

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.1 - phase: 0
(935 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
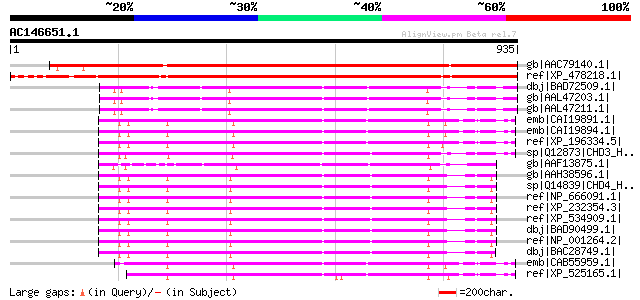
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC79140.1| human Mi-2 autoantigen-like protein [Arabidopsis ... 1239 0.0
ref|XP_478218.1| chromodomain helicase DNA binding protein-like ... 1153 0.0
dbj|BAD72509.1| chromatin-remodeling factor CHD3 [Oryza sativa (... 555 e-156
gb|AAL47203.1| chromatin-remodeling factor CHD3 [Oryza sativa (i... 555 e-156
gb|AAL47211.1| chromatin-remodeling factor CHD3 [Oryza sativa] 555 e-156
emb|CAI19891.1| chromodomain helicase DNA binding protein 5 [Hom... 538 e-151
emb|CAI19894.1| chromodomain helicase DNA binding protein 5 [Hom... 538 e-151
ref|XP_196334.5| PREDICTED: chromodomain helicase DNA binding pr... 537 e-151
sp|Q12873|CHD3_HUMAN Chromodomain helicase-DNA-binding protein 3... 536 e-150
gb|AAF13875.1| chromatin remodeling factor CHD3 [Arabidopsis tha... 533 e-150
gb|AAH38596.1| CHD4 protein [Homo sapiens] 530 e-149
sp|Q14839|CHD4_HUMAN Chromodomain helicase-DNA-binding protein 4... 530 e-149
ref|NP_666091.1| chromodomain helicase DNA binding protein 4 [Mu... 530 e-149
ref|XP_232354.3| PREDICTED: chromodomain helicase DNA binding pr... 530 e-149
ref|XP_534909.1| PREDICTED: similar to Mi-2 protein [Canis famil... 530 e-149
dbj|BAD90499.1| mKIAA4075 protein [Mus musculus] 530 e-149
ref|NP_001264.2| chromodomain helicase DNA binding protein 4 [Ho... 530 e-149
dbj|BAC28749.1| unnamed protein product [Mus musculus] 529 e-148
emb|CAB55959.1| hypothetical protein [Homo sapiens] 525 e-147
ref|XP_525165.1| PREDICTED: similar to chromodomain helicase DNA... 518 e-145
>gb|AAC79140.1| human Mi-2 autoantigen-like protein [Arabidopsis thaliana]
gi|9758384|dbj|BAB08833.1| helicase-like protein
[Arabidopsis thaliana] gi|15241584|ref|NP_199293.1|
chromodomain-helicase-DNA-binding family protein / CHD
family protein [Arabidopsis thaliana]
Length = 2228
Score = 1239 bits (3205), Expect = 0.0
Identities = 624/876 (71%), Positives = 728/876 (82%), Gaps = 21/876 (2%)
Query: 74 ATDDLGSCARDGT----DQDDYAVSDEQLEKENDKLETEENLNVVLRGDGNSKLPNNCEM 129
++ + G +RD D DD A+ E + + +++ +E+ N L + + E
Sbjct: 433 SSSETGKSSRDSRLRDKDMDDSALGTEGMVEVKEEMLSEDISNATLSRHVDDEDMKVSET 492
Query: 130 HDSLE----------TKQKEVVLEKGMGSSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNS 179
H S+E T +K V ++ + K D IGE VSYEFLVKWV KS+IHN+
Sbjct: 493 HVSVERELLEEAHQETGEKSTVADEEIEEPVAAKTSDLIGETVSYEFLVKWVDKSNIHNT 552
Query: 180 WISESHLKVIAKRKLENYKAKYGTATINICEEQWKNPERLLAIRTSKQGTSEAFVKWTGK 239
WISE+ LK +AKRKLENYKAKYGTA INICE++WK P+R++A+R SK+G EA+VKWTG
Sbjct: 553 WISEAELKGLAKRKLENYKAKYGTAVINICEDKWKQPQRIVALRVSKEGNQEAYVKWTGL 612
Query: 240 PYNECTWESLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQNDIVNLLEQP 299
Y+ECTWESL+EP+L++SSHLI F+ +E TLER SK N T++ + ++V L EQP
Sbjct: 613 AYDECTWESLEEPILKHSSHLIDLFHQYEQKTLERN-SKGNPTRE----RGEVVTLTEQP 667
Query: 300 KELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPC 359
+ELRGG+LF HQLEALNWLR+CW+KS+NVILADEMGLGKT+SA AF+SSLYFEF V+RPC
Sbjct: 668 QELRGGALFAHQLEALNWLRRCWHKSKNVILADEMGLGKTVSASAFLSSLYFEFGVARPC 727
Query: 360 LVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKF 419
LVLVPL TM NWL+EF+LWAP +NVV+YHG AK RAIIR YEWHA + +G KK +YKF
Sbjct: 728 LVLVPLSTMPNWLSEFSLWAPLLNVVEYHGSAKGRAIIRDYEWHAKNSTGTTKKPTSYKF 787
Query: 420 NVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPL 479
NVLLT+YEMVLAD SH RGVPWEVL+VDEGHRLKNSESKLFSLLN+ SFQHRVLLTGTPL
Sbjct: 788 NVLLTTYEMVLADSSHLRGVPWEVLVVDEGHRLKNSESKLFSLLNTFSFQHRVLLTGTPL 847
Query: 480 QNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQN 539
QNN+GEMYNLLNFLQP+SFPSLS+FEERF+DLTSAEKV+ELKKLV+PHMLRRLKKDAMQN
Sbjct: 848 QNNIGEMYNLLNFLQPSSFPSLSSFEERFHDLTSAEKVEELKKLVAPHMLRRLKKDAMQN 907
Query: 540 IPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYL 599
IPPKTERMVPVEL+SIQAEYYRAMLTKNYQILRNIGKG+AQQSMLNIVMQLRKVCNHPYL
Sbjct: 908 IPPKTERMVPVELTSIQAEYYRAMLTKNYQILRNIGKGVAQQSMLNIVMQLRKVCNHPYL 967
Query: 600 IPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLN 659
IPGTEP+SGS+EFLH+MRIKASAKLTLLHSMLK+L+KEGHRVLIFSQMTKLLDILEDYLN
Sbjct: 968 IPGTEPESGSLEFLHDMRIKASAKLTLLHSMLKVLHKEGHRVLIFSQMTKLLDILEDYLN 1027
Query: 660 IEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDS 719
IEFGPKT+ERVDGSV+V DRQ AIARFNQDK+RFVFLLSTR+CGLGINLATADTVIIYDS
Sbjct: 1028 IEFGPKTFERVDGSVAVADRQAAIARFNQDKNRFVFLLSTRACGLGINLATADTVIIYDS 1087
Query: 720 DFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQK 779
DFNPHADIQAMNRAHRIGQS RLLVYRLVVRASVEERILQLAKKKLMLDQLF KSGSQK
Sbjct: 1088 DFNPHADIQAMNRAHRIGQSKRLLVYRLVVRASVEERILQLAKKKLMLDQLFVNKSGSQK 1147
Query: 780 EVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCT 839
E EDIL+WGTEELFNDS N KDT+E+N + + + ++E K RK+ GGLGDVY+DKCT
Sbjct: 1148 EFEDILRWGTEELFNDSAGENKKDTAESNGNL--DVIMDLESKSRKKGGGLGDVYQDKCT 1205
Query: 840 DNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEEHVEGES 899
+ + KI+WD+ AI+KLLDRSNLQ ASTD A+ + +NDMLGS+K +EWN+E EE V ES
Sbjct: 1206 EGNGKIVWDDIAIMKLLDRSNLQSASTDAADTELDNDMLGSVKPVEWNEETAEEQVGAES 1265
Query: 900 PPHGADDMCTQNSEKKEDNAVIGGEENEWDRLLRLR 935
P DD +SE+K+D+ V EENEWDRLLR+R
Sbjct: 1266 PALVTDDTGEPSSERKDDDVVNFTEENEWDRLLRMR 1301
>ref|XP_478218.1| chromodomain helicase DNA binding protein-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 2259
Score = 1153 bits (2982), Expect = 0.0
Identities = 603/934 (64%), Positives = 718/934 (76%), Gaps = 34/934 (3%)
Query: 2 SENQTRLAEDNSACDNDLDGEIAENLVHDPQNVKSSDEGELHNTDRVEKIHVYRRSITKE 61
S + T + ED AC ++L + EN N+ SD + N + H ++ S+ +
Sbjct: 399 SNSDTNVTED--ACADELANDGGEN------NLDCSDAQKESNV----RSHGHKESLNAK 446
Query: 62 SKNGNLLNSLSKATDDLGSCARDGTDQDDYAVSDEQLEKENDKLETEENLNVVLRGDGNS 121
++N+ S + D +D Y + E E EE ++
Sbjct: 447 E----IMNTASACSADQIVTVKDAGAVQTYVTASVNGEYETVTDIPEEK--------NDT 494
Query: 122 KLPNNCEMHDSLETKQKEVVLEKGMGSSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNSWI 181
K P + + + TKQ+ K G + ++++ G +YEFLVKWVGKS+IHNSWI
Sbjct: 495 KHPVS-KADTEVHTKQEHTPDSKLHGKIQETELKEHDG--TTYEFLVKWVGKSNIHNSWI 551
Query: 182 SESHLKVIAKRKLENYKAKYGTATINICEEQWKNPERLLAIRTSKQGTSEAFVKWTGKPY 241
SES LK +AKRKLENYKAKYGT INIC+EQW P+R++A+RTS EA +KW PY
Sbjct: 552 SESELKALAKRKLENYKAKYGTGLINICKEQWCQPQRVIALRTSLDEIEEALIKWCALPY 611
Query: 242 NECTWESLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQNDIVNLLEQPKE 301
+ECTWE LDEP + +HL+T+F FE+ L+++ K S K + Q + L+EQPKE
Sbjct: 612 DECTWERLDEPTMVKYAHLVTQFKKFESQALDKD--KGGSHAKPREHQ-EFNMLVEQPKE 668
Query: 302 LRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
L+GG LFPHQLEALNWLRKCWYKS+NVILADEMGLGKT+SACAF+SSL E+K++ PCLV
Sbjct: 669 LQGGMLFPHQLEALNWLRKCWYKSKNVILADEMGLGKTVSACAFLSSLCCEYKINLPCLV 728
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNV 421
