: 52 QSSKPLCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSV 111
SS PL +EL G YYLLP++ S ATA+ Q
Sbjct: 61 HSSPPLLHGEELFPGNIYYLLPLS----------SSAAATAQLDSSDQ------------ 98
Query: 112 PTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIW 171
L PYRMS K+ I ++ S G+W
Sbjct: 99 ----LSTPYRMSF----------------------------GKTPIMAA--LSGGGCGVW 124
Query: 172 KVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLS 231
KV+L I+PE+L EIL+++V T+ L+ESVR VAKCG GCG + SDQ S++
Sbjct: 125 KVRLVISPEQLAEILAEDVETEALVESVRTVAKCG-----GYGCGGGVHSRANSDQLSVT 179
Query: 232 SS 233
SS
Sbjct: 180 SS 181
>emb|CAB85509.1| putative protein [Arabidopsis thaliana] gi|9758018|dbj|BAB08615.1|
unnamed protein product [Arabidopsis thaliana]
gi|15237558|ref|NP_196009.1| hypothetical protein
[Arabidopsis thaliana] gi|11289338|pir||T48416
hypothetical protein F8F6.100 - Arabidopsis thaliana
Length = 179
Score = 58.2 bits (139), Expect = 2e-07
Identities = 36/110 (32%), Positives = 59/110 (52%), Gaps = 10/110 (9%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQF 60
MGNCL+ +IK++ DG ++E+ PI+V+ I +F GH+I N L
Sbjct: 1 MGNCLVMEKK----VIKIVRDDGKVLEYREPISVHHILTQFSGHSISHNNTH----LLPD 52
Query: 61 DELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQS 110
+L++G+ YYLLP T+ K++ T A E G +++R+ S+S
Sbjct: 53 AKLLSGRLYYLLPTTM--TKKKVNKKVTFANPEVEGDERLLREEEDSSES 100
>dbj|BAC42468.1| unknown protein [Arabidopsis thaliana] gi|28372890|gb|AAO39927.1|
At3g10120 [Arabidopsis thaliana]
gi|30681157|ref|NP_187623.2| expressed protein
[Arabidopsis thaliana]
Length = 173
Score = 38.9 bits (89), Expect = 0.13
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 4/73 (5%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQF 60
MGNCL+ +IK++ DG ++E+ P+ V+ I +F H + ++ L
Sbjct: 1 MGNCLVMEKK----VIKIMRNDGKVVEYRGPMKVHHILTQFSPHYSLFDSLTNNCHLHPQ 56
Query: 61 DELVAGQSYYLLP 73
+L+ G+ YYLLP
Sbjct: 57 AKLLCGRLYYLLP 69
>gb|AAF98436.1| Hypothetical protein [Arabidopsis thaliana]
gi|15217852|ref|NP_174140.1| expressed protein
[Arabidopsis thaliana] gi|25373104|pir||H86407 F3H9.15
protein - Arabidopsis thaliana
Length = 266
Score = 36.2 bits (82), Expect = 0.85
Identities = 22/85 (25%), Positives = 41/85 (47%), Gaps = 4/85 (4%)
Query: 16 IKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKPLCQFDELVAGQSYYL 71
IK++ DG++ + P+ V+ +T +F H I ++ Q + L + + L G +Y+L
Sbjct: 39 IKLVRSDGSLQVYDRPVVVSELTKDFPKHKICRSDLLYIGQKTPVLSETETLKLGLNYFL 98
Query: 72 LPMTVLSPNNKIDYPSTCATAETGG 96
LP + +T T + GG
Sbjct: 99 LPSDFFKNDLSFLTIATLKTPQNGG 123
>dbj|BAC43473.1| unknown protein [Arabidopsis thaliana]
Length = 266
Score = 36.2 bits (82), Expect = 0.85
Identities = 22/85 (25%), Positives = 41/85 (47%), Gaps = 4/85 (4%)
Query: 16 IKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKPLCQFDELVAGQSYYL 71
IK++ DG++ + P+ V+ +T +F H I ++ Q + L + + L G +Y+L
Sbjct: 39 IKLVRSDGSLQVYDRPVVVSELTKDFPKHKICRSDLLYIGQKTPVLSETETLKLGLNYFL 98
Query: 72 LPMTVLSPNNKIDYPSTCATAETGG 96
LP + +T T + GG
Sbjct: 99 LPSDFFKNDLSFLTIATLKTPQNGG 123
>gb|EAL30699.1| GA18776-PA [Drosophila pseudoobscura]
Length = 200
Score = 36.2 bits (82), Expect = 0.85
Identities = 33/103 (32%), Positives = 51/103 (49%), Gaps = 4/103 (3%)