LVPL TM NW+AEFA WAP +NVV+YHG A+AR+IIRQYEWH D S + K +++KFNV
Sbjct: 729 LVPLSTMPNWMAEFASWAPHLNVVEYHGSARARSIIRQYEWHEGDASQMGKIKKSHKFNV 788
Query: 422 LLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQN 481
LLT+YEMVL D ++ R V WEVLIVDEGHRLKNS SKLFSLLN++SFQHRVLLTGTPLQN
Sbjct: 789 LLTTYEMVLVDAAYLRSVSWEVLIVDEGHRLKNSSSKLFSLLNTLSFQHRVLLTGTPLQN 848
Query: 482 NLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIP 541
N+GEMYNLLNFLQPASFPSL++FEE+FNDLT+ EKV+ELK LV+PHMLRRLKKDAMQNIP
Sbjct: 849 NIGEMYNLLNFLQPASFPSLASFEEKFNDLTTTEKVEELKNLVAPHMLRRLKKDAMQNIP 908
Query: 542 PKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIP 601
PKTERMVPVEL+SIQAEYYRAMLTKNYQ+LRNIGKG A QS+LNIVMQLRKVCNHPYLIP
Sbjct: 909 PKTERMVPVELTSIQAEYYRAMLTKNYQVLRNIGKGGAHQSLLNIVMQLRKVCNHPYLIP 968
Query: 602 GTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIE 661
GTEP+SGS EFLHEMRIKASAKLTLLHSMLKIL+K+GHRVLIFSQMTKLLDILEDYL E
Sbjct: 969 GTEPESGSPEFLHEMRIKASAKLTLLHSMLKILHKDGHRVLIFSQMTKLLDILEDYLTWE 1028
Query: 662 FGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDF 721
FGPKT+ERVDGSVSV +RQ AIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDF
Sbjct: 1029 FGPKTFERVDGSVSVAERQAAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDF 1088
Query: 722 NPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEV 781
NPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERIL LAKKKLMLDQLF KS SQKEV
Sbjct: 1089 NPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILHLAKKKLMLDQLFVNKSESQKEV 1148
Query: 782 EDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDN 841
EDI++WGTEELF +S + KD +E + + D VAEVE KH+++TGGLGDVYED+C D
Sbjct: 1149 EDIIRWGTEELFRNS-DVAVKDNNEASGAKND--VAEVEFKHKRKTGGLGDVYEDRCADG 1205
Query: 842 SSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEEHVEGESPP 901
S+K +WDENAI KLLDRSN+ + +GD +NDMLG++K+++WNDE ++ E P
Sbjct: 1206 SAKFIWDENAITKLLDRSNVPSTVAESTDGDLDNDMLGTVKSIDWNDELNDDPGATEDIP 1265
Query: 902 HGADDMCTQNSEKKEDNAVIGGEENEWDRLLRLR 935
+ +D C Q SE K+D A EENEWD+LLR+R
Sbjct: 1266 NIDNDGCEQASEAKQD-AANRVEENEWDKLLRVR 1298
>dbj|BAD72509.1| chromatin-remodeling factor CHD3 [Oryza sativa (japonica
cultivar-group)] gi|55771379|dbj|BAD72546.1|
chromatin-remodeling factor CHD3 [Oryza sativa (japonica
cultivar-group)]
Length = 1354
Score = 555 bits (1431), Expect = e-156
Identities = 332/802 (41%), Positives = 476/802 (58%), Gaps = 101/802 (12%)
Query: 166 FLVKWVGKSHIHNSWISESHLKVIAK------RKLENYKAKYGTAT-----INICEEQWK 214
+L+KW G SH+H +W+SES AK +L N+ + + + +W
Sbjct: 137 YLIKWKGISHLHCTWVSESEYLETAKIYPRLKTRLNNFHKQMDSTDKSDDDYSAIRPEWT 196
Query: 215 NPERLLAIRTSKQGTSEAFVKWTGKPYNECTWES-LDEPVLQNSSHLITRFNMFETLTLE 273
+R+LA R S G E +VKW Y+ECTWE+ D V Q I RFN E
Sbjct: 197 TVDRILATRKSSTGEREYYVKWKELTYDECTWENDSDIAVFQPQ---IERFN-------E 246
Query: 274 REASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADE 333
++ ++ ST K +I E PK L GG+L P+QLE LN+LR WY ++ VIL DE
Sbjct: 247 IQSRRKKSTDKCKSVTREIRQYKESPKFLSGGTLHPYQLEGLNFLRYSWYHNKRVILGDE 306
Query: 334 MGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKA 393
MGLGKTI + AF+ SL+ + K+ P LV+ PL T+ NW EFA WAP +NVV Y G A +
Sbjct: 307 MGLGKTIQSIAFLGSLFVD-KLG-PHLVVAPLSTLRNWEREFATWAPQMNVVMYFGSAAS 364
Query: 394 RAIIRQYEWH------------ASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPW 441
R IIR+YE++ S PS +KK KF+VLLTSYEM+ D + + + W
Sbjct: 365 REIIRKYEFYYPKEKPKKLKKKKSSPSNEDKKQSRIKFDVLLTSYEMINMDSTVLKTIEW 424
Query: 442 EVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSL 501
E +IVDEGHRLKN +SKLF L +HRVLLTGTP+QNNL E++ L++FL+ SF S+
Sbjct: 425 ECMIVDEGHRLKNKDSKLFGQLKEYHTKHRVLLTGTPVQNNLDELFMLMHFLEGDSFGSI 484
Query: 502 SAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYR 561
+ +E F D+ ++V++L ++ PH+LRR KKD M+ +PPK E ++ VEL+S Q EYY+
Sbjct: 485 ADLQEEFKDINQDKQVEKLHGMLKPHLLRRFKKDVMKELPPKKELILRVELTSKQKEYYK 544
Query: 562 AMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKAS 621
A+LTKNY++L G S++N+VM+LRK+C H ++ E + S E L + +++S
Sbjct: 545 AILTKNYEVLTRRSGG--HVSLINVVMELRKLCCHAFMTDEPEEPANSEEALRRL-LESS 601
Query: 622 AKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQT 681
K+ LL M+ L ++GHRVLI+SQ +LD+LEDYL+ + +YER+DG + +RQ
Sbjct: 602 GKMELLDKMMVKLKEQGHRVLIYSQFQHMLDLLEDYLS--YRKWSYERIDGKIGGAERQI 659
Query: 682 AIARFN-QDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
I RFN ++ +RF FLLSTR+ GLGINLATADTVIIYDSD+NPHAD+QAM RAHR+GQ++
Sbjct: 660 RIDRFNAKNSTRFCFLLSTRAGGLGINLATADTVIIYDSDWNPHADLQAMARAHRLGQTS 719
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLD-----QLFKGKSGSQKEVEDILKWGTEELFND 795
++++YRLV R ++EER++QL KKK++L+ +L KG + Q+E++DI++ G++ELF+D
Sbjct: 720 KVMIYRLVSRGTIEERMMQLTKKKMVLEHLVVGRLTKGTNIVQEELDDIIRHGSKELFDD 779
Query: 796 SCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKL 855
EN+ + K S +I +D+ AI +L
Sbjct: 780 ----------ENDEAGK-----------------------------SCQIHYDDAAIDRL 800
Query: 856 LDRSNLQDASTDIAEGDSENDMLGSMKA--LEWNDEPTEEHVEGESPPHGADDMCTQNSE 913
LDR Q + E + E++ L K E+ DE + E + +
Sbjct: 801 LDRD--QADGEEPVEDEEEDEFLKGFKVANFEYIDEAKALAAKEE-----------EARK 847
Query: 914 KKEDNAVIGGEENEWDRLLRLR 935
K E A N WD+LL+ R
Sbjct: 848 KAEAEAANSDRANFWDKLLKDR 869
>gb|AAL47203.1| chromatin-remodeling factor CHD3 [Oryza sativa (indica
cultivar-group)]
Length = 1111
Score = 555 bits (1431), Expect = e-156
Identities = 332/802 (41%), Positives = 476/802 (58%), Gaps = 101/802 (12%)
Query: 166 FLVKWVGKSHIHNSWISESHLKVIAK------RKLENYKAKYGTAT-----INICEEQWK 214
+L+KW G SH+H +W+SES AK +L N+ + + + +W
Sbjct: 143 YLIKWKGISHLHCTWVSESEYLETAKIYPRLKTRLNNFHKQMDSTDKSDDDYSAIRPEWT 202
Query: 215 NPERLLAIRTSKQGTSEAFVKWTGKPYNECTWES-LDEPVLQNSSHLITRFNMFETLTLE 273
+R+LA R S G E +VKW Y+ECTWE+ D V Q I RFN E
Sbjct: 203 TVDRILATRKSSTGEREYYVKWKELTYDECTWENDSDIAVFQPQ---IERFN-------E 252
Query: 274 REASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADE 333
++ ++ ST K +I E PK L GG+L P+QLE LN+LR WY ++ VIL DE
Sbjct: 253 IQSRRKKSTDKCKSVTREIRQYKESPKFLSGGTLHPYQLEGLNFLRYSWYHNKRVILGDE 312
Query: 334 MGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKA 393
MGLGKTI + AF+ SL+ + K+ P LV+ PL T+ NW EFA WAP +NVV Y G A +
Sbjct: 313 MGLGKTIQSIAFLGSLFVD-KLG-PHLVVAPLSTLRNWEREFATWAPQMNVVMYFGSAAS 370
Query: 394 RAIIRQYEWH------------ASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPW 441
R IIR+YE++ S PS +KK KF+VLLTSYEM+ D + + + W
Sbjct: 371 REIIRKYEFYYPKEKPKKLKKKKSSPSNEDKKQSRIKFDVLLTSYEMINMDSTVLKTIEW 430
Query: 442 EVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSL 501
E +IVDEGHRLKN +SKLF L +HRVLLTGTP+QNNL E++ L++FL+ SF S+
Sbjct: 431 ECMIVDEGHRLKNKDSKLFGQLKEYHTKHRVLLTGTPVQNNLDELFMLMHFLEGDSFGSI 490
Query: 502 SAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYR 561
+ +E F D+ ++V++L ++ PH+LRR KKD M+ +PPK E ++ VEL+S Q EYY+
Sbjct: 491 ADLQEEFKDINQDKQVEKLHGMLKPHLLRRFKKDVMKELPPKKELILRVELTSKQKEYYK 550
Query: 562 AMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKAS 621