Query: 73 PMTVLSP-NNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQG 131
P+T L+P + ++ T GG + R+GH +S+ ++ L P R S + +
Sbjct: 14 PLTPLTPLSTQVFKFDTKEELTPTGGDTMGRRGHGEFRSLTSSRLTVPTRFSQSFLRTRS 73
Query: 132 VGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVK 174
V ST + S + S+ +T SSS R+SRKST KVK
Sbjct: 74 VFSPNSQSTANGS---SYSAGGGTTRSSSSRTSRKSTFSTKVK 113
>emb|CAC28825.2| related to a-agglutinin core protein AGA1 [Neurospora crassa]
gi|32404922|ref|XP_323074.1| hypothetical protein
[Neurospora crassa] gi|28922658|gb|EAA31883.1|
hypothetical protein [Neurospora crassa]
Length = 464
Score = 35.4 bits (80), Expect = 1.5
Identities = 42/168 (25%), Positives = 68/168 (40%), Gaps = 13/168 (7%)
Query: 75 TVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGVGL 134
T+ S + S ++A+ +Q S S + +P P + +
Sbjct: 159 TIASSTQQPTTSSASSSAQATSSSSSAQQSTTSSSS---SSVPTPEPSTTTQAPPVTITT 215
Query: 135 LKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKE 194
TST S T ++ KST SS+ ++S STG + E+ L ++S S
Sbjct: 216 TSTTSTTSTIPTTTSTTSIKSTSSSAPQASPSSTGTSVSE----SERTLVLISTSSSASS 271
Query: 195 LIESVRIVA------KCGVTAASSGGCGISSTTSIVSDQWSLSSSGRS 236
+ S V+ + GV++ +S G SST+S S SSS S
Sbjct: 272 VSSSASSVSSSEPSSEAGVSSTTSSSSGSSSTSSRSSSSLEPSSSSTS 319
>emb|CAB81192.1| hypothetical protein [Arabidopsis thaliana]
gi|4539365|emb|CAB40059.1| hypothetical protein
[Arabidopsis thaliana] gi|21553841|gb|AAM62934.1|
unknown [Arabidopsis thaliana]
gi|26453086|dbj|BAC43619.1| unknown protein [Arabidopsis
thaliana] gi|28827298|gb|AAO50493.1| unknown protein
[Arabidopsis thaliana] gi|18413395|ref|NP_567368.1|
expressed protein [Arabidopsis thaliana]
gi|7459223|pir||T04286 hypothetical protein F25I24.120 -
Arabidopsis thaliana
Length = 136
Score = 35.4 bits (80), Expect = 1.5
Identities = 17/50 (34%), Positives = 31/50 (62%)
Query: 163 SSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASS 212
S ++ G+WK K+ I ++L EIL+ E +T LI+ +R A + +++S
Sbjct: 69 SPQRRNGVWKAKVVIGSKQLEEILAVEGNTHALIDQLRFAAAEALVSSTS 118
>gb|AAX70878.1| hypothetical protein, conserved [Trypanosoma brucei]
Length = 739
Score = 35.0 bits (79), Expect = 1.9
Identities = 31/104 (29%), Positives = 38/104 (35%), Gaps = 11/104 (10%)
Query: 73 PMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGV 132
P T P N ST ATA T G + G+ S T PAP + G+
Sbjct: 507 PATSALPTNPFSPGSTTATAPTAGKPTTLFGGYATGDSSAATSAPAPGSAF----SFGGI 562
Query: 133 GLLKRTSTESFS-------CRTNRSSVNKSTISSSRRSSRKSTG 169
G ST FS T+ + VN + SR S TG
Sbjct: 563 GTTAPASTVGFSFGGMPKVAETSTAGVNTTISPFSRAGSTAVTG 606
>ref|NP_198600.1| expressed protein [Arabidopsis thaliana]
gi|48310396|gb|AAT41812.1| At5g37840 [Arabidopsis
thaliana] gi|46518385|gb|AAS99674.1| At5g37840
[Arabidopsis thaliana]
Length = 214
Score = 35.0 bits (79), Expect = 1.9
Identities = 22/90 (24%), Positives = 40/90 (44%), Gaps = 9/90 (10%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQS-----SK 55
MGN ++ + +KV+ DG+I P+T + T E+ G + + +K
Sbjct: 1 MGNTIMVRRNK----VKVMKIDGDIFRLKTPVTASDATKEYPGFVLLDSETVKRLGVRAK 56
Query: 56 PLCQFDELVAGQSYYLLPMTVLSPNNKIDY 85
PL L +Y+L+ + + NK+ Y
Sbjct: 57 PLEPNQILKPNHTYFLVDLPPVDKRNKLPY 86
>ref|XP_640327.1| hypothetical protein DDB0218364 [Dictyostelium discoideum]