A+LTKNY++L G S++N+VM+LRK+C H ++ E + S E L + +++S
Sbjct: 551 AILTKNYEVLTRRSGG--HVSLINVVMELRKLCCHAFMTDEPEEPANSEEALRRL-LESS 607
Query: 622 AKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQT 681
K+ LL M+ L ++GHRVLI+SQ +LD+LEDYL+ + +YER+DG + +RQ
Sbjct: 608 GKMELLDKMMVKLKEQGHRVLIYSQFQHMLDLLEDYLS--YRKWSYERIDGKIGGAERQI 665
Query: 682 AIARFN-QDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
I RFN ++ +RF FLLSTR+ GLGINLATADTVIIYDSD+NPHAD+QAM RAHR+GQ++
Sbjct: 666 RIDRFNAKNSTRFCFLLSTRAGGLGINLATADTVIIYDSDWNPHADLQAMARAHRLGQTS 725
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLD-----QLFKGKSGSQKEVEDILKWGTEELFND 795
++++YRLV R ++EER++QL KKK++L+ +L KG + Q+E++DI++ G++ELF+D
Sbjct: 726 KVMIYRLVSRGTIEERMMQLTKKKMVLEHLVVGRLTKGTNIVQEELDDIIRHGSKELFDD 785
Query: 796 SCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKL 855
EN+ + K S +I +D+ AI +L
Sbjct: 786 ----------ENDEAGK-----------------------------SCQIHYDDAAIDRL 806
Query: 856 LDRSNLQDASTDIAEGDSENDMLGSMKA--LEWNDEPTEEHVEGESPPHGADDMCTQNSE 913
LDR Q + E + E++ L K E+ DE + E + +
Sbjct: 807 LDRD--QADGEEPVEDEEEDEFLKGFKVANFEYIDEAKALAAKEE-----------EARK 853
Query: 914 KKEDNAVIGGEENEWDRLLRLR 935
K E A N WD+LL+ R
Sbjct: 854 KAEAEAANSDRANFWDKLLKDR 875
>gb|AAL47211.1| chromatin-remodeling factor CHD3 [Oryza sativa]
Length = 1360
Score = 555 bits (1431), Expect = e-156
Identities = 332/802 (41%), Positives = 476/802 (58%), Gaps = 101/802 (12%)
Query: 166 FLVKWVGKSHIHNSWISESHLKVIAK------RKLENYKAKYGTAT-----INICEEQWK 214
+L+KW G SH+H +W+SES AK +L N+ + + + +W
Sbjct: 143 YLIKWKGISHLHCTWVSESEYLETAKIYPRLKTRLNNFHKQMDSTDKSDDDYSAIRPEWT 202
Query: 215 NPERLLAIRTSKQGTSEAFVKWTGKPYNECTWES-LDEPVLQNSSHLITRFNMFETLTLE 273
+R+LA R S G E +VKW Y+ECTWE+ D V Q I RFN E
Sbjct: 203 TVDRILATRKSSTGEREYYVKWKELTYDECTWENDSDIAVFQPQ---IERFN-------E 252
Query: 274 REASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADE 333
++ ++ ST K +I E PK L GG+L P+QLE LN+LR WY ++ VIL DE
Sbjct: 253 IQSRRKKSTDKCKSVTREIRQYKESPKFLSGGTLHPYQLEGLNFLRYSWYHNKRVILGDE 312
Query: 334 MGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKA 393
MGLGKTI + AF+ SL+ + K+ P LV+ PL T+ NW EFA WAP +NVV Y G A +
Sbjct: 313 MGLGKTIQSIAFLGSLFVD-KLG-PHLVVAPLSTLRNWEREFATWAPQMNVVMYFGSAAS 370
Query: 394 RAIIRQYEWH------------ASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPW 441
R IIR+YE++ S PS +KK KF+VLLTSYEM+ D + + + W
Sbjct: 371 REIIRKYEFYYPKEKPKKLKKKKSSPSNEDKKQSRIKFDVLLTSYEMINMDSTVLKTIEW 430
Query: 442 EVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSL 501
E +IVDEGHRLKN +SKLF L +HRVLLTGTP+QNNL E++ L++FL+ SF S+
Sbjct: 431 ECMIVDEGHRLKNKDSKLFGQLKEYHTKHRVLLTGTPVQNNLDELFMLMHFLEGDSFGSI 490
Query: 502 SAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYR 561
+ +E F D+ ++V++L ++ PH+LRR KKD M+ +PPK E ++ VEL+S Q EYY+
Sbjct: 491 ADLQEEFKDINQDKQVEKLHGMLKPHLLRRFKKDVMKELPPKKELILRVELTSKQKEYYK 550
Query: 562 AMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKAS 621
A+LTKNY++L G S++N+VM+LRK+C H ++ E + S E L + +++S
Sbjct: 551 AILTKNYEVLTRRSGG--HVSLINVVMELRKLCCHAFMTDEPEEPANSEEALRRL-LESS 607
Query: 622 AKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQT 681
K+ LL M+ L ++GHRVLI+SQ +LD+LEDYL+ + +YER+DG + +RQ
Sbjct: 608 GKMELLDKMMVKLKEQGHRVLIYSQFQHMLDLLEDYLS--YRKWSYERIDGKIGGAERQI 665
Query: 682 AIARFN-QDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
I RFN ++ +RF FLLSTR+ GLGINLATADTVIIYDSD+NPHAD+QAM RAHR+GQ++
Sbjct: 666 RIDRFNAKNSTRFCFLLSTRAGGLGINLATADTVIIYDSDWNPHADLQAMARAHRLGQTS 725
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLD-----QLFKGKSGSQKEVEDILKWGTEELFND 795
++++YRLV R ++EER++QL KKK++L+ +L KG + Q+E++DI++ G++ELF+D
Sbjct: 726 KVMIYRLVSRGTIEERMMQLTKKKMVLEHLVVGRLTKGTNIVQEELDDIIRHGSKELFDD 785
Query: 796 SCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKL 855
EN+ + K S +I +D+ AI +L
Sbjct: 786 ----------ENDEAGK-----------------------------SCQIHYDDAAIDRL 806
Query: 856 LDRSNLQDASTDIAEGDSENDMLGSMKA--LEWNDEPTEEHVEGESPPHGADDMCTQNSE 913
LDR Q + E + E++ L K E+ DE + E + +
Sbjct: 807 LDRD--QADGEEPVEDEEEDEFLKGFKVANFEYIDEAKALAAKEE-----------EARK 853
Query: 914 KKEDNAVIGGEENEWDRLLRLR 935
K E A N WD+LL+ R
Sbjct: 854 KAEAEAANSDRANFWDKLLKDR 875
>emb|CAI19891.1| chromodomain helicase DNA binding protein 5 [Homo sapiens]
gi|56203111|emb|CAI19450.1| chromodomain helicase DNA
binding protein 5 [Homo sapiens]
gi|19773960|gb|AAL98962.1| chromodomain helicase DNA
binding protein 5 [Homo sapiens]
gi|51701343|sp|Q8TDI0|CHD5_HUMAN Chromodomain
helicase-DNA-binding protein 5 (CHD-5)
gi|24308089|ref|NP_056372.1| chromodomain helicase DNA
binding protein 5 [Homo sapiens]
Length = 1954
Score = 538 bits (1385), Expect = e-151
Identities = 351/845 (41%), Positives = 474/845 (55%), Gaps = 105/845 (12%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
EF VKW G S+ H SW+ E L++ NY+ K YG+ + E+ K
Sbjct: 510 EFFVKWAGLSYWHCSWVKELQLELYHTVMYRNYQRKNDMDEPPPFDYGSGDEDGKSEKRK 569
Query: 215 NPE-----------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWE--SL 249
N + R+L K+G +KW PY++CTWE +
Sbjct: 570 NKDPLYAKMEERFYRYGIKPEWMMIHRILNHSFDKKGDVHYLIKWKDLPYDQCTWEIDDI 629
Query: 250 DEPVLQNSSHLI---TRFNMFETLTLEREASKENSTKKSSDRQ---------NDIVNLLE 297
D P N + E L + K+ K D+Q + V +
Sbjct: 630 DIPYYDNLKQAYWGHRELMLGEDTRLPKRLLKKGK-KLRDDKQEKPPDTPIVDPTVKFDK 688
Query: 298 QP--KELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKV 355
QP + GG+L P+QLE LNWLR W + + ILADEMGLGKT+ F+ SLY E
Sbjct: 689 QPWYIDSTGGTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHS 748
Query: 356 SRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD---PSGLN- 411
P LV PL T+ NW EF +WAPD VV Y G ++R++IR+ E+ D SG
Sbjct: 749 KGPYLVSAPLSTIINWEREFEMWAPDFYVVTYTGDKESRSVIRENEFSFEDNAIRSGKKV 808
Query: 412 ---KKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISF 468
KK KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LNS
Sbjct: 809 FRMKKEVQIKFHVLLTSYELITIDQAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSYKI 868
Query: 469 QHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHM 528
+++LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHM
Sbjct: 869 DYKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHM 928
Query: 529 LRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVM 588
LRRLK D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M
Sbjct: 929 LRRLKADVFKNMPAKTELIVRVELSQMQKKYYKFILTRNFEALNSKGGG-NQVSLLNIMM 987
Query: 589 QLRKVCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFS 645
L+K CNHPYL P P + + +K+S KL LL MLK L EGHRVLIFS
Sbjct: 988 DLKKCCNHPYLFPVAAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFS 1047
Query: 646 QMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGL 704
QMTK+LD+LED+L E+ YER+DG ++ RQ AI RFN +F FLLSTR+ GL
Sbjct: 1048 QMTKMLDLLEDFL--EYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGL 1105
Query: 705 GINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKK 764
GINLATADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AK+K
Sbjct: 1106 GINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRK 1165
Query: 765 LMLDQL-----FKGKSGS--QKEVEDILKWGTEELFND---SCALNGK----DTSENNNS 810
+ML L KSGS ++E++DILK+GTEELF D G+ + +S
Sbjct: 1166 MMLTHLVVRPGLGSKSGSMTKQELDDILKFGTEELFKDDVEGMMSQGQRPVTPIPDVQSS 1225
Query: 811 NKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAE 870
A + KH G ++K ++SS I +D+ AI KLLDR+ QDA TD E
Sbjct: 1226 KGGNLAASAKKKHGSTPPG-----DNKDVEDSSVIHYDDAAISKLLDRN--QDA-TDDTE 1277
Query: 871 GDSENDMLGSMKALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEW 928
+ N+ L S K ++ +E E VE E K+E+N + + W
Sbjct: 1278 LQNMNEYLSSFKVAQYVVREEDGVEEVERE-------------IIKQEENV----DPDYW 1320
Query: 929 DRLLR 933
++LLR
Sbjct: 1321 EKLLR 1325
>emb|CAI19894.1| chromodomain helicase DNA binding protein 5 [Homo sapiens]
gi|56203112|emb|CAI19451.1| chromodomain helicase DNA
binding protein 5 [Homo sapiens]
Length = 1195
Score = 538 bits (1385), Expect = e-151
Identities = 351/845 (41%), Positives = 474/845 (55%), Gaps = 105/845 (12%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
EF VKW G S+ H SW+ E L++ NY+ K YG+ + E+ K
Sbjct: 26 EFFVKWAGLSYWHCSWVKELQLELYHTVMYRNYQRKNDMDEPPPFDYGSGDEDGKSEKRK 85
Query: 215 NPE-----------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWE--SL 249
N + R+L K+G +KW PY++CTWE +
Sbjct: 86 NKDPLYAKMEERFYRYGIKPEWMMIHRILNHSFDKKGDVHYLIKWKDLPYDQCTWEIDDI 145
Query: 250 DEPVLQNSSHLI---TRFNMFETLTLEREASKENSTKKSSDRQ---------NDIVNLLE 297
D P N + E L + K+ K D+Q + V +
Sbjct: 146 DIPYYDNLKQAYWGHRELMLGEDTRLPKRLLKKGK-KLRDDKQEKPPDTPIVDPTVKFDK 204
Query: 298 QP--KELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKV 355
QP + GG+L P+QLE LNWLR W + + ILADEMGLGKT+ F+ SLY E
Sbjct: 205 QPWYIDSTGGTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHS 264
Query: 356 SRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD---PSGLN- 411
P LV PL T+ NW EF +WAPD VV Y G ++R++IR+ E+ D SG
Sbjct: 265 KGPYLVSAPLSTIINWEREFEMWAPDFYVVTYTGDKESRSVIRENEFSFEDNAIRSGKKV 324
Query: 412 ---KKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISF 468
KK KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LNS
Sbjct: 325 FRMKKEVQIKFHVLLTSYELITIDQAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSYKI 384
Query: 469 QHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHM 528
+++LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHM
Sbjct: 385 DYKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHM 444
Query: 529 LRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVM 588
LRRLK D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M
Sbjct: 445 LRRLKADVFKNMPAKTELIVRVELSQMQKKYYKFILTRNFEALNSKGGG-NQVSLLNIMM 503
Query: 589 QLRKVCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFS 645
L+K CNHPYL P P + + +K+S KL LL MLK L EGHRVLIFS
Sbjct: 504 DLKKCCNHPYLFPVAAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFS 563
Query: 646 QMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGL 704
QMTK+LD+LED+L E+ YER+DG ++ RQ AI RFN +F FLLSTR+ GL
Sbjct: 564 QMTKMLDLLEDFL--EYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGL 621
Query: 705 GINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKK 764
GINLATADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AK+K
Sbjct: 622 GINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRK 681
Query: 765 LMLDQL-----FKGKSGS--QKEVEDILKWGTEELFND---SCALNGK----DTSENNNS 810
+ML L KSGS ++E++DILK+GTEELF D G+ + +S
Sbjct: 682 MMLTHLVVRPGLGSKSGSMTKQELDDILKFGTEELFKDDVEGMMSQGQRPVTPIPDVQSS 741
Query: 811 NKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAE 870
A + KH G ++K ++SS I +D+ AI KLLDR+ QDA TD E
Sbjct: 742 KGGNLAASAKKKHGSTPPG-----DNKDVEDSSVIHYDDAAISKLLDRN--QDA-TDDTE 793
Query: 871 GDSENDMLGSMKALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEW 928
+ N+ L S K ++ +E E VE E K+E+N + + W
Sbjct: 794 LQNMNEYLSSFKVAQYVVREEDGVEEVERE-------------IIKQEENV----DPDYW 836
Query: 929 DRLLR 933
++LLR
Sbjct: 837 EKLLR 841
>ref|XP_196334.5| PREDICTED: chromodomain helicase DNA binding protein 5 [Mus musculus]
Length = 1906
Score = 537 bits (1384), Expect = e-151
Identities = 346/844 (40%), Positives = 474/844 (55%), Gaps = 103/844 (12%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
EF VKW G S+ H SW+ E L++ NY+ K YG+ + E+ K
Sbjct: 466 EFFVKWAGLSYWHCSWVKELQLELYHTVMYRNYQRKNDMDEPPPFDYGSGDEDGKSEKRK 525
Query: 215 NPE-----------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWE--SL 249
N + R+L K+G +KW PY++CTWE +
Sbjct: 526 NKDPLYAKMEERFYRYGIKPEWMMVHRILNHSFDKKGDIHYLIKWKDLPYDQCTWEIDEI 585
Query: 250 DEPVLQNSSHLI---TRFNMFETLTLEREASKENSTKKSSDRQ--------NDIVNLLEQ 298
D P N + E L + K+ K ++ + V +Q
Sbjct: 586 DIPYYDNLKQAYWGHRELMLGEDARLPKRLVKKGKKLKDDKQEKPPDTPIVDPTVKFDKQ 645
Query: 299 P--KELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVS 356
P + GG+L P+QLE LNWLR W + + ILADEMGLGKT+ F+ SLY E
Sbjct: 646 PWYIDATGGTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHSK 705
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLN----- 411
P LV PL T+ NW EF +WAPD VV Y G ++R++IR+ E+ D +
Sbjct: 706 GPYLVSAPLSTIINWEREFEMWAPDFYVVTYTGDKESRSVIRENEFSFEDNAIRGGKKVF 765
Query: 412 --KKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQ 469
KK KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LNS
Sbjct: 766 RMKKEVQIKFHVLLTSYELITIDQAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSYKID 825
Query: 470 HRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHML 529
+++LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHML
Sbjct: 826 YKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHML 885
Query: 530 RRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQ 589
RRLK D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M
Sbjct: 886 RRLKADVFKNMPAKTELIVRVELSQMQKKYYKFILTRNFEALNSKGGG-NQVSLLNIMMD 944
Query: 590 LRKVCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQ 646
L+K CNHPYL P P + + +K+S KL LL MLK L EGHRVLIFSQ
Sbjct: 945 LKKCCNHPYLFPVAAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFSQ 1004
Query: 647 MTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLG 705
MTK+LD+LED+L E+ YER+DG ++ RQ AI RFN +F FLLSTR+ GLG
Sbjct: 1005 MTKMLDLLEDFL--EYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGLG 1062
Query: 706 INLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKL 765
INLATADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AK+K+
Sbjct: 1063 INLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRKM 1122
Query: 766 MLDQL-----FKGKSGS--QKEVEDILKWGTEELFNDS----CALNGKDTS---ENNNSN 811
ML L KSGS ++E++DILK+GTEELF D + + T+ + ++
Sbjct: 1123 MLTHLVVRPGLGSKSGSMTKQELDDILKFGTEELFKDDVEGMMSQGQRPTTPIPDIQSTK 1182
Query: 812 KDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEG 871