gi|60468363|gb|EAL66370.1| hypothetical protein
DDB0218364 [Dictyostelium discoideum]
Length = 867
Score = 35.0 bits (79), Expect = 1.9
Identities = 33/140 (23%), Positives = 58/140 (40%), Gaps = 19/140 (13%)
Query: 35 NFITNEFQGHAIFPTNDQSSKPLCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAET 94
+FITNE + ++P + + KPL F +LV + N I S+C ++E
Sbjct: 34 DFITNELR--KVYP--EYNRKPLKGFKQLVEK-----------AMNQIIKNLSSCESSEN 78
Query: 95 GGGGQIIRQGHVRSQSVPTTPLPAP----YRMSLDYQYYQGVGLLKRTSTESFSCRTNRS 150
G + + + + PLP P L QY Q + T+T + + NR
Sbjct: 79 EDGDNKMEESDGDDVEIISPPLPPPSSNILNEQLTSQYKQNIKNTPPTTTTTTPAKRNRD 138
Query: 151 SVNKSTISSSRRSSRKSTGI 170
S ++S+ ++ + I
Sbjct: 139 ENIPSNVNSNNNNNNNNNAI 158
>gb|AAS07910.1| oxidoreductase, short-chain dehydrogenase/reductase family
[uncultured bacterium 463]
Length = 314
Score = 35.0 bits (79), Expect = 1.9
Identities = 19/58 (32%), Positives = 31/58 (52%), Gaps = 1/58 (1%)
Query: 191 STKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPSKVDALVLDI 248
ST E+IE V + KC + +S G G+ ++ S+ S ++ R A SK+D V +
Sbjct: 9 STSEVIEGVDLKGKCALVTGASSGLGVETSRSLASAGAAVIMVARDA-SKLDTAVAQV 65
>emb|CAA87091.1| secreted acid phosphatase 2 (SAP2) [Leishmania mexicana]
gi|11359724|pir||T46726 secreted acid phosphatase 2
precursor [imported] - Leishmania mexicana
Length = 888
Score = 34.3 bits (77), Expect = 3.2
Identities = 36/135 (26%), Positives = 57/135 (41%), Gaps = 9/135 (6%)
Query: 112 PTTPLPAPYRMS-LDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISS--SRRSSRKST 168
P+ P Y +S +D+Q Y G + TS+ S T SS +T SS + SS + T
Sbjct: 425 PSKACPDSYILSAVDHQCYPGPDVTNPTSSSSSEGTTTSSSEGTATSSSDVTTTSSSEGT 484
Query: 169 GIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQW 228
+ + S + +T ++ + G T++S S T+ SD
Sbjct: 485 ATSSSDATTSSSEGTATSSSDATTSSSSDATTTSSSEGTTSSS------SDATTSSSDAT 538
Query: 229 SLSSSGRSAPSKVDA 243
+ SSS +A S DA
Sbjct: 539 TTSSSEGTATSSSDA 553
>ref|ZP_00421276.1| Haemagluttinin motif:Hep_Hag [Burkholderia vietnamiensis G4]
gi|67535522|gb|EAM32254.1| Haemagluttinin motif:Hep_Hag
[Burkholderia vietnamiensis G4]
Length = 2463
Score = 34.3 bits (77), Expect = 3.2
Identities = 37/114 (32%), Positives = 50/114 (43%), Gaps = 13/114 (11%)
Query: 138 TSTESFSCRTNRSSVNKSTISSSRRSSRK----STGIWKVKLCITPEKLLEILSQEVST- 192
TST S T SS N S +S S +S STG+ +T LS ST
Sbjct: 474 TSTGISSLSTGLSSTNSSVVSLSTSTSTGISSLSTGLSSTNSSVTS------LSSSTSTG 527
Query: 193 -KELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSG-RSAPSKVDAL 244
S+ ++ T SS G+SST S V+ S +S+G SA S + +L
Sbjct: 528 LSSATSSITSLSTSTSTGISSLSTGLSSTNSSVTSLSSSTSTGLSSATSSITSL 581
>gb|AAF04412.1| unknown protein [Arabidopsis thaliana]
Length = 167
Score = 34.3 bits (77), Expect = 3.2
Identities = 18/59 (30%), Positives = 33/59 (55%)
Query: 15 LIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQFDELVAGQSYYLLP 73
+IK++ DG ++E+ P+ V+ I +F H + ++ L +L+ G+ YYLLP
Sbjct: 5 VIKIMRNDGKVVEYRGPMKVHHILTQFSPHYSLFDSLTNNCHLHPQAKLLCGRLYYLLP 63
>ref|XP_324370.1| predicted protein [Neurospora crassa] gi|28917339|gb|EAA27046.1|
predicted protein [Neurospora crassa]
Length = 1165
Score = 34.3 bits (77), Expect = 3.2