A + KH G ++K ++SS I +D+ AI KLLDR+ QDA TD E
Sbjct: 1183 GGSLTAGAKKKHGSTPPG-----DNKDVEDSSVIHYDDAAISKLLDRN--QDA-TDDTEL 1234
Query: 872 DSENDMLGSMKALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEWD 929
+ N+ L S K ++ +E E VE E K+E+N + + W+
Sbjct: 1235 QNMNEYLSSFKVAQYVVREEDGVEEVERE-------------VIKQEENV----DPDYWE 1277
Query: 930 RLLR 933
+LLR
Sbjct: 1278 KLLR 1281
>sp|Q12873|CHD3_HUMAN Chromodomain helicase-DNA-binding protein 3 (CHD-3) (Mi-2 autoantigen
240 kDa protein) (Mi2-alpha) gi|2645433|gb|AAB87383.1|
CHD3 [Homo sapiens]
Length = 1944
Score = 536 bits (1380), Expect = e-150
Identities = 343/834 (41%), Positives = 461/834 (55%), Gaps = 124/834 (14%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEE--- 211
EF VKWVG S+ H SW E L++ NY+ K YG+ + +
Sbjct: 549 EFFVKWVGLSYWHCSWAKELQLEIFHLVMYRNYQRKNDMDEPPPLDYGSGEDDGKSDKRK 608
Query: 212 --------------------QWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDE 251
+W R++ K+G VKW PY++ TWE +
Sbjct: 609 VKDPHYAEMEEKYYRFGIKPEWMTVHRIINHSVDKKGNYHYLVKWRDLPYDQSTWEEDEM 668
Query: 252 PVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQND----------IVNLLEQPKE 301
+ + H + + E + E A KK + Q D V QP+
Sbjct: 669 NIPEYEEHKQSYWRHRELIMGEDPAQPRKYKKKKKELQGDGPPSSPTNDPTVKYETQPRF 728
Query: 302 LR--GGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPC 359
+ GG+L +QLE LNWLR W + + ILADEMGLGKTI F+ SLY E P
Sbjct: 729 ITATGGTLHMYQLEGLNWLRFSWAQGTDTILADEMGLGKTIQTIVFLYSLYKEGHTKGPF 788
Query: 360 LVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPS------GLNKK 413
LV PL T+ NW EF +WAP VV Y G +RAIIR+ E+ D + K
Sbjct: 789 LVSAPLSTIINWEREFQMWAPKFYVVTYTGDKDSRAIIRENEFSFEDNAIKGGKKAFKMK 848
Query: 414 TEAY-KFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRV 472
EA KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LN H++
Sbjct: 849 REAQVKFHVLLTSYELITIDQAALGSIRWACLVVDEAHRLKNNQSKFFRVLNGYKIDHKL 908
Query: 473 LLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRL 532
LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHMLRRL
Sbjct: 909 LLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHMLRRL 968
Query: 533 KKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRK 592
K D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M L+K
Sbjct: 969 KADVFKNMPAKTELIVRVELSPMQKKYYKYILTRNFEALNSRGGG-NQVSLLNIMMDLKK 1027
Query: 593 VCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTK 649
CNHPYL P P S + IK+S KL LL ML+ L ++GHRVLIFSQMTK
Sbjct: 1028 CCNHPYLFPVAAMESPKLPSGAYEGGALIKSSGKLMLLQKMLRKLKEQGHRVLIFSQMTK 1087
Query: 650 LLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINL 708
+LD+LED+L+ E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINL
Sbjct: 1088 MLDLLEDFLDYE-GYK-YERIDGGITGALRQEAIDRFNAPGAQQFCFLLSTRAGGLGINL 1145
Query: 709 ATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLD 768
ATADTVII+DSD+NPH DIQA +RAHRIGQ+N++++YR V RASVEERI Q+AK+K+ML
Sbjct: 1146 ATADTVIIFDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRASVEERITQVAKRKMMLT 1205
Query: 769 QLF-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEH 821
L K S S++E++DILK+GTEELF D EN NK+E
Sbjct: 1206 HLVVRPGLGSKAGSMSKQELDDILKFGTEELFKD----------ENEGENKEE------- 1248
Query: 822 KHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSM 881
+SS I +D AI +LLDR+ QDA+ D + + N+ L S
Sbjct: 1249 -------------------DSSVIHYDNEAIARLLDRN--QDATED-TDVQNMNEYLSSF 1286
Query: 882 KALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEWDRLLR 933
K ++ +E E +E E K+E+N + + W++LLR
Sbjct: 1287 KVAQYVVREEDKIEEIERE-------------IIKQEENV----DPDYWEKLLR 1323
>gb|AAF13875.1| chromatin remodeling factor CHD3 [Arabidopsis thaliana]
gi|6318930|gb|AAF07084.1| GYMNOS/PICKLE [Arabidopsis
thaliana] gi|25453560|pir||T52301 GYMNOS/PICKLE protein
[imported] - Arabidopsis thaliana
gi|18400745|ref|NP_565587.1| chromatin remodeling factor
CHD3 (PICKLE) [Arabidopsis thaliana]
Length = 1384
Score = 533 bits (1374), Expect = e-150
Identities = 321/765 (41%), Positives = 460/765 (59%), Gaps = 89/765 (11%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVI------AKRKLENYKAKYGTATINICEE------- 211
++LVKW G S++H SW+ E + K ++ N+ + + N E+
Sbjct: 130 QYLVKWKGLSYLHCSWVPEKEFQKAYKSNHRLKTRVNNFHRQM--ESFNNSEDDFVAIRP 187
Query: 212 QWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFETLT 271
+W +R+LA R + G E VK+ Y+EC WES E + + I RF + T
Sbjct: 188 EWTTVDRILACR-EEDGELEYLVKYKELSYDECYWES--ESDISTFQNEIQRFKDVNSRT 244
Query: 272 LEREASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILA 331
SK+ K++ D P+ L+G L P+QLE LN+LR W K +VILA
Sbjct: 245 ---RRSKDVDHKRNP---RDFQQFDHTPEFLKG-LLHPYQLEGLNFLRFSWSKQTHVILA 297
Query: 332 DEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCA 391
DEMGLGKTI + A ++SL+ E + P LV+ PL T+ NW EFA WAP +NVV Y G A
Sbjct: 298 DEMGLGKTIQSIALLASLFEENLI--PHLVIAPLSTLRNWEREFATWAPQMNVVMYFGTA 355
Query: 392 KARAIIRQYEWHAS-DPSGLNKKTEAY----------KFNVLLTSYEMVLADYSHFRGVP 440
+ARA+IR++E++ S D + KK KF+VLLTSYEM+ D + + +
Sbjct: 356 QARAVIREHEFYLSKDQKKIKKKKSGQISSESKQKRIKFDVLLTSYEMINLDSAVLKPIK 415
Query: 441 WEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPS 500
WE +IVDEGHRLKN +SKLFS L S HR+LLTGTPLQNNL E++ L++FL F S
Sbjct: 416 WECMIVDEGHRLKNKDSKLFSSLTQYSSNHRILLTGTPLQNNLDELFMLMHFLDAGKFGS 475
Query: 501 LSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYY 560
L F+E F D+ E++ L K+++PH+LRR+KKD M+++PPK E ++ V+LSS+Q EYY
Sbjct: 476 LEEFQEEFKDINQEEQISRLHKMLAPHLLRRVKKDVMKDMPPKKELILRVDLSSLQKEYY 535
Query: 561 RAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKA 620
+A+ T+NYQ+L KG AQ S+ NI+M+LRKVC HPY++ G EP + +++
Sbjct: 536 KAIFTRNYQVLTK--KGGAQISLNNIMMELRKVCCHPYMLEGVEPVIHDANEAFKQLLES 593
Query: 621 SAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQ 680
KL LL M+ L ++GHRVLI++Q +LD+LEDY + YER+DG V +RQ
Sbjct: 594 CGKLQLLDKMMVKLKEQGHRVLIYTQFQHMLDLLEDYCTHK--KWQYERIDGKVGGAERQ 651
Query: 681 TAIARFN-QDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQS 739
I RFN ++ ++F FLLSTR+ GLGINLATADTVIIYDSD+NPHAD+QAM RAHR+GQ+
Sbjct: 652 IRIDRFNAKNSNKFCFLLSTRAGGLGINLATADTVIIYDSDWNPHADLQAMARAHRLGQT 711
Query: 740 NRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGK----SGSQKEVEDILKWGTEELFND 795
N++++YRL+ R ++EER++QL KKK++L+ L GK + +Q+E++DI+++G++ELF
Sbjct: 712 NKVMIYRLINRGTIEERMMQLTKKKMVLEHLVVGKLKTQNINQEELDDIIRYGSKELF-- 769
Query: 796 SCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKL 855
SE++ + K S KI +D+ AI KL
Sbjct: 770 --------ASEDDEAGK-----------------------------SGKIHYDDAAIDKL 792
Query: 856 LDRSNLQDASTDIAEGDSENDMLGSMKA--LEWNDEPTEEHVEGE 898
LDR +L +A + + EN L + K E+ DE +E +
Sbjct: 793 LDR-DLVEAEEVSVDDEEENGFLKAFKVANFEYIDENEAAALEAQ 836
>gb|AAH38596.1| CHD4 protein [Homo sapiens]
Length = 1937
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 539 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 598
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 599 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 658
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 659 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 717
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 718 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 777
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 778 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 837
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 838 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 897
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 898 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 957
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 958 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1016
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1017 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1076
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1077 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1134
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1135 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1194
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1195 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1238
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1239 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1279
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1280 AQYVVREEEMGEEEEVERE 1298
>sp|Q14839|CHD4_HUMAN Chromodomain helicase-DNA-binding protein 4 (CHD-4) (Mi-2 autoantigen
218 kDa protein) (Mi2-beta) gi|1107696|emb|CAA60384.1|
Mi-2 protein [Homo sapiens]
Length = 1912
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 542 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 601
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 602 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 661
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 662 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 720
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 721 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 780
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 781 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 840
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 841 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 900
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 901 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 960
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 961 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1019
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1020 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1079
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1080 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1137
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1138 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1197
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1198 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1241
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1242 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1282
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1283 AQYVVREEEMGEEEEVERE 1301
>ref|NP_666091.1| chromodomain helicase DNA binding protein 4 [Mus musculus]
gi|35193271|gb|AAH58578.1| Chromodomain helicase DNA
binding protein 4 [Mus musculus]
gi|51701319|sp|Q6PDQ2|CHD4_MOUSE Chromodomain
helicase-DNA-binding protein 4 (CHD-4)
Length = 1915
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 535 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 594
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 595 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 654
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 655 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 713
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 714 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 773
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 774 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 833
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 834 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 893
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 894 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 953
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 954 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1012
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1013 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1072
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1073 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1130
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1131 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1190
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1191 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1234
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1235 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1275
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1276 AQYVVREEEMGEEEEVERE 1294
>ref|XP_232354.3| PREDICTED: chromodomain helicase DNA binding protein 4 [Rattus
norvegicus]
Length = 1960
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 535 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 594
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 595 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 654
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 655 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 713
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 714 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 773
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 774 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 833
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 834 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 893
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 894 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 953
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 954 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1012
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1013 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1072
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1073 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1130
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1131 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1190
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1191 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1234
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1235 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1275