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Query: 66 GQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLD 125
GQ L T LS ++ +D+ ++ +T+ + I + H R Q PT+P + + +++D
Sbjct: 1038 GQLGPLSHATTLSLSDPLDHTNSASTSSS------ISKSHYRQQQKPTSPTKSDFSLAVD 1091
Query: 126 -YQYYQGVGL 134
Y Y G G+
Sbjct: 1092 NYNKYNGFGM 1101
>ref|ZP_00319709.1| COG1705: Muramidase (flagellum-specific) [Oenococcus oeni PSU-1]
Length = 390
Score = 33.9 bits (76), Expect = 4.2
Identities = 31/103 (30%), Positives = 47/103 (45%), Gaps = 5/103 (4%)
Query: 138 TSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIE 197
TST + S ++ SSV+ S+ +SS SS S+ T S S+ +
Sbjct: 182 TSTSTSSAASSASSVSSSSATSSSASSAASSASSVSSSSATSSS----ASSAASSASSVS 237
Query: 198 SVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPSK 240
S + +AASS +SS+++ S S SSS S+ SK
Sbjct: 238 SSSATSSSASSAASSAS-SVSSSSATSSSASSTSSSSTSSSSK 279
>ref|NP_609495.1| CG4751-PA [Drosophila melanogaster] gi|7297828|gb|AAF53077.1|
CG4751-PA [Drosophila melanogaster]
gi|15292267|gb|AAK93402.1| LD44770p [Drosophila
melanogaster]
Length = 1412
Score = 33.5 bits (75), Expect = 5.5
Identities = 54/225 (24%), Positives = 87/225 (38%), Gaps = 37/225 (16%)
Query: 45 AIFPT-NDQSSKPLCQFDELVAGQ---SYYLLPMTVLSPNNKIDYPST------CATAET 94
A PT ND + L Q +AGQ ++ + L + DY S+ A A+
Sbjct: 757 ACLPTANDLMAASLAQ----LAGQLPPNFLQGDLAALFQQQRKDYGSSSLNQLAAAAAKV 812
Query: 95 GGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSS-VN 153
GG Q A Y S + GVG+ ++ + S ++S +
Sbjct: 813 GGSKQSNASASSGLNDPNVAAAVAAYSNSFNMPLPTGVGIGGSGNSNAHSSSKSKSERSS 872
Query: 154 KSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSG 213
KS+ SSS +S ++ +K KL ++L E+ + + ELI S A A + G
Sbjct: 873 KSSSSSSSSNSSSTSNSYKTKLM---KELDELKNDPLKMSELIRSPEYAALLLQQAEALG 929
Query: 214 -------------------GCGISSTTSIVSDQWSLSSSGRSAPS 239
G G+ + ++ Q S SSS +S+ S
Sbjct: 930 ATTLGTLGFGSDYSYLTGAGLGVPAANALTGGQSSNSSSSKSSKS 974
>gb|AAM62831.1| unknown [Arabidopsis thaliana] gi|26451664|dbj|BAC42928.1| unknown
protein [Arabidopsis thaliana]
gi|28973267|gb|AAO63958.1| unknown protein [Arabidopsis
thaliana] gi|18420002|ref|NP_568020.1| expressed protein
[Arabidopsis thaliana]
Length = 168
Score = 33.5 bits (75), Expect = 5.5
Identities = 29/107 (27%), Positives = 48/107 (44%), Gaps = 10/107 (9%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKP 56
MG C ++ T K+I DG +MEF P+ V ++ ++ I ++ D +
Sbjct: 1 MGICSSSESTQVATA-KLILQDGRMMEFANPVKVGYVLLKYPMCFICNSDDMDFDDAVAA 59
Query: 57 LCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAET-----GGGG 98
+ +EL GQ Y+ LP+ L K + + A + GGGG
Sbjct: 60 ISADEELQLGQIYFALPLCWLRQPLKAEEMAALAVKASSALMRGGGG 106
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 418,642,492
Number of Sequences: 2540612
Number of extensions: 16513321
Number of successful extensions: 34465
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 34385
Number of HSP's gapped (non-prelim): 70
length of query: 248
length of database: 863,360,394
effective HSP length: 125
effective length of query: 123
effective length of database: 545,783,894
effective search space: 67131418962
effective search space used: 67131418962
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146664.11