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1276 AQYVVREEEMGEEEEVERE 1294
>ref|XP_534909.1| PREDICTED: similar to Mi-2 protein [Canis familiaris]
Length = 2933
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 550 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 609
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 610 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 669
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 670 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 728
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 729 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 788
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 789 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 848
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 849 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 908
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 909 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 968
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 969 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1027
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1028 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1087
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1088 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1145
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1146 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1205
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1206 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1249
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1250 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1290
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1291 AQYVVREEEMGEEEEVERE 1309
>dbj|BAD90499.1| mKIAA4075 protein [Mus musculus]
Length = 1945
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 563 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 622
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 623 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 682
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 683 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 741
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 742 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 801
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 802 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 861
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 862 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 921
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 922 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 981
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 982 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1040
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1041 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1100
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1101 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1158
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1159 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1218
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1219 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1262
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1263 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1303
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1304 AQYVVREEEMGEEEEVERE 1322
>ref|NP_001264.2| chromodomain helicase DNA binding protein 4 [Homo sapiens]
Length = 1912
Score = 530 bits (1366), Expect = e-149
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 542 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 601
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 602 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 661
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 662 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 720
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 721 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 780
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 781 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 840
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 841 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 900
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 901 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 960
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 961 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1019
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1020 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1079
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1080 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1137
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1138 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1197
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1198 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1241
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1242 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1282
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1283 AQYVVREEEMGEEEEVERE 1301
>dbj|BAC28749.1| unnamed protein product [Mus musculus]
Length = 1045
Score = 529 bits (1363), Expect = e-148
Identities = 336/797 (42%), Positives = 447/797 (55%), Gaps = 104/797 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 287 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 346
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 347 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 406
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 407 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 465
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 466 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 525
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 526 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 585
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 586 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 645
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 646 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 705
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 706 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 764
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 765 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 824
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 825 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 882
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 883 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 942
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 943 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 986
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 987 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1027
Query: 884 LEW----NDEPTEEHVE 896
++ + EE VE
Sbjct: 1028 AQYVVREEEMGEEEEVE 1044
>emb|CAB55959.1| hypothetical protein [Homo sapiens]
Length = 1388
Score = 525 bits (1351), Expect = e-147
Identities = 336/784 (42%), Positives = 454/784 (57%), Gaps = 78/784 (9%)
Query: 193 KLENYKAKYGTATINICEEQWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWE--SLD 250
K+E +YG + +W R+L K+G +KW PY++CTWE +D
Sbjct: 11 KMEERFYRYGI------KPEWMMIHRILNHSFDKKGDVHYLIKWKDLPYDQCTWEIDDID 64
Query: 251 EPVLQNSSHLI---TRFNMFETLTLEREASKENSTKKSSDRQ---------NDIVNLLEQ 298
P N + E L + K+ K D+Q + V +Q
Sbjct: 65 IPYYDNLKQAYWGHRELMLGEDTRLPKRLLKKGK-KLRDDKQEKPPDTPIVDPTVKFDKQ 123
Query: 299 PKEL--RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVS 356
P + GG+L P+QLE LNWLR W + + ILADEMGLGKT+ F+ SLY E
Sbjct: 124 PWYIDSTGGTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHSK 183
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDP---SGLN-- 411
P LV PL T+ NW EF +WAPD VV Y G ++R++IR+ E+ D SG
Sbjct: 184 GPYLVSAPLSTIINWEREFEMWAPDFYVVTYTGDKESRSVIRENEFSFEDNAIRSGKKVF 243
Query: 412 --KKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQ 469
KK KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LNS
Sbjct: 244 RMKKEVQIKFHVLLTSYELITIDQAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSYKID 303
Query: 470 HRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHML 529
+++LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHML
Sbjct: 304 YKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHML 363
Query: 530 RRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQ 589
RRLK D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M
Sbjct: 364 RRLKADVFKNMPAKTELIVRVELSQMQKKYYKFILTRNFEALNSKGGG-NQVSLLNIMMD 422
Query: 590 LRKVCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQ 646
L+K CNHPYL P P + + +K+S KL LL MLK L EGHRVLIFSQ
Sbjct: 423 LKKCCNHPYLFPVAAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFSQ 482
Query: 647 MTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLG 705
MTK+LD+LED+L E+ YER+DG ++ RQ AI RFN +F FLLSTR+ GLG
Sbjct: 483 MTKMLDLLEDFL--EYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGLG 540
Query: 706 INLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKL 765
INLATADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AK+K+
Sbjct: 541 INLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRKM 600
Query: 766 MLDQL-----FKGKSGS--QKEVEDILKWGTEELFND---SCALNGK----DTSENNNSN 811
ML L KSGS ++E++DILK+GTEELF D G+ + +S
Sbjct: 601 MLTHLVVRPGLGSKSGSMTKQELDDILKFGTEELFKDDVEGMMSQGQRPVTPIPDVQSSK 660
Query: 812 KDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEG 871
A + KH G ++K ++SS I +D+ AI KLLDR+ QDA TD E
Sbjct: 661 GGNLAASAKKKHGSTPPG-----DNKDVEDSSVIHYDDAAISKLLDRN--QDA-TDDTEL 712
Query: 872 DSENDMLGSMKALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEWD 929
+ N+ L S K ++ +E E VE E K+E+N + + W+
Sbjct: 713 QNMNEYLSSFKVAQYVVREEDGVEEVERE-------------IIKQEENV----DPDYWE 755
Query: 930 RLLR 933
+LLR
Sbjct: 756 KLLR 759
>ref|XP_525165.1| PREDICTED: similar to chromodomain helicase DNA binding protein 5
[Pan troglodytes]
Length = 2473
Score = 518 bits (1333), Expect = e-145
Identities = 332/779 (42%), Positives = 446/779 (56%), Gaps = 90/779 (11%)
Query: 216 PERLLAIRTSKQGTSEAFVKWTGKPYNECTWE--SLDEPVLQNSSHLI---TRFNMFETL 270
P R + K+G +KW PY++CTWE +D P N + E
Sbjct: 1011 PPRHCCVGFDKKGDVHYLIKWKDLPYDQCTWEIDDIDIPYYDNLKQAYWGHRELMLGEDT 1070
Query: 271 TLEREASKENSTKKSSDRQ---------NDIVNLLEQPKEL--RGGSLFPHQLEALNWLR 319
L + K+ K D+Q + V +QP + GG+L P+QLE LNWLR
Sbjct: 1071 RLPKRLLKKGK-KLRDDKQEKPPDTPIVDPTVKFDKQPWYIDSTGGTLHPYQLEGLNWLR 1129
Query: 320 KCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWA 379
W + + ILADEMGLGKT+ F+ SLY E P LV PL T+ NW EF +WA
Sbjct: 1130 FSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHSKGPYLVSAPLSTIINWEREFEMWA 1189
Query: 380 PDVNVVQYHGCAKARAIIRQYEWHASDP---SGLN----KKTEAYKFNVLLTSYEMVLAD 432
PD VV Y G ++R++IR+ E+ D SG KK KF+VLLTSYE++ D
Sbjct: 1190 PDFYVVTYTGDKESRSVIRENEFSFEDNAIRSGKKVFRMKKEVQIKFHVLLTSYELITID 1249
Query: 433 YSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNF 492
+ + W L+VDE HRLKN++SK F +LNS +++LLTGTPLQNNL E+++LLNF
Sbjct: 1250 QAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSYKIDYKLLLTGTPLQNNLEELFHLLNF 1309
Query: 493 LQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVEL 552
L P F +L F E F D++ +++ +L L+ PHMLRRLK D +N+P KTE +V VEL
Sbjct: 1310 LTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHMLRRLKADVFKNMPAKTELIVRVEL 1369
Query: 553 SSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIP-----GTEPDS 607
S +Q +YY+ +LT+N++ L + G G Q S+LNI+M L+K CNHPYL P G
Sbjct: 1370 SQMQKKYYKFILTRNFEALNSKGGG-NQVSLLNIMMDLKKCCNHPYLFPVAAVVGFPGHH 1428
Query: 608 GSV-------------EFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDIL 654
GS + +K+S KL LL MLK L EGHRVLIFSQMTK+LD++
Sbjct: 1429 GSFSMTAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFSQMTKMLDLV 1488
Query: 655 EDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLATADT 713
ED+L E+ YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLATADT
Sbjct: 1489 EDFL--EYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATADT 1546
Query: 714 VIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL--- 770
VIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AK+K+ML L
Sbjct: 1547 VIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRKMMLTHLVVR 1606
Query: 771 --FKGKSGS--QKEVEDILKWGTEELFNDSCALNGKDTSEN----------NNSNKDEAV 816
KSGS ++E++DILK+GTEELF D + S+ +S
Sbjct: 1607 PGLGSKSGSMTKQELDDILKFGTEELFKDDVRVGLCMMSQGQRPVTPIPDVQSSKGGNLA 1666
Query: 817 AEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSEND 876
A + KH G ++K ++SS I +D+ AI KLLDR+ QDA TD E + N+
Sbjct: 1667 ASAKKKHGSTPPG-----DNKDVEDSSVIHYDDAAISKLLDRN--QDA-TDDTELQNMNE 1718
Query: 877 MLGSMKALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEWDRLLR 933
L S K ++ +E E VE E K+E+N + + W++LLR
Sbjct: 1719 YLSSFKVAQYVVREEDGVEEVERE-------------IIKQEENV----DPDYWEKLLR 1760
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,582,771,552
Number of Sequences: 2540612
Number of extensions: 68080952
Number of successful extensions: 185865
Number of sequences better than 10.0: 2177
Number of HSP's better than 10.0 without gapping: 1691
Number of HSP's successfully gapped in prelim test: 495
Number of HSP's that attempted gapping in prelim test: 174750
Number of HSP's gapped (non-prelim): 4869
length of query: 935
length of database: 863,360,394
effective HSP length: 138
effective length of query: 797
effective length of database: 512,755,938
effective search space: 408666482586
effective search space used: 408666482586
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146651.1


